Abstract
Photosynthetic organisms need copper for cytochrome oxidase and for plastocyanin in the fundamental processes of respiration and photosynthesis. However, excess of free copper is detrimental inside the cells and therefore organisms have developed homeostatic mechanisms to tightly regulate its acquisition, sequestration, and efflux. Herein we show that the CopRS two-component system (also known as Hik31-Rre34) is essential for copper resistance in Synechocystis sp. PCC 6803. It regulates expression of a putative heavy-metal efflux-resistance nodulation and division type copper efflux system (encoded by copBAC) as well as its own expression (in the copMRS operon) in response to the presence of copper in the media. Mutants in this two-component system or the efflux system render cells more sensitive to the presence of copper in the media and accumulate more intracellular copper than the wild type. Furthermore, CopS periplasmic domain is able to bind copper, suggesting that CopS could be able to detect copper directly. Both operons (copMRS and copBAC) are also induced by the photosynthetic inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone but this induction requires the presence of copper in the media. The reduced response of two mutant strains to copper, one lacking plastocyanin and a second one impaired in copper transport to the thylakoid, due to the absence of the PI-type ATPases PacS and CtaA, suggests that CopS can detect intracellular copper. In addition, a tagged version of CopS with a triple HA epitope localizes to both the plasma and the thylakoid membranes, suggesting that CopS could be involved in copper detection in both the periplasm and the thylakoid lumen.
Copper is an element required for essential biological processes such as respiration, through the cytochrome oxidase, or in photosynthesis through the electron transfer protein plastocyanin in plants, some algae, and cyanobacteria. It is also used as a metal cofactor of different enzymes including oxidases, monooxygenases, dioxygenases, and superoxide dismutases. The ability of copper to alternate between its cuprous Cu(I) and cupric Cu(II) oxidation states makes it an excellent biological cofactor. However, when unbound within a cell redox cycling means copper is toxic, largely due to its ability to catalyze Fenton-like reaction, causing the production of highly reactive hydroxyl radicals that damage biomolecules such as DNA, proteins, and lipids (Imlay, 2003). An alternative copper toxicity mechanism has been also demonstrated in some bacteria in which copper interferes with the formation of catalytic iron-sulfur clusters, damaging essential enzymatic activities and also leading to the generation of reactive oxygen species (Macomber and Imlay, 2009; Chillappagari et al., 2010; Tottey et al., 2012). As a result, microorganisms have developed diverse mechanisms for the control of copper homeostasis.
Copper homeostasis is a complex process involving acquisition, sequestration, and efflux of the metal ion. In bacteria, active efflux is one of the key mechanisms for copper resistance and three nonrelated families of export system have been implicated in copper resistance and homeostasis: PI-type ATPases, such as Escherichia coli CopA (Rensing et al., 2000; Grass and Rensing, 2001; Rensing and Grass, 2003), heavy-metal efflux-resistance nodulation and division (HME-RND) efflux systems, such as CusBAC (Grass and Rensing, 2001), and membrane proteins such as CopB and CopD from Pseudomonas syringae (Mills et al., 1993; Osman and Cavet, 2008). Periplasmic copper metabolism also has an important role in copper homeostasis, since most copper-containing proteins are periplasmic or plasma membrane proteins. In fact, copper homeostasis systems usually contain periplasmic copper-binding proteins, and in some cases, copper oxidases, which oxidize Cu(I) to the less toxic Cu(II) (Osman and Cavet, 2008; Kim et al., 2010). In addition, some bacteria contain intracellular copper chaperones, which deliver intracellular copper to target proteins (Robinson and Winge, 2010). These copper resistance systems are, in general, regulated by metalloregulatory proteins that are able to bind the metal. Two unrelated families of copper-responsive repressors have been described: CopY, a winged helix DNA-binding protein, and CsoR, which belongs to a new family of transcriptional repressors (Solioz et al., 2010). In addition, two other regulatory systems that work as activators have been also described: CueR, a MerR family copper-dependent activator (Outten et al., 2000), and CopRS, a two-component copper-responsive system (Osman and Cavet, 2008). CueR, CopY, and CsoR detect cytoplasmic copper levels, while CopRS is thought to detect periplasmic copper.
Photosynthetic organisms have high intracellular copper requirements, mainly for the photosynthetic electron transfer protein plastocyanin, and they have adapted to accommodate variable copper concentrations in the environment. In plants, copper import requires the action of several transporters at different locations in the plant. The import of copper in the roots is mediated by the CTR and ZIP families of transporters while the PI-type ATPases PAA1 and PAA2 are involved in copper transport into the chloroplast (Pilon et al., 2006, 2009; Puig and Peñarrubia, 2009). Copper transport systems from roots to shoots are much less characterized (Puig and Peñarrubia, 2009). As in other organisms, copper chaperones assist the trafficking and loading of copper to proteins in the cytosol (ATX1, CCH1, CCS1), the mitochondria (COX17), or the chloroplast (CCS1; Puig et al., 2007b). Most of these genes are regulated at the transcriptional level after copper excess. Thus, transporters such as COPT1-2 and COPT4, ZIP2 and 4, and PAA1, PAA2, and HMA1 are down-regulated, while copper chaperones are induced (del Pozo et al., 2010). Under copper-deficiency conditions, photosynthetic organisms express alternative isoenzymes that use different metal cofactors to copper and also induce copper import proteins (Yamasaki et al., 2009; Castruita et al., 2011; Bernal et al., 2012) to save copper for plastocyanin that is strictly required for photosynthesis in plants (Puig et al., 2007a). Some algae and cyanobacteria can also express an alternative electron transfer protein: a heme-containing cytochrome c6 (Merchant and Bogorad, 1986; Zhang et al., 1992; Merchant et al., 2006). This response is regulated by homologous transcriptional factors in eukaryotic photosynthetic organisms: CRR1 in Chlamydomonas reinhardtii (Kropat et al., 2005) and SPL7 in Arabidopsis (Arabidopsis thaliana; Yamasaki et al., 2009; Bernal et al., 2012). In contrast, very little is known about copper gene regulation in cyanobacteria despite the early discovery of the switch in gene expression between plastocyanin (encoded by petE) and cytochrome c6 (encoded by petJ) depending on copper availability (Zhang et al., 1992). In cyanobacteria, copper metabolism has been analyzed mainly in Synechocystis sp. PCC 6803 (hereafter Synechocystis). Copper import is mediated by two PI-type ATPases, CtaA and PacS, a small soluble copper metallochaperone, Atx1 (SynAtx1; Tottey et al., 2002), and a periplasmic iron-containing protein, FutA2 (Waldron et al., 2007). These proteins are required for normal photosynthetic electron transfer via plastocyanin and for the activity of a second thylakoid-located copper protein, a caa3-type cytochrome oxidase (Tottey et al., 2001, 2002, 2012; Waldron et al., 2007), although the exact role of the periplasmic protein FutA2 is not completely clear (Waldron et al., 2007). Copper is imported inside the cell by CtaA, which delivers it to SynAtx1, which is then thought to transfer it to PacS, which in turn transports it to the thylakoid lumen. Recently, glutathione has been shown to cooperate with SynAtx1 to buffer cytoplasmic copper levels, preventing deleterious side reactions (Tottey et al., 2012).
Here we present evidence that the Hik31/Rre34 two-component system (designated CopRS here) is involved in copper resistance in Synechocystis by directly regulating an HME-RND export system, CopBAC (encoded by open reading frames [ORFs] slr6042, slr6043, and slr6044), and a protein of unknown function, CopM (encoded by ORFs sll0788 and slr6039). Although responsive to copper, CopRS is neither involved in the regulation of copper import system nor in the switch between petE and petJ. Furthermore, using a combination of different genetic and molecular biology approaches, we show that CopS is able to bind copper and partially localizes to thylakoid membranes. copMRS is also induced by conditions that alter the electron transport rate around PSI, which indicates that these genes are under redox control. Under these conditions, plastocyanin protein levels decrease, and this mirrors copMRS induction. This induction strictly requires the presence of copper in the media and CopRS. Furthermore, induction of copMRS after a low copper addition is diminished in mutants with reduced levels of plastocyanin, suggesting that part of the signal detected by CopS needs copper to be incorporated into plastocyanin.
RESULTS
CopRS Is Involved in Copper Resistance
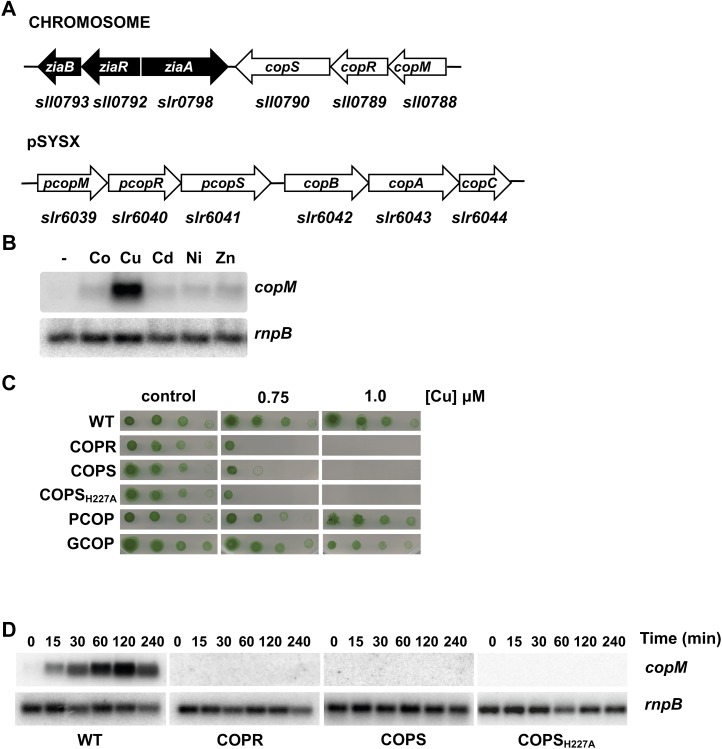
A gene cluster involved in metal resistance in Synechocystis was previously characterized (Thelwell et al., 1998; Rutherford et al., 1999; García-Domínguez et al., 2000). The two-component system hik31-rre34 (sll0789 and sll0790) is located next to the metal resistance cluster, downstream of ziaA (Fig. 1A), and code for the closest homolog to the NrsRS two-component system in Synechocystis (46% identity; 64% similarity). Upstream of these two genes there is an additional ORF (sll0788) that contains two DUF305 domains of unknown function and likely forms an operon with them. These three genes are repeated in one of the Synechocystis endogenous plasmids (Kaneko et al., 2003), pSYSX (slr6039, slr6040, and slr6041 with a 93% identity at the nucleotide level, including 71 pb before the starting GTG for sll0788 and slr6039, and 95% at the amino acid sequence level). We have named these genes copMRS and pcopMRS, respectively. Their location and homology led us to study its putative role in metal resistance. As a first step we have analyzed their expression in response to different metals in the media. We analyzed expression of both copMRS and pcopMRS since their high sequence homology did not allow us to distinguish between them (therefore we will refer to both copies simply as copMRS when analyzing gene expression). As shown in Figure 1B, copM expression was induced in the presence of an excess of copper (3 μm CuSO4), but induction by other metals was negligible (Fig. 1B). Furthermore, northern and reverse-transcription-PCR analysis confirmed that copM was cotranscribed with copR and copS and therefore the three genes form an operon (Supplemental Fig. S1; Summerfield et al., 2011). To further study their role in metal homeostasis we analyzed growth of mutant strains lacking one or both copies of these genes (Supplemental Table S1) in the presence of different metals in the media. Mutants lacking a functional copy of copMRS (GCOP strain) or pcopMRS (PCOP strain; Fig. 1C) are indistinguishable from the wild type. In contrast, double mutants lacking functional copies of both copR and pcopR (COPR strain), copS and pcopS (COPS strain), or carrying a mutation in the catalytic His (COPSH227A) showed reduced growth at 0.75 μm of copper and failed to grow at 1 μm of the metal (Fig. 1C), showing that this two-component system is essential for copper resistance, but not to other metals (Supplemental Fig. S2). Moreover, COPR cells accumulated about twice the amount of copper than wild-type cells (576 ± 43 versus 339 ± 14 μg copper mg−1 dry weight) after a 5-h exposure to 3 μm of copper, suggesting that CopRS controls a copper resistance system.
Figure 1.
CopRS is involved in copper resistance. A, Schematic representation of copMRS and pcopMRS-copBAC genomic regions. B, Northern-blot analysis of the expression of copM. Total RNA was isolated from wild-type cells grown in BG11C-Cu medium and exposed for 90 min to 3 μm of the indicated metal ions. Control cells were not exposed to added metals (−).The filter was hybridized with a copM probe and subsequently stripped and rehybridized with an rnpB probe as a control. C, Phenotypic characterization of mutants in copRS. Tolerance of wild-type, COPR, COPS, COPSH227A, PCOP, and GCOP strains to copper was examined. Ten-fold serial dilutions of a suspension of 1 μg chlorophyll mL−1 cells were spotted onto BG11C-Cu supplemented with the indicated copper concentrations. Plates were photographed after 5 d of growth. D, Loss of copM induction in COPR, COPS, and COPSH227A strains. Total RNA was isolated from wild-type, COPR, COPS, and COPSH227A strains grown in BG11C-Cu medium after addition of 3 μm of copper. Samples were taken at the indicated times. The filter was hybridized with a copM probe and subsequently stripped and rehybridized with an rnpB probe as a control. [See online article for color version of this figure.]
Two-component systems are often autoregulated in a positive feedback loop, and to test whether CopRS regulated its own expression we analyzed copM expression in the COPR, COPS, and COPSH227A strains. copM mRNA levels increased (75-fold induction) at least during the first 2 h after addition of 3 μm of copper in wild-type cells, but this induction was completely lost in the COPR, COPS, and COPSH227A strains (Fig. 1D), suggesting that CopRS controls its own induction in response to copper.
CopRS Controls the Expression of an HME-RND Efflux System Involved in Copper Resistance
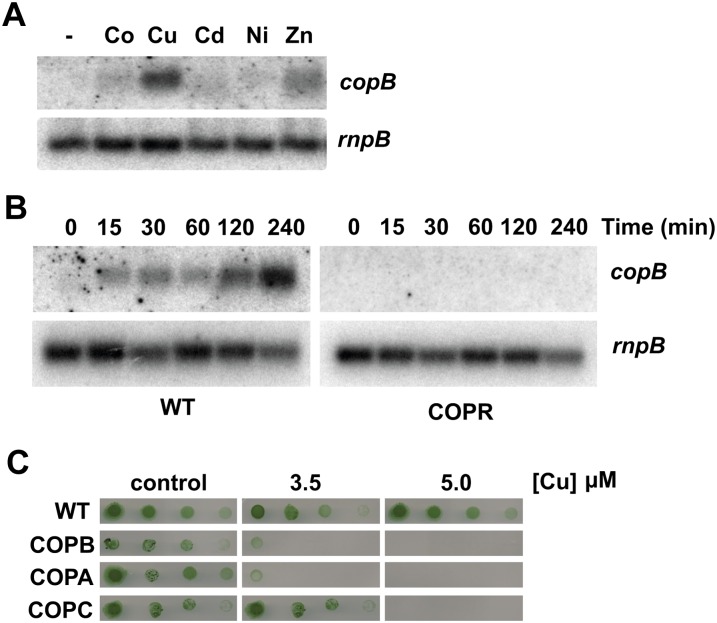
Downstream of pcopMRS, in the plasmid pSYSX, there are three ORFs (slr6042, slr6043, and slr6044) that code for a putative HME-RND transport system (Fig. 1A). These three ORFs code for proteins with homology to a membrane fusion protein, an RND protein, and an outer membrane protein, respectively. We have designated these three genes as copB, copA, and copC. To test if copBAC was involved in metal resistance, we analyzed its expression in response to the presence of different metals in the media. copB was induced in response to the presence of copper, and, to a lesser extent, zinc, while induction by other metals was negligible (Fig. 2A). Northern and reverse transcription-PCR analysis showed that copA and copC were also induced by copper, composing a single transcriptional unit with copB (Supplemental Fig. S3). Since they were induced by copper, we wanted to test if they were regulated by the CopRS system. copBAC expression increased (14-fold induction) after an addition of 3 μm of copper during at least the first 4 h, although with delayed kinetics when compared with copMRS. This induction was lost in the COPR strain (Fig. 2B), showing that CopRS is involved in copBAC induction in response to copper. To further clarify their role in metal homeostasis, we constructed mutants in all three genes (Supplemental Table S1) and tested their sensitivity to different metals. These strains were sensitive to the presence of copper, but its tolerance to other metals was not drastically different from the wild type (Supplemental Fig. S2). COPB and COPA strains presented growth defects in the presence of 3.5 μm or higher copper concentrations (Fig. 2C). However, the COPC strain showed lower sensitivity to copper, because it was able to grow on 3.5 μm of copper and only at 5 μm of copper was its growth fully inhibited (Fig. 2C). We also analyzed the copper content of COPB cells (which lack expression of copBAC) in liquid media and these cells also accumulated 20% more intracellular copper than wild-type cells (400 ± 8 versus 339 ± 14 μg copper mg−1 dry weight) when challenged with 3 μm of copper for 5 h, although to a lesser extent than COPR cells, which is in agreement with the lower sensitivity of COPB cells to copper in our plate assay.
Figure 2.
A new RND system involved in copper resistance. A, Northern-blot analysis of the expression of copB. Total RNA was isolated from wild-type cells grown in BG11C-Cu medium and exposed for 90 min to 3 μm of the indicated metal ions. Control cells were not exposed to added metals (−).The filter was hybridized with a copB probe and subsequently stripped and rehybridized with an rnpB probe as a control. B, Loss of copB induction in the COPR strain. Total RNA was isolated from wild-type and COPR strains grown in BG11C-Cu medium after addition of 3 μm of copper. Samples were taken at the indicated times. The filter was hybridized with a copB probe subsequently stripped and rehybridized with an rnpB probe as a control. C, Phenotypic characterization of copBAC mutants. Tolerance of wild-type, COPB, COPA, and COPC strains to copper was examined. Ten-fold serial dilutions of a 1 μg chlorophyll mL−1 cells suspension were spotted onto BG11C-Cu supplemented with the indicated copper concentrations. Plates were photographed after 5 d of growth. [See online article for color version of this figure.]
CopR Binds to copMRS and copBAC Promoters
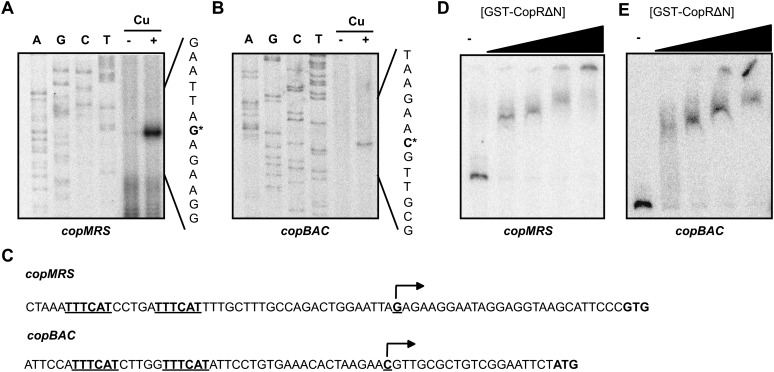
The transcription start points were determined by primer extension to establish the location of copMRS and copBAC promoters. Both copMRS and pcopMRS transcripts start 27 nucleotides upstream of the predicted copM or pcopM starting codon (Fig. 3A), since these sequences are identical and we could not distinguish between them. copBAC transcripts start 19 nucleotides upstream of the putative copB starting codon (Fig. 3B). No consensus −10 and −35 boxes could be identified in these promoters but two repeats, in the form of TTTCAT separated by 5 bp, are present in both promoters, replacing −35 boxes (Fig. 3C). CopR belongs to the OmpR family of response regulators that binds to direct repeats around the −35 boxes in promoters to activate transcription (Kenney, 2002; Blanco et al., 2011). To test whether CopR binds to these promoters, we purified a truncated version lacking the amino terminal receiver domain fused to glutathione S-transferase (GST; CopRΔN; as we were unable to obtain a soluble full-length protein preparation) and used it in electrophoretic mobility shift assays. CopRΔN was able to bind to probes containing copM and copB promoters (Fig. 3, D and E) and therefore the repeated sequences found in copMRS and copBAC promoters are likely to be CopR binding sites to regulate their transcription.
Figure 3.
CopR regulates directly copMRS and copBAC promoters. A, Primer extension of copMRS and pcopMRS transcripts from wild-type cells grown in BG11C-Cu medium and exposed to copper 3 μm for 1 h. Sequencing ladders generated with the same oligonucleotide used for primer extension is also shown. B, Primer extension of copBAC transcript from wild-type cells grown in BG11C-Cu medium and exposed to copper 3 μm for 1 h. Sequencing ladders generated with the same oligonucleotide used for primer extension is also shown. C, Sequences of the copMRS and copBAC promoters. Transcriptional start sites are marked with an arrow and direct repeated sequences are underlined. D, Band-shift assay of the copMRS promoter region with increasing quantities GST-CopRΔN. E, Band-shift assay of the copBAC promoter region with increasing quantities GST-CopRΔN.
CopS Periplasmic Domain Binds Metals
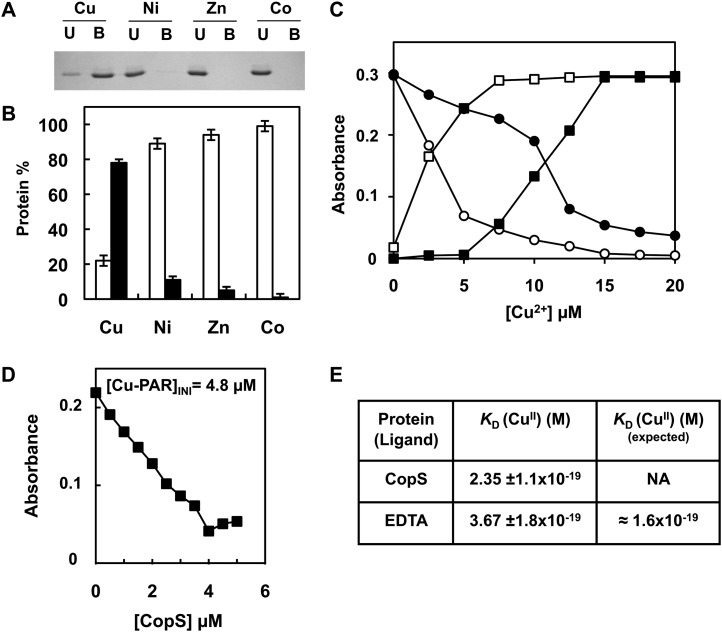
CopS is composed of two protein domains: a carboxy-terminal domain containing the His kinase catalytic site and amino-terminal sensor domain. This sensor domain contains two putative transmembrane segments (residues 15–37 and 185–207) and a putative periplasmic region. To test if the periplasmic region was able to bind metals, we have expressed and purified the region between the transmembrane segments (expanding from residues 38–183) fused to a strep-tag to facilitate its purification (CopS38-183). We tested whether CopS38-183 was able to bind metals using metal chromatography. The protein was retained by beads charged with 0.5 mm of Cu2+ but not by Zn2+, Ni2+, and Co2+ charged beads (Fig. 4, A and B). To further analyze CopS38-183 interaction with copper we used ligand competition with apo-4-(2-pyridylazo)-resorcinol (PAR). CopS38-183 was able to extract one equivalent of Cu2+ from PAR, suggesting that one atom of copper binds to one molecule of CopS38-183 (Fig. 4C). Titration of Cu2+-loaded PAR with increasing amounts of CopS38-183 revealed a concentration-dependent decrease in PAR-Cu2+ concentration (Fig. 4D; Supplemental Fig. S4), and allowed us to calculate an apparent dissociation constant (KDapp) for CopS38-183 of 2.3·10−19 after calibration of the assay with EDTA (Fig. 4, D and E). These data demonstrated that CopS periplasmic region is able to bind copper with high affinity in vitro.
Figure 4.
CopS periplasmic domain binds copper. A, Analysis of CopS(38-183) protein interaction with metals. His-Bind resin columns were loaded with 0.5 mm CuSO4, NiSO4, ZnSO4, and CoCl2. About 10 μg of purified CopS(38-183) protein was applied to the columns. Unbound (U lanes) and bound (B lanes) fractions were analyzed by 15% SDS-PAGE and Coomassie Blue staining. B, Quantification of CopS in bound and unbound fractions. Coomassie-stained gel was scanned and the intensity of the bands was quantified using ImageJ program; the graph represents the average of two experiments. Unbound fraction (white), bound fraction (black). C, Titration of PAR, which absorbs at 410 nm (circles), to its copper form absorbing at 500 nm (squares), in the absence (white symbols) and presence (black symbols) of 10 μm CopS. D, Determination of the Cu2+ dissociation constant, KD, of CopS by titration into a solution of 10 μm PAR. The graph shows the decrease at 500 nm relative to CopS additions for a [Cu-PAR]TOTAL of 0.9 μm. E, Apparent KD CopS and EDTA at pH 7.5 derived from competition titration using Cu2+-PAR. Dissociation constant, KD, was estimated as described in “Materials and Methods” from four independent experiments like the one shown in D. NA, Not available.
Redox Induction of copMRS Depends on the Presence of Copper
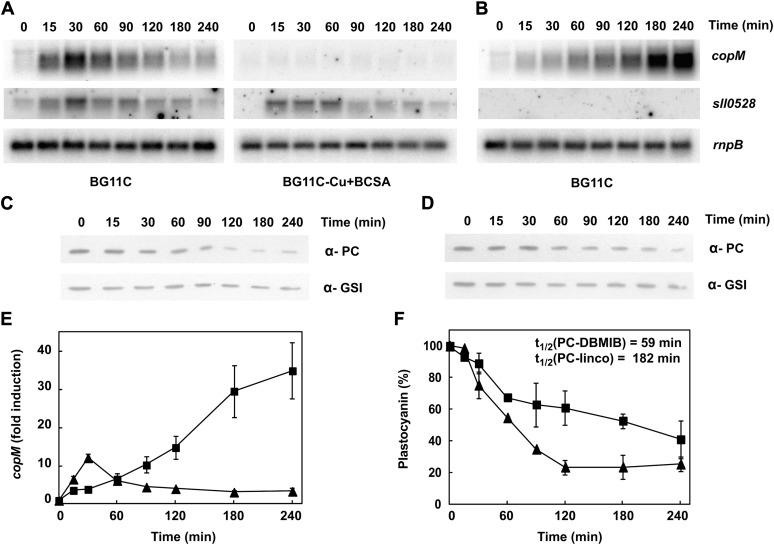
Previous microarray studies have shown that copMRS operon is highly induced by 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB; which blocks electron transfer from the plastoquinone pool to the cytochrome b6f), but not by 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU; which blocks electron transfer from PSII to the platoquinone pool; Hihara et al., 2003), suggesting that these genes were controlled by the redox state of the plastoquinone pool. Having established that copMRS had a role in copper homeostasis, we wanted to investigate if there was any interaction between copper metabolism and DBMIB induction of copMRS. First, we confirmed that the addition of 10 μm DBMIB to a Synechocystis culture induced expression of copMRS (Fig. 5A; Supplemental Fig. S5A) and copBAC (Supplemental Fig. S5A), but these genes were not induced by the addition of 10 μm DCMU (Supplemental Fig. S6). Second, when the DBMIB treatment was performed in a medium without copper (BG11C-Cu) plus bathocuproinedisulfonic acid (BCSA), a copper chelator, to avoid any residual copper in the media (Durán et al., 2004), neither copMRS nor copBAC operons were induced, as determined by copM and copB expression (Fig. 5A; Supplemental Fig. S5A). DBMIB treatment in the COPR strain was also ineffective at inducing the expression of both copM and copB (Supplemental Fig. S7). However, sll0528, another gene induced by DBMIB in the microarray analysis (Hihara et al., 2003), was still fully induced in both cases (Fig. 5A; Supplemental Fig. S5 and Supplemental Fig. S6). These results suggested that induction after DBMIB treatment of both copMRS and copBAC was related to copper metabolism, rather than a direct effect of the redox state of the plastoquinone pool, and that it was dependent on CopRS.
Figure 5.
CopS responds to plastocyanin protein levels. A, Northern-blot analysis of the expression of copM and sll0528 after DBMIB addition. Total RNA was isolated from wild-type cells grown in BG11C or BG11C-Cu + BCSA medium after addition of DBMIB 10 μm. Samples were taken at the indicated times. The filters were hybridized with copM and sll0528 probes and subsequently stripped and rehybridized with an rnpB probe as a control. B, Northern-blot analysis of the expression of copM and sll0528 after lincomycin addition. Total RNA was isolated from wild-type cells grown in BG11C medium after addition of lincomycin 250 μg mL−1. Samples were taken at the indicated times. The filters were hybridized with copM and sll0528 probes and subsequently stripped and rehybridized with an rnpB probe as a control. C, Western-blot analysis of plastocyanin levels after DBMIB addition. Wild-type cells were grown in BG11C medium and exposed for 4 h to DBMIB 10 μm. Cells were harvested at the indicated times, and 5 μg of total protein from soluble extracts was separated by 15% SDS-PAGE and subjected to western blot to detect plastocyanin or glutamine synthetase type I (GSI). D, Western-blot analysis of plastocyanin levels after lincomycin addition. Wild-type cells were grown in BG11C medium and exposed for 4 h to lincomycin 250 μg mL−1. Cells were harvested at the indicated times, and 5 μg of total protein from soluble extracts was separated by 15% SDS-PAGE and subjected to western blot to detect plastocyanin or GSI. E, Quantification of relative mRNA levels of copM, in response to DBMIB and lincomycin addition in wild-type strain. Radioactive signals of three independent experiments for each strain were quantified and averaged. RNA levels were normalized with the rnpB signal. Plots of relative mRNA levels versus time were drawn; error bars represent se. DBMIB treatment (triangles), lincomycin treatment (squares). F, Quantification of plastocyanin levels, in response to DBMIB and lincomycin addition in wild-type strain. Western-blot signal of three independent experiments were quantified using Image J program. Plastocyanin levels were normalized with the GSI signal. Error bars represent se. DBMIB treatment (triangles), lincomycin treatment (squares). Half-life, t1/2, of plastocyanin was estimated as described in “Materials and Methods” from three independent experiments.
The Response of CopS to Plastocyanin Protein Levels
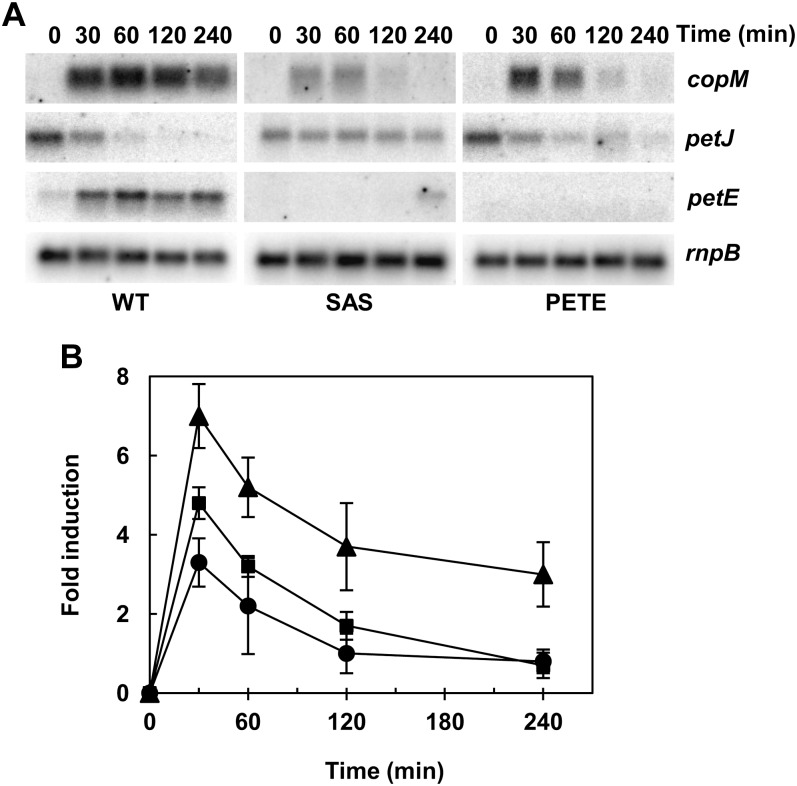
Photosynthetic electron transport between cytochrome b6f complex and PSI is mediated by plastocyanin or cytochrome c6 depending on the availability of copper in Synechocystis (Zhang et al., 1992; Waldron et al., 2007). DBMIB blocks the electron transfer between the plastoquinone pool and cytochrome b6f, and therefore impairs the plastocyanin and cytochrome c6 reduction, causing their accumulation in the oxidized form (Trebst, 2007). Plastocyanin is the main copper-containing protein in Synechocystis cells and it is confined to the thylakoid lumen (Waldron et al., 2007). To test if DBMIB treatment induces plastocyanin degradation, we analyzed plastocyanin protein levels by western blot. As shown in Figure 5C the amount of plastocyanin rapidly declined after DBMIB treatment. To test whether reduction of plastocyanin levels were responsible for copM induction, lincomycin, a protein synthesis inhibitor, was added to Synechocystis cells growing in copper-containing medium. Induction of copM and copB and the decrease in plastocyanin levels occurred in parallel after lincomycin treatment (Fig. 5, B and D; Supplemental Fig. S5), but with delayed time course respective of the DBMIB treatment (Fig. 5). In agreement to this, plastocyanin half-life was 3 times longer in lincomycin-treated cells (t1/2=182 min) compared with DBMIB-treated cells (t1/2=59 min; Fig. 5F). Similar to the DBMIB treatment, no induction of copM and copB expression was observed when lincomycin was added to cells growing in medium without copper + BCSA (Supplemental Fig. S5B). Furthermore, we analyzed whether plastocyanin was required for copM and copB induction. For that, a Synechocystis mutant lacking plastocyanin was constructed (PETE) and copM induction was followed after the addition of 200 nm of copper, since higher copper concentrations were toxic to the PETE strain. As shown in Figure 6, copM expression was lower in the PETE strain (about 60% of the wild-type induction), although it followed the same kinetics of the wild-type strain (Fig. 6B), suggesting that part of the signal sensed by CopS depends on the presence of plastocyanin in the thylakoid lumen. Copper is delivered to plastocyanin by the sequential action of two PI-type ATPases, CtaA and PacS, and mutant strains lacking these genes have reduced levels of plastocyanin (Tottey et al., 2001, 2012). We constructed a double mutant lacking both ATPases (SAS strain) to test whether copper import was needed for CopS activation. After the addition of 200 nm of copper to the SAS strain, copM induction was also lower (about 50% of the wild-type induction), similar to the PETE strain behavior and with the same kinetics as the wild-type strain (Fig. 6B). Although the behavior of both strains was similar, they accumulated different amounts of intracellular copper after this treatment: The PETE strain accumulated only 60% of the wild-type copper (42.7 ± 1.8 versus 70 ± 14 μg mg−1 dry weight), while the SAS strain accumulated the same amount as the wild type (71.4 ± 9.1 μg mg−1 dry weight). Even more, the SAS strain failed to do the switch from petJ to petE expression after this low copper addition, unlike the wild-type and PETE strains (Fig. 6). Single mutants in these two ATPases have been shown to accumulate similar copper contents but reduced copper-loaded plastocyanin (Tottey et al., 2001, 2012), and our double mutant (SAS strain) did not express petE, reinforcing that copper loading into plastocyanin is needed for activation of CopS.
Figure 6.
CopS responds to intracellular copper. A, Northern-blot analysis of the expression of copM, petE, and petJ in response to copper addition in wild-type, SAS, and PETE strains. Total RNA was isolated from wild-type, SAS, and PETE cells grown in BG11C-Cu medium after addition of copper 200 nm. Samples were taken at the indicated times. The filters were hybridized with copM, petE, and petJ probes and subsequently stripped and rehybridized with an rnpB probe as a control. B, Quantification of relative mRNA levels of copM in response to copper addition in wild-type, SAS, and PETE strains. Radioactive signals of three independent experiments for each strain were quantified and averaged. RNA levels were normalized with the rnpB signal in all strains. Plots of relative mRNA levels versus time were drawn; error bars represent se. Wild-type strain (triangles), SAS strain (circles), PETE strain (squares).
CopS Is Localized to Both Plasma and Thylakoid Membranes
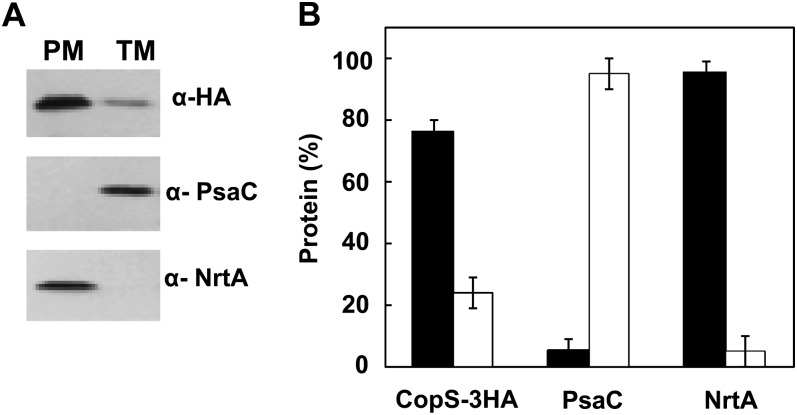
All of the aforementioned results demonstrated that CopS could detect signals both at the periplasmic space and at the thylakoid lumen (where plastocyanin is located). In that way, CopS would need to be inserted into both the plasma and thylakoid membranes. With the aim of determining the subcellular localization of CopS, we constructed a strain (COPSHA) that expresses CopS fused to a triple HA epitope (CopS-3HA) under control of the nrsBACD promoter that is induced by nickel (García-Domínguez et al., 2000; López-Maury et al., 2002). After the addition of 2 μm of nickel for 4 h to the COPSHA strain, thylakoid and plasma membrane fractions were prepared by Suc density gradient centrifugation and aqueous polymer two-phase partitioning (Norling et al., 1998). As shown in Figure 7, a single protein band of the corresponding molecular mass of CopS-3HA (56 kD) was detected in both thylakoids (about 25% of total fraction) and plasma membranes, while marker proteins PsaC (a PSI protein; Kruip et al., 1997) and NtrA (a plasma membrane attached protein; Norling et al., 1998) were exclusively detected in thylakoid fraction and plasma membrane fraction, respectively. This result shows that CopS is localized to both thylakoid and plasma membranes and therefore could perceive signals in both compartments.
Figure 7.
CopS is localized to plasma and thylakoid membranes. A, Membrane localization of CopS. Membrane fractions from COPSHA strain induced for 4 h with 2 μm of nickel were prepared by Suc density gradient and aqueous polymer two-phase partitioning. Five micrograms of total protein were loaded and separated by SDS-PAGE. CopS-3HA, NrtA, and PsaC proteins were detected by western blot. PM, Plasma membrane; TM, thylakoid membrane. B, Quantification of CopS in different membrane fractions. Western-blot signal of three independent experiments were quantified using Image J program and averaged; error bars represent se. Plasma membrane (black); thylakoid membrane (white).
DISCUSSION
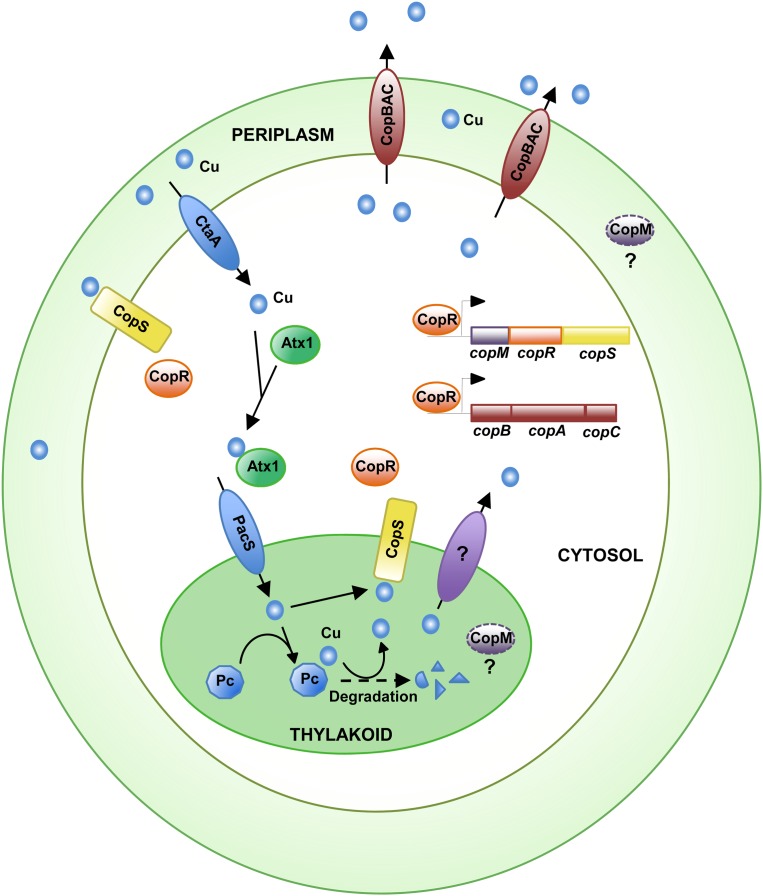
This work shows the existence of a copper resistance system in Synechocystis comprised of a two-component system (CopRS), an HME-RND transport system (CopBAC), and a protein of an unknown function, CopM. CopRS is essential for the expression of both copBAC and copMRS operons. The system responds specifically to the presence of copper but not to other metals (Figs. 1 and 2). Mutant strains affecting the regulatory system (COPR, COPS, COPSH227A) are more sensitive to the presence of copper in the media than strains lacking components of the CopBAC transport system (Figs. 1C and 2C), suggesting that CopRS might control more genes involved in copper homeostasis. These strains lack expression of both copBAC and copM (Figs. 1D and 2B), and therefore the more likely candidate to be involved in copper resistance is CopM. CopM contains an uncharacterized Duf305 domain that is present in conserved proteins in several other cyanobacteria (Nagarajan et al., 2012) and bacteria, but the function of proteins containing the Duf305 domain has not been reported. CopM contained an elevated number of Met and His residues and a signal peptide that will target it to the periplasmic and/or thylakoid compartment. In other copper resistance systems, periplasmic proteins with an elevated number of these residues work as copper chaperones, acting either as a buffer and/or transferring periplasmic copper to RND transport systems (Loftin et al., 2005; Bagai et al., 2008; Chong et al., 2009; Mealman et al., 2011) that efflux it outside the cell. Attempts to delete copM without affecting copRS expression have been unsuccessful, and for that reason we could not determine the contribution of CopM to copper resistance. The fact that COPR strains accumulate more copper than wild-type or COPB cells suggests that either CopM contributes to copper extrusion or that CopRS controls other genes involved in copper transport. Other obvious candidates to be controlled by CopRS are genes that code for proteins required for copper import (ctaA, pacS, atx1), cytochrome c6 (petJ), and plastocyanin (petE), all of which are regulated by the presence of copper in the media. We tested whether the expression of these genes was under CopRS control but they behaved similarly in wild-type and COPR strains (Supplemental Fig. S8). On the other hand, mutant strains in copB or copA tolerated up to 3.5 μm of copper, while mutant strains in copC resisted up to 5 μm. copC codes for an outer membrane protein, which in other HME-RND systems connects the RND protein to the outer membrane and allows extrusion of metals outside of the cells. In this regard, recent structural and functional studies show that the E. coli CusBA complex could be able to transport copper from the cytosol to the periplasm in the absence of CusC, the homolog of Synechocystis CopC (Franke et al., 2003; Su et al., 2011), where it could be buffered by CopM.
The CopRS two-component system (previously known as Hik31-Rre34) was reported as affecting Synechocystis cell growth under mixotrophic and heterotrophic conditions (Kahlon et al., 2006; Nagarajan et al., 2012), and also in the regulation of the response to low-oxygen conditions (Summerfield et al., 2011). Even more, their mutants lack the expression of icfG, a gene essential for Glc metabolism (Kahlon et al., 2006). In our hands, the COPR strain is able to grow in the presence of Glc and expresses the icfG gene to levels similar to the wild type, both in the presence and absence of Glc (Supplemental Fig. S9). It has been previously shown that differences in strain genetic background affect Glc sensitivity in Synechocystis (Kahlon et al., 2006), and this could explain these discrepancies. Nagarajan et al. also showed that their single and double mutants of the copMRS genes presented different metal sensitivities to nickel, cobalt, zinc, and cadmium (Nagarajan et al., 2012), but our mutants in both copRS or copBAC were as resistant as the wild type to all metals except copper (Figs. 1 and 2; Supplemental Fig. S2).
In contrast to most bacteria, cyanobacteria have high intracellular copper requirements in the form of the electron transfer protein plastocyanin (Waldron et al., 2007). This protein is localized to the thylakoid lumen and is essential for electron transfer reaction during photosynthesis in copper-containing media (Durán et al., 2004). copMRS has been described to be highly induced by different conditions, all of which alter the photosynthetic electron transport, such as treatment with DBMIB (Fig. 5; Hihara et al., 2003), nitrogen starvation (Supplemental Fig. S10; Osanai et al., 2006), and sulfur starvation (Zhang et al., 2008). We have shown here that induction in DBMIB-treated cells (Fig. 5) and nitrogen-starved cells (Supplemental Fig. S10) is dependent on the presence of copper in the media, thus establishing that this induction is related to copper metabolism and not to other factors. All of these conditions have in common a general decrease in photosynthetic electron flux (or a complete blockage, in the case of DBMIB) that will probably lead to accumulation of oxidized plastocyanin. We have shown that under these conditions plastocyanin protein levels are reduced in vivo (Fig. 5; Supplemental Fig. S10). This reduction in plastocyanin protein levels leads to activation of CopS (Fig. 5). Further support for this comes from the induction of copM and copB after treatment with the translation inhibitor lincomycin, which also causes a reduction in plastocyanin protein levels (Fig. 5D). In both cases, induction of these genes correlates with plastocyanin degradation, although the response is maintained after lincomycin treatment since it completely blocks translation and therefore cells are not able to respond to this treatment. Furthermore, these results are reinforced with our genetic data about copM and copB induction using the PETE and SAS strains (Fig. 6). Both of these mutants lack copper plastocyanin in the thylakoid lumen (Fig. 6; Waldron et al., 2007; Tottey et al., 2012) and show a reduced induction of the copMRS operon, even if they accumulate less (PETE mutant), or the same amount of (SAS mutant), copper than the wild-type strain. These data strongly suggest that copper needs to be incorporated into plastocyanin to be detected by CopS. Plastocyanin degradation will probably release copper into the thylakoid lumen and this copper could be detected by CopS. In addition, we have shown that the CopS periplasmic domain is able to bind one atom of Cu2+ with comparable affinity to the recently described MAP kinase (Turski et al., 2012), supporting that CopS detects copper directly (Fig. 4). All of these data, together with the localization of CopS to both plasma and thylakoid membranes (Fig. 7), showed that this protein responds to copper (probably by direct binding to it) in both the periplasm and the thylakoid lumen (Fig. 8). Since plastocyanin levels have been estimated to be in the high micromolar range inside the thylakoid (Durán et al., 2004; Finazzi et al., 2005), even a small decrease in plastocyanin levels could generate large amounts of free copper ions in the thylakoid lumen. This copper could be enough to activate CopS, even if it is present at low levels in the thylakoid membrane (Fig. 7). Why is CopS detecting thylakoid copper levels? The thylakoid lumen contains numerous proteins that are highly sensitive to oxidative damage (Nishiyama et al., 2001), and therefore copper will be highly toxic in this compartment. CopS activation will induce copMRS and copBAC. CopBAC efflux system is unlikely to be able to detoxify copper from the thylakoid lumen, but it will at least export the surplus of copper that could be accumulated in the periplasm and the cytosol, creating a positive concentration gradient for copper efflux from the thylakoid. In addition, CopM could have an unidentified role in detoxifying thylakoid copper, preventing damage in this compartment. Finally, we cannot rule out that CopRS controls other unknown genes involved in copper homeostasis.
Figure 8.
Simplified model showing the copper transport proteins and its relation to CopRS and CopBAC resistance systems. [See online article for color version of this figure.]
Whether the responses described here are conserved in photosynthetic eukaryotes is unknown, but copper trafficking in the chloroplast is also mediated by PI-type ATPases, homologous to CtaA and PacS, and copper chaperones (Puig et al., 2007a). Therefore, it seems reasonable to expect that the drastic reduction in the photosynthetic electron flux that leads to accumulation of oxidized plastocyanin could lead to its degradation, releasing free copper in the thylakoid lumen. It is also anticipated that this excess of free copper could be detected and a response similar the one observed here could be launched to detoxify this copper. The proteins studied here are only conserved in some cyanobacteria (Nagarajan et al., 2012; J. Giner-Lamia, L. López-Maury, and F.J. Florencio, unpublished data), and therefore the response in photosynthetic eukaryotes is likely mediated by a different set of regulatory proteins and effectors, in the same way the petE to petJ switch is conserved between cyanobacteria and Chlamydomonas but the regulatory mechanisms are not (Merchant and Bogorad, 1986; Zhang et al., 1992; Merchant et al., 2006).
CONCLUSION
In summary, we have shown that the CopRS two-component system is essential for copper resistance in Synechocystis by regulating expression of copMRS and copBAC operons in response to copper. CopS is probably detecting copper directly, as its putative periplasmic sensor domain is able to bind copper in vitro. We also present evidence that redox induction of copMRS is strictly dependent on the presence of copper and that this induction is probably related to plastocyanin degradation. Furthermore, we show that CopS localized to both plasma and thylakoid membranes and therefore could respond to copper both in the periplasm and in the thylakoid lumen. Whether CopRS controls additional mechanisms involved in thylakoid copper detoxification remains to be elucidated. To our knowledge, CopS is the first His kinase detecting events directly inside the thylakoid lumen in cyanobacteria, despite the extensive regulation mediated by changes that occur in this compartment in photosynthetic organisms.
MATERIALS AND METHODS
Strains and Culture Conditions
Synechocystis cells were grown photoautotrophically on BG11C, BG11C-Cu (lacking CuSO4), and BG11C-N (lacking NaNO3) medium (Rippka et al., 1979) at 30°C under continuous illumination (50 μE m−2 s−1) and bubbled with a stream of 1% (v/v) CO2 in air. For Supplemental Figure S5 to Supplemental Figure S7 and Supplemental Figure S10, BG11C-Cu or BG11C-Cu-N was supplemented with 300 μm BCSA as a chelating agent to eliminate any traces of copper (Durán et al., 2004). For plate cultures, medium was supplemented with 1% (w/v) agar. Kanamycin, chloramphenicol, and spectinomycin were added to a final concentration of 50 μg mL−1, 20 μg mL−1, and 5 μg mL−1, respectively. BG11C-Cu medium was supplemented with different concentrations of CuSO4, NiSO4, ZnSO4, CdCl2, and CoCl2 when indicated. Experiments were performed using cultures from the midlogarithmic phase (3–5 μg chlorophyll mL−1). Glc, DBMIB, DCMU, and lincomycin were added to a final concentration of 5 mm, 10 μm, 10 μm, and 250 μg mL−1, respectively. Synechocystis strains and their relevant genotypes are described in Supplemental Table S1. Escherichia coli DH5α or BL21 cells were grown in Luria broth medium and supplemented with 100 μg mL−1 ampicillin, 50 μg mL−1 kanamycin, 20 μg mL−1 chloramphenicol, and 100 μg mL−1 spectinomycin when required.
Construction of Synechocystis Strains
Synechocystis cells were transformed as described in Ferino and Chauvat (1989). Plasmid construction is detailed in Supplemental Materials and Methods S1. All the oligonucleotides used in this work are described in Supplemental Table S2.
RNA Isolation and Northern-Blot Analysis
Total RNA was isolated from 30-mL samples of Synechocystis cultures in the midexponential growth phase (3–5 μg chlorophyll mL−1). Extractions were performed by vortexing cells in the presence of phenol chloroform and acid-washed baked glass beads (0.25–0.3 mm diameter) as previously described (García-Domínguez and Florencio, 1997). Five micrograms of total RNA was loaded per lane and electrophoresed in 1.2% agarose denaturing formaldehyde gels (Sambrook et al., 1989) and transferred to nylon membranes (Hybond N-Plus; Amersham). Prehybridization, hybridization, and washes were in accordance with Amersham instruction manuals. All probes were synthesized by PCR and oligonucleotide pairs used are described in Supplemental Table S3. Hybridization signals were quantified with a Cyclone Phosphor system (Packard).
Determination of Cellular Copper Content
The cellular copper contents were determined from 800 mL of exponentially growing cells that were treated with 200 nm of copper for 1 h (wild type, SAS, and PETE strains) or 3 μm of copper for 5 h (wild type, COPR, and COPB strains). Cells were centrifuged at 5,000g, washed twice with BG11C-Cu, and dried overnight in an oven at 85°C. One hundred micrograms of dried cells was microwave digested, dissolved in suprapure HNO3, and analyzed by induced coupled plasma (ICP) in an ICP-OES Varian ICP 720-ES (Tottey et al., 2001; Andrés-Colás et al., 2006). Data shown represent the average ± se.
Primer Extension Analysis of copMRS and copBAC Transcripts
Oligonucleotides NIY3 and COPA3, end labeled with T4 polynucleotide kinase and [γ-32P]-dATP (3,000 Ci mmol−1) following standard procedures (Sambrook et al., 1989), were used for primer extension analysis of copMRS or copBAC promoters, respectively. For annealing, a 10-μL mixture containing 0.15 m KCl, 10 mm Tris HCl pH 8.0, 1 mm EDTA, 20 μg of total RNA, and about 2 pmol of oligonucleotides (106 cpm) was prepared. The annealing mixture was heated for 2 min at 90°C in a water bath and cooled slowly to 50°C. For extension, a 10-μL mixture was prepared with half of the annealing mixture: 10 mm dithiothreitol, 0.5 mm each dNTP, 2 mg mL−1 of actinomycin D, 50 mm Tris HCl (pH 8.3), 75 mm KCl, 3 mm MgCl2, and 100 units of Superscript II RNase H-Reverse Transcriptase (Invitrogen). The mixture was incubated for 45 min at 45°C, and the reaction was stopped by adding 4 μL of formamide-loading buffer. Half of the reaction was electrophoresed on a 6% polyacrylamide sequencing gel together with a sequencing reaction of the copMRS or copBAC promoter regions using the same oligonucleotides.
Cloning and Purification of CopRΔN
The complete DNA-binding domain from copR was cloned from Synechocystis DNA after PCR amplification with oligonucleotides COPR3 and NIY2 and cloned into BamHI-SalI pGEX6P. GST-CopRΔN fusion protein was expressed in E. coli DH5α. Two-hundred milliliters of culture was grown in Luria broth medium to an optical density at 600 nm of 0.6, induced with 0.5 mm isopropyl-b-d-thiogalactopyranoside for 2.5 h, harvested by centrifugation, and resuspended in 5 mL of phosphate-buffered saline buffer (150 mm NaCl, 16 mm Na2HPO4, 4 mm NaH2PO4, 4 mm phenylmethylsulfonyl fluoride, 7 mm β-mercaptoethanol) supplemented with 0.1% Triton X-100. Cells were broken by sonication on ice, and insoluble debris were pelleted by centrifugation. Extracts were mixed with 1 mL of glutathione agarose beads (Amersham) and incubated for 2 h at 4°C with gentle agitation. Then beads were transferred to a column and washed extensively with phosphate-buffered saline buffer until no more protein was eluted from the column. GST fusion proteins were eluted with 3 mL of 50 mm Tris HCl (pH 8) containing 10 mm of reduced glutathione.
Gel Retardation Assays
Probes were PCR synthesized using oligonucleotides NIY4 and NIY5, for copMRS promoter, and COPA4 and COPA5 for copBAC promoter, which introduce an NcoI restriction site in both cases. The resulting DNA was digested with NcoI and end labeled with [α-32P]-dCTP (3,000 Ci mmol−1) using Klenow fragment. The binding reaction was carried out in a final volume of 25 μL containing 4 ng of labeled DNA and 4 μg salmon sperm DNA in 20 mm Tris HCl (pH 8.0), 150 mm KCl, 10 mm spermidine, 10 mm dithiothreitol, 1 mm EDTA, 10% glycerol, and different amounts (from 0.2–1 μg) of partially purified GST-CopRΔN. The mixtures were incubated for 25 min at 4°C and loaded on a nondenaturing 6% polyacrylamide gel. Electrophoresis was carried out at 4°C and 200 V in 0.25× Tris-borate/EDTA. Gels were transferred to a Whatman 3 MM paper, dried, and autoradiographed.
Cloning, Purification, and Metal-Binding Assays of CopS Periplasmic Domain (CopS38-183)
A 462-pb band coding for the CopS periplasmic domain was PCR amplified from genomic DNA with oligonucleotides CopSperiF2-CopSperiR2, digested with BamHI and SacI, and cloned into pET51 digested with the same enzymes. CopS38-183 was expressed in E. coli BL21. A total of 1.5 L of culture was grown in Luria broth medium to an optical density at 600 nm of 0.6, induced with 0.2 mm isopropyl-b-d-thiogalactopyranoside, and incubated for 6 h at 25°C; cells were harvested by centrifugation and frozen at −20°C. Frozen pellets were resuspended in 40 mL of 100 mm Tris HCl (pH 8), 150 mm NaCl, 1 mm BCSA, 1 mm EDTA, and 2 mm Tris(2-carboxyethyl)-phosphine (buffer S) and broken by sonication. The suspension was centrifuged 30 min at 30,000g at 4°C and the supernatant was loaded into a 5-mL streptavidin beads (IBA GmbH) column equilibrated in buffer S. Beads were washed with 50 mL of buffer S and CopS38-183 was eluted with 10 mL of 1× Strep-Tag elution buffer (IBA GmbH). CopS38-183 was further purified by gel filtration in a Hi-Load Superdex 75 (GE-Healthcare) column equilibrated with 20 mm Tris HCl (pH 8), 150 mm NaCl. The purified protein was concentrated using a 3K Vivaspin concentrator.
Interaction of CopS38-183 with Cu2+, Ni2+, Zn2+, and Co2+ was investigated by immobilized metal ion affinity chromatography. A 100-μL aliquot of His-bind resin (Novagen) was loaded with 0.5 mL of 0.5 mm of CuSO4, NiSO4, ZnSO4, or CoCl2 in water and then equilibrated in 25 mm Tris HCl (pH 8), 500 mm NaCl (buffer A). About 10 μg of purified CopS38-183 were applied to the columns. Unbound proteins were removed by washing with 2 mL of buffer A. Bound proteins were eluted with 100 μL of 0.4 m imidazole in buffer A. Fifteen microliters of the imidazole eluted and flowthrough fractions were analyzed by SDS-PAGE and Coomassie Blue staining. Quantities of bound and unbound proteins were determined by densitometry.
Analysis of CopS38-183 Cu2+ binding was obtained via colorimetric titration similar to that described previously with the divalent metal ligand PAR (Tottey et al., 2008). PAR (10 μm) in 20 mm Tris HCl (pH 7.5), 50 mm NaCl (buffer B) was titrated against copper (0–20 μm) measuring absorbance in the 350 to 600 nm range. Absorbance of PAR (410 nm) and Cu2+-PAR (500 nm) were plotted against [Cu2+]. Titration was repeated in the same way but with the addition of 10 μm apo-CopS38-183. The apparent dissociation constant (KD) of CopS38-183 for Cu2+ was estimated using competition experiments as described previously (Turski et al., 2012). The quantitative release of the 1:1 Cu2+/PAR complex upon titration of apoCopS38-183 was monitored spectrophotometrically at 500 nm in buffer B. The samples were equilibrated for 5 min at room temperature before the measure. The affinity of Cu2+-PAR complex (formation constant [β]) is 3.2 × 1017, and the Cu2+ binding affinity was calibrated using a spectroscopically silent ligand EDTA, with a known affinity for Cu2+ of 1.6 × 10−19 (Turski et al., 2012).
Membrane Fractionation and Western Blotting
Thylakoid and plasma membranes were prepared from Synechocystis as described previously (Norling et al., 1998). For western-blot analysis, proteins were fractionated on SDS-PAGE and immunoblotted (Sambrook et al., 1989) with antibodies against: HA (1:1,000; Sigma catalog number H9658), NrtA (1:10,000; Omata et al., 1989), PsaC (1:3,000; Mata-Cabana et al., 2007), plastocyanin (1:12,000; Durán et al., 2004), or Synechococcus sp. PCC 6301 Gln synthetase I (1:20,000; Mérida et al., 1990). The ECL Plus immunoblotting system (Amersham) was used to detect the different antigens with anti-rabbit or anti-mouse secondary antibodies conjugated to horseradish peroxidase (1:10,000). Films were scanned and quantified using Image J software.
The genes named in this article can be found in the Cyanobase database (http://genome.kazusa.or.jp/cyanobase/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. copMRS are expressed as a single transcriptional unit.
Supplemental Figure S2. Mutants in cop genes are not differentially affected with respect to the wild-type strain by Ni2+, Co2+, and Zn2+.
Supplemental Figure S3. copBAC are expressed as a single transcriptional unit.
Supplemental Figure S4. Spectral changes of the Cu2+-PAR complex on the CopS(38-183) titration.
Supplemental Figure S5. Redox induction of copMRS and copBAC expression depends on the presence of copper in the medium.
Supplemental Figure S6. copMRS and copBAC expression is not induced after DCMU treatment.
Supplemental Figure S7. copM and copB induction depends on CopR after DBMIB treatment.
Supplemental Figure S8. CopRS do not control copper-related genes.
Supplemental Figure S9. Growth of COPR is not affected by Glc.
Supplemental Figure S10. Nitrogen starvation leads to copM, copB induction, and plastocyanin degradation.
Supplemental Table S1. Synechocystis strains used in this work.
Supplemental Table S2. Oligonucleotides used in this work.
Supplemental Table S3. Oligonucleotides pairs used to synthesize probes used for northern-blot analysis.
Supplemental Materials and Methods S1. Insertional mutagenesis and reverse transcription-PCR.
Supplementary Material
Acknowledgments
We thank Tatsuo Omata, Raúl V. Durán, and Marika Lindhal for kindly providing antibodies against NrtA, plastocyanin, and PsaC. We thank Jose Luis Crespo and Maria José Huertas for critical reading of the manuscript.
Glossary
- HME-RND
heavy-metal efflux-resistance nodulation and division
- DBMIB
2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone
- ORFs
open reading frames
- PAR
apo-4-(2-pyridylazo)-resorcinol
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- BCSA
bathocuproinedisulfonic acid
References
- Andrés-Colás N, Sancenón V, Rodríguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Peñarrubia L. (2006) The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J 45: 225–236 [DOI] [PubMed] [Google Scholar]
- Bagai I, Rensing C, Blackburn NJ, McEvoy MM. (2008) Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry 47: 11408–11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, et al. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24: 738–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco AG, Canals A, Bernués J, Solà M, Coll M. (2011) The structure of a transcription activation subcomplex reveals how σ(70) is recruited to PhoB promoters. EMBO J 30: 3776–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, et al. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. (2010) Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol 192: 2512–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong LX, Ash MR, Maher MJ, Hinds MG, Xiao Z, Wedd AG. (2009) Unprecedented binding cooperativity between Cu(I) and Cu(II) in the copper resistance protein CopK from Cupriavidus metallidurans CH34: implications from structural studies by NMR spectroscopy and x-ray crystallography. J Am Chem Soc 131: 3549–3564 [DOI] [PubMed] [Google Scholar]
- del Pozo T, Cambiazo V, González M. (2010) Gene expression profiling analysis of copper homeostasis in Arabidopsis thaliana. Biochem Biophys Res Commun 393: 248–252 [DOI] [PubMed] [Google Scholar]
- Durán RV, Hervás M, De La Rosa MA, Navarro JA. (2004) The efficient functioning of photosynthesis and respiration in Synechocystis sp. PCC 6803 strictly requires the presence of either cytochrome c6 or plastocyanin. J Biol Chem 279: 7229–7233 [DOI] [PubMed] [Google Scholar]
- Ferino F, Chauvat F. (1989) A promoter-probe vector-host system for the cyanobacterium, Synechocystis PCC6803. Gene 84: 257–266 [DOI] [PubMed] [Google Scholar]
- Finazzi G, Sommer F, Hippler M. (2005) Release of oxidized plastocyanin from photosystem I limits electron transfer between photosystem I and cytochrome b6f complex in vivo. Proc Natl Acad Sci USA 102: 7031–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke S, Grass G, Rensing C, Nies DH. (2003) Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol 185: 3804–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Domínguez M, Florencio FJ. (1997) Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 35: 723–734 [DOI] [PubMed] [Google Scholar]
- García-Domínguez M, Lopez-Maury L, Florencio FJ, Reyes JC. (2000) A gene cluster involved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 182: 1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G, Rensing C. (2001) Genes involved in copper homeostasis in Escherichia coli. J Bacteriol 183: 2145–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Sonoike K, Kanehisa M, Ikeuchi M. (2003) DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 185: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. (2003) Pathways of oxidative damage. Annu Rev Microbiol 57: 395–418 [DOI] [PubMed] [Google Scholar]
- Kahlon S, Beeri K, Ohkawa H, Hihara Y, Murik O, Suzuki I, Ogawa T, Kaplan A. (2006) A putative sensor kinase, Hik31, is involved in the response of Synechocystis sp. strain PCC 6803 to the presence of glucose. Microbiology 152: 647–655 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Sasamoto S, Watanabe A, Kohara M, Matsumoto M, Shimpo S, Yamada M, Tabata S. (2003) Structural analysis of four large plasmids harboring in a unicellular cyanobacterium, Synechocystis sp. PCC 6803. DNA Res 10: 221–228 [DOI] [PubMed] [Google Scholar]
- Kenney LJ. (2002) Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol 5: 135–141 [DOI] [PubMed] [Google Scholar]
- Kim EH, Rensing C, McEvoy MM. (2010) Chaperone-mediated copper handling in the periplasm. Nat Prod Rep 27: 711–719 [DOI] [PubMed] [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S. (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruip J, Chitnis PR, Lagoutte B, Rögner M, Boekema EJ. (1997) Structural organization of the major subunits in cyanobacterial photosystem 1: localization of subunits PsaC, -D, -E, -F, and -J. J Biol Chem 272: 17061–17069 [DOI] [PubMed] [Google Scholar]
- Loftin IR, Franke S, Roberts SA, Weichsel A, Héroux A, Montfort WR, Rensing C, McEvoy MM. (2005) A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry 44: 10533–10540 [DOI] [PubMed] [Google Scholar]
- López-Maury L, García-Domínguez M, Florencio FJ, Reyes JC. (2002) A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol Microbiol 43: 247–256 [DOI] [PubMed] [Google Scholar]
- Macomber L, Imlay JA. (2009) The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci USA 106: 8344–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Cabana A, Florencio FJ, Lindahl M. (2007) Membrane proteins from the cyanobacterium Synechocystis sp. PCC 6803 interacting with thioredoxin. Proteomics 7: 3953–3963 [DOI] [PubMed] [Google Scholar]
- Mealman TD, Bagai I, Singh P, Goodlett DR, Rensing C, Zhou H, Wysocki VH, McEvoy MM. (2011) Interactions between CusF and CusB identified by NMR spectroscopy and chemical cross-linking coupled to mass spectrometry. Biochemistry 50: 2559–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S, Bogorad L. (1986) Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol Cell Biol 6: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Allen MD, Kropat J, Moseley JL, Long JC, Tottey S, Terauchi AM. (2006) Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim Biophys Acta 1763: 578–594 [DOI] [PubMed] [Google Scholar]
- Mérida A, Leurentop L, Candau P, Florencio FJ. (1990) Purification and properties of glutamine synthetases from the cyanobacteria Synechocystis sp. strain PCC 6803 and Calothrix sp. strain PCC 7601. J Bacteriol 172: 4732–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills SD, Jasalavich CA, Cooksey DA. (1993) A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J Bacteriol 175: 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan S, Sherman DM, Shaw I, Sherman LA. (2012) Functions of the duplicated hik31 operons in central metabolism and responses to light, dark, and carbon sources in Synechocystis sp. strain PCC 6803. J Bacteriol 194: 448–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N. (2001) Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J 20: 5587–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norling B, Zak E, Andersson B, Pakrasi H. (1998) 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 436: 189–192 [DOI] [PubMed] [Google Scholar]
- Omata T, Ohmori M, Arai N, Ogawa T. (1989) Genetically engineered mutant of the cyanobacterium Synechococcus PCC 7942 defective in nitrate transport. Proc Natl Acad Sci USA 86: 6612–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai T, Imamura S, Asayama M, Shirai M, Suzuki I, Murata N, Tanaka K. (2006) Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res 13: 185–195 [DOI] [PubMed] [Google Scholar]
- Osman D, Cavet JS. (2008) Copper homeostasis in bacteria. Adv Appl Microbiol 65: 217–247 [DOI] [PubMed] [Google Scholar]
- Outten FW, Outten CE, Hale J, O’Halloran TV. (2000) Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J Biol Chem 275: 31024–31029 [DOI] [PubMed] [Google Scholar]
- Pilon M, Abdel-Ghany SE, Cohu CM, Gogolin KA, Ye H. (2006) Copper cofactor delivery in plant cells. Curr Opin Plant Biol 9: 256–263 [DOI] [PubMed] [Google Scholar]
- Pilon M, Cohu CM, Ravet K, Abdel-Ghany SE, Gaymard F. (2009) Essential transition metal homeostasis in plants. Curr Opin Plant Biol 12: 347–357 [DOI] [PubMed] [Google Scholar]
- Puig S, Andrés-Colás N, García-Molina A, Peñarrubia L. (2007a) Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ 30: 271–290 [DOI] [PubMed] [Google Scholar]
- Puig S, Mira H, Dorcey E, Sancenón V, Andrés-Colás N, Garcia-Molina A, Burkhead JL, Gogolin KA, Abdel-Ghany SE, Thiele DJ, et al. (2007b) Higher plants possess two different types of ATX1-like copper chaperones. Biochem Biophys Res Commun 354: 385–390 [DOI] [PubMed] [Google Scholar]
- Puig S, Peñarrubia L. (2009) Placing metal micronutrients in context: transport and distribution in plants. Curr Opin Plant Biol 12: 299–306 [DOI] [PubMed] [Google Scholar]
- Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. (2000) CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci USA 97: 652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Grass G. (2003) Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev 27: 197–213 [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herman M, Stanier RY. (1979) Generic assignment, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–61 [Google Scholar]
- Robinson NJ, Winge DR. (2010) Copper metallochaperones. Annu Rev Biochem 79: 537–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford JC, Cavet JS, Robinson NJ. (1999) Cobalt-dependent transcriptional switching by a dual-effector MerR-like protein regulates a cobalt-exporting variant CPx-type ATPase. J Biol Chem 274: 25827–25832 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York [Google Scholar]
- Solioz M, Abicht HK, Mermod M, Mancini S. (2010) Response of gram-positive bacteria to copper stress. J Biol Inorg Chem 15: 3–14 [DOI] [PubMed] [Google Scholar]
- Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. (2011) Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470: 558–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield TC, Nagarajan S, Sherman LA. (2011) Gene expression under low-oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and -independent responses. Microbiology 157: 301–312 [DOI] [PubMed] [Google Scholar]
- Thelwell C, Robinson NJ, Turner-Cavet JS. (1998) An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc Natl Acad Sci USA 95: 10728–10733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S, Patterson CJ, Banci L, Bertini I, Felli IC, Pavelkova A, Dainty SJ, Pernil R, Waldron KJ, Foster AW, et al. (2012) Cyanobacterial metallochaperone inhibits deleterious side reactions of copper. Proc Natl Acad Sci USA 109: 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S, Rich PR, Rondet SA, Robinson NJ. (2001) Two Menkes-type atpases supply copper for photosynthesis in Synechocystis PCC 6803. J Biol Chem 276: 19999–20004 [DOI] [PubMed] [Google Scholar]
- Tottey S, Rondet SA, Borrelly GP, Robinson PJ, Rich PR, Robinson NJ. (2002) A copper metallochaperone for photosynthesis and respiration reveals metal-specific targets, interaction with an importer, and alternative sites for copper acquisition. J Biol Chem 277: 5490–5497 [DOI] [PubMed] [Google Scholar]
- Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, Cheek TR, Gray J, Banfield MJ, Dennison C, et al. (2008) Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 455: 1138–1142 [DOI] [PubMed] [Google Scholar]
- Trebst A. (2007) Inhibitors in the functional dissection of the photosynthetic electron transport system. Photosynth Res 92: 217–224 [DOI] [PubMed] [Google Scholar]
- Turski ML, Brady DC, Kim HJ, Kim BE, Nose Y, Counter CM, Winge DR, Thiele DJ. (2012) A novel role for copper in Ras/mitogen-activated protein kinase signaling. Mol Cell Biol 32: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron KJ, Tottey S, Yanagisawa S, Dennison C, Robinson NJ. (2007) A periplasmic iron-binding protein contributes toward inward copper supply. J Biol Chem 282: 3837–3846 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. (2009) SQUAMOSA promoter binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, McSpadden B, Pakrasi HB, Whitmarsh J. (1992) Copper-mediated regulation of cytochrome c553 and plastocyanin in the cyanobacterium Synechocystis 6803. J Biol Chem 267: 19054–19059 [PubMed] [Google Scholar]
- Zhang Z, Pendse ND, Phillips KN, Cotner JB, Khodursky A. (2008) Gene expression patterns of sulfur starvation in Synechocystis sp. PCC 6803. BMC Genomics 9: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.