Abstract
Freshwater habitats have been identified as one of the largest reservoirs of archaeal genetic diversity, with specific lineages of ammonia-oxidizing archaea (AOA) populations different from soils and seas. The ecology and biology of lacustrine AOA is, however, poorly known. In the present study, vertical changes in archaeal abundance by CARD-FISH, quantitative PCR (qPCR) analyses and identity by clone libraries were correlated with environmental parameters in the deep glacial high-altitude Lake Redon. The lake is located in the central Spanish Pyrenees where atmospheric depositions are the main source of reactive nitrogen. Strong correlations were found between abundance of thaumarchaeotal 16S rRNA gene, archaeal amoA gene and nitrite concentrations, indicating an ammonium oxidation potential by these microorganisms. The bacterial amoA gene was not detected. Three depths with potential ammonia-oxidation activity were unveiled along the vertical gradient, (i) on the top of the lake in winter–spring (that is, the 0 oC slush layers above the ice-covered sheet), (ii) at the thermocline and (iii) the bottom waters in summer—autumn. Overall, up to 90% of the 16S rRNA gene sequences matched Thaumarchaeota, mostly from both the Marine Group (MG) 1.1a (Nitrosoarchaeum-like) and the sister clade SAGMGC−1 (Nitrosotalea-like). Clone-libraries analysis showed the two clades changed their relative abundances with water depth being higher in surface and lower in depth for SAGMGC−1 than for MG 1.1a, reflecting a vertical phylogenetic segregation. Overall, the relative abundance and recurrent appearance of SAGMGC−1 suggests a significant environmental role of this clade in alpine lakes. These results expand the set of ecological and thermal conditions where Thaumarchaeota are distributed, unveiling vertical positioning in the water column as a key factor to understand the ecology of different thaumarchaeotal clades in lacustrine environments.
Keywords: Archae, AOA, lakes, alpine
Introduction
Freshwater habitats have recently been proposed as one of the largest reservoirs of archaeal genetic diversity (Llirós et al., 2008; Auguet et al., 2010). Some of the representative freshwater lineages identified may represent potential key archaeal groups in terms of distribution and ecological processes (Auguet et al., 2010; Barberán et al., 2011). Unfortunately, at present, only a few examples provide enough information to gain clues on the ecological significance of these uncultured microorganisms, such as the ammonia-oxidizing populations within the thaumarchaeotal Marine Group (MG) 1.1a. The genomes of widespread and abundant representatives of the domain Archaea encode for the ammonia mono-oxygenase enzyme (Venter et al., 2004; Treusch et al., 2005). The isolated Thaumarchaeota Nitrosopumilus maritimus (Konneke et al., 2005) have shown chemolithoautotrophic growth fueled with ammonia as energy source, initiating further research on the role of archaea in the global nitrogen cycle.
In aquatic ecosystems, the phylogenies of both amoA and 16S rRNA genes indicate that most ammonia-oxidizing archaea (AOA) belong to a few specific lineages, mainly the MG 1.1a and the deeply branching thaumarchaeotal clade pSL12 (Mincer et al., 2007). These lineages are distantly related to those associated with soil or sediment (Auguet et al., 2011). Despite the fact that only a handful of freshwater environments have been investigated so far, AOA have been consistently detected in rivers (Galand et al., 2006; Herfort et al., 2009; Merbt et al., 2011), the Great African Lake Kivu (Llirós et al., 2010), groundwater (Reed et al., 2009), drinking waters (van der Wielen et al., 2009), freshwater sediments (Herrmann et al., 2008) and alpine lakes (Auguet et al., 2011). These studies have significantly expanded the archaeal amoA phylogenetic tree showing lacustrine amoA gene sequences grouped in coherent phylogenetic clusters, and suggesting that freshwater habitats contain archaeal nitrifiers different from soil and marine habitats.
Spatio-temporal changes in the distribution of planktonic freshwater AOA are, however, poorly documented (Pouliot et al., 2009; Llirós et al., 2010; Auguet et al., 2011) in contrast to what is known in the marine realm. The characteristic AOA abundance peak observed in the oxygen-minimum zone and dimly lit oceanic waters are often associated to maxima in nitrite concentrations, suggesting active ammonia oxidation in these areas. Significant correlations found between archaeal amoA transcripts abundance and nitrite maxima in the oxygen-minimum zone of the Black Sea (Lam et al., 2007), the Peruvian upwelling (Lam et al., 2009), and upwelled waters north of the equator (Church et al., 2010) have been reported. Consistent differences have been observed in the sea for vertical AOA abundance, identity and activity (Francis et al., 2005; Hallam et al., 2006; Mincer et al., 2007; Beman et al., 2008; Agogué et al., 2008; Church et al., 2010). In an interannual study of surface waters in the Mediterranean sea, the abundance of AOA showed a consistent seasonal pattern, and amoA gene relative abundance was positively correlated to nitrate concentrations (Galand et al., 2010). From the few studies carried out in planktonic freshwater, both strong seasonal dynamics in AOA observed in surface waters of shallow alpine lakes, which were well explained by temporal changes in ammonium and nitrite concentrations (Auguet et al., 2011), and the maximal abundance of AOA in the oxycline/nitracline of chemically stratified lakes (Pouliot et al., 2009; Llirós et al., 2010), suggest a close link between freshwater Thaumarchaeota and the nitrogen cycling. In a recent investigation (Auguet et al., 2011), we studied the temporal changes in the archaeal assemblages inhabiting the surface waters of three connected shallow and well-mixed Pyrenean lakes (Redó AiguesTortes (AT), Llong and Llebreta in the AiguesTortes valley) with a very low water-residence time (see Table 1 in Auguet and Casamayor, 2008). In the present study, we followed the vertical changes of AOA along the water column in the deep glacial stratified alpine Lake Redon (three orders of magnitude higher water-residence time, Auguet and Casamayor, 2008) during one year unveiling potential archaeal nitrification hotspots along the water column and vertical phylotype segregation.
Table 1. Diversity indicators of archaeal assemblages in Lake Redon based on OTUs defined by the 16S rRNA (at 3% cutoff) and amoA (5% cutoff) genes.
| Clone libraries | Clone numbers | OTU | Coverage(%) | Chao1 | PD |
|---|---|---|---|---|---|
|
16S rRNA
| |||||
| Slush-April | 24 | 4 | 87 | 7 | 0.78±0.05 |
| 20m-April | 25 | 8 | 86 | 10 | 0.92±0.08 |
| 60m-April | 26 | 13 | 73 | 18 | 1.03±0.11 |
| SML-Sep | 29 | 8 | 89 | 9 | 0.48±0.02 |
| 20m-Sep | 29 | 13 | 69 | 31 | 1.05±0.10 |
| 60m-Sep | 29 | 16 | 59 | 49 | 1.46±0.15 |
|
amoA
| |||||
| Slush-April | 24 | 6 | 91 | 6 | 0.86±0.09 |
| 20m-April | 18 | 5 | 89 | 6 | 0.79±0.05 |
| 60m-April | 30 | 10 | 83 | 13 | 0.64±0.06 |
| SML-Sep | 5 | 4 | 40 | 5 | 0.59±0.10 |
| 20m-Sep | 27 | 6 | 92 | 6 | 0.35±0.08 |
| 60m-Sep | 27 | 5 | 96 | 5 | 0.39±0.11 |
Abbreviations: OTUs, operational taxonomic units; PD, phylogenetic diversity; rRNA, ribosomal RNA; SML, surface microlayer.
Materials and methods
Study site, sampling and general analyses
Lake Redon is a deep-glacial cirque lake located in the Conangles valley, central Spanish Pyrenees (42º38′34″N, 0º46′13″E, altitude 2240, m, maximum depth 73 m, surface 0.24 km2) extensively studied in the last 30 years by limnologists (see Catalan et al., 2006 for more details) and recently by bacterial ecologists (Hervàs and Casamayor 2009; Llorens-Marès et al., 2012). This lake is completely different from Lake Redó AT located in the Aigües Tortes National Park, a shallow (12 m depth) well-mixed lake with a very low water-residence time (see more details in Auguet and Casamayor, 2008; Auguet et al., 2011). Local precipitation is the sole source of water for Lake Redon, as there are no permanent rivers in the basin, and the lake has a small catchment area of c. 1.5 km2. The lake is usually ice-covered for about 6 months of the year. The thickness of the ice cover reaches several meters during its maximum in the month of April with dynamic changes in composition of the ice bacterial assemblages (Llorens-Marès et al., 2012). Lake Redon has a dimictic regime, with mixing periods in spring and in autumn. The lake is oligotrophic owing to the sparse vegetation and poorly developed soils in the small catchment area, and atmospheric deposition is the main source of reactive nitrogen (Catalan et al., 1994).
Samples for this study were collected monthly between May 2007 and April 2008 from six selected depths (air–water surface microlayer (SML), 2, 10, 20, 35 and 60 m) at the deepest point of the lake using a 2-l Niskin bottle. These depths covered different limnological characteristics of the lake. The lake was frozen from November to April and inaccessible during January and February (data for these two months are missing in the graphs). The first 400 μm of the SML were collected with a nylon screen sampler (Auguet and Casamayor, 2008) during the ice-free period. In March and April 2008, we sampled the slush (that is, mixture layer of snow and lake water formed within the ice cover) as previously reported (Llorens-Marès et al., 2012) and samples were processed in the laboratory within 1–2 h.
Samples for dissolved organic carbon (DOC) were filtered at the sampling site through a pre-combusted Whatman GF/F filter (Whatman Ltd, Kent, UK) and stored in the dark at 4 °C until analyzed (between 3 and 5 h after sampling) by high temperature combustion/direct injection into a Shimadzu TOC 5000 IR gas analyzer (Shimadzu Co., Kyoto, Japan). Sample stability and blank tests were run to check that the storage did not affect DOC measurements. Ammonium was colorimetrically determined according to Solórzano (1969). Nitrate was measured by capillary electrophoresis (model Quanta-4000, Waters Co., Milford, MA, USA) (Doble and Haddad, 1999). Nitrite was determined spectrophotometrically at 560 nm after the reaction in H3PO4 acid medium with a solution of sulfanilamide and N-(1-naphthyl) ethylene diamine dihydrochloride. Chlorophyll a was spectrophotometrically determined using water samples (0.5 ml) collected on Whatmann GF/F glass fiber filters and extracted with acetone (90%, v/v). Prokaryotic cells were collected in triplicate on 0.2-μm pore-size polycarbonate filters and processed for DAPI and CARD-FISH counts with probes EUB338 for bacterial and ARCH915 for archaeal cells as previously reported (Auguet and Casamayor, 2008).
Genetic analyses
Water samples were pre-filtered in situ through a 40 μm pore-size net, to exclude large zooplankton and algae, and 300–500 ml were subsequently filtered on 0.2 μm polycarbonate membranes (47 mm diameter, Nuclepore, Whatman Ltd). The filter was stored at –20 °C until further processing. Filters were incubated with lysozyme, proteinase K, and sodium dodecyl sulfate in lysis buffer (40 mℳ EDTA, 50 mℳ Tris pH 8.3, 0.75 ℳ sucrose), and DNA was phenol extracted as previously reported (Demergasso et al., 2008).
We initially used denaturing gradient gel fingerprinting (DGGE) on the whole samples set for a rapid screening on the vertical shifts in archaeal 16S rRNA gene diversity (see Auguet et al., 2011 for specific PCR and DGGE conditions). Next, according to the shifts observed in the fingerprints, we selected six samples to describe the phylogenetic composition and vertical changes by PCR cloning of both the 16S rRNA (general archaeal primer set 21f-958r, 937 bp fragment length) and the archaeal amoA (primer set Arch-amoAF – Arch-amoAR, 635 bp fragment length) genes following previously reported conditions (Auguet et al., 2011 and see also the original references for the primers therein). PCR products were purified with the QIA quick PCR Purification kit (Qiagen, Hilden, Germany) and cloned with the TOPO TA cloning kit (Invitrogen, Life Technologies Co., Carlsbad, CA, USA) following the manufacturer's instructions. The presence of inserts was checked by ampicillin resistance and blue/white selection on LB plates supplemented with ampicillin 100 μg ml−1 and X-gal 40 μg ml−1 as previously reported (Ferrera et al., 2004). Single-strand sequencing reactions were carried out using external facilities (http://www.macrogen.com).
Phylogenetic analysis
The 16S rRNA gene sequences were automatically aligned with the NAST aligner (DeSantis et al., 2006), clustered at 97% identity threshold, and imported into the Greengenes database (http://greengenes.lbl.gov/) based on the ARB package (Ludwig et al., 2004; http://www.arb-home.de). The Archaea base frequency ARB filter was applied to exclude highly variable positions before sequences were added using the ARB parsimony insertion tool to the original Greengenes tree provided by default.
The amoA gene sequences were manually checked with BioEdit (Hall, 1999) and submitted for matching in the protein database using translated nucleotide sequences (BLASTX, www..nlm.nih.gov; Altschul et al., 1990) to check for protein identity. Next, sequences were clustered at 99% identity in nucleotides with CD-hit (Li and Godzik, 2006). Multiple sequence alignment, phylogenetic inference by maximum likelihood and calculation of amoA gene identity matrices was carried out as recently reported (Auguet et al., 2011).
Quantitative PCR (qPCR)
Abundances of thaumarchaeotal (both MG 1.1a and SAGMGC−1) 16S rRNA and archaeal amoA genes were determined by quantitative real-time PCR amplification. The qPCR assays were run in triplicate on 96 well white qPCR plates with adhesive seals (Bio-Rad, Hercules, CA, USA) in a DNA Engine thermal cycler (Bio-Rad) equipped with a Chromo 4 Real-Time Detector (Bio-Rad) using primers sets (MCGI-391f/MCGI-554r and AOA-amoA-f/AOA-amoA-r, respectively) and conditions previously reported (Auguet et al., 2011). Primers MCGI-391f/MCGI-554r targeted both MG 1.1a and SAGMGC-1. Additionally, the bacterial amoA gene was targeted with primers amoA-1F (5′-GGGTTTCTACTGGTGGT-3′) and amoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (Rotthauwe et al., 1997). However, no amplification for the amoA gene of bacteria was obtained with the set of samples tested. Overall, average efficiencies for all quantification reactions ranged from 86% for the archaeal amoA to 91% for the 16S rRNA gene with R values>0.99. Specificity of reactions was confirmed by melting curve analyses and after looking for unspecific PCR products such as primer dimers or gene fragments of unexpected length in agarose gel electrophoresis.
Diversity indices and statistical analyses
Phylogenetic diversity (PD) was calculated as the sum of the branch lengths associated with the 16S rRNA gene sequences for each sample (Faith, 1992). To correct the unequal number of sequences, we calculated the mean phylogenetic diversity of 1000 randomized subsamples (Barberán and Casamayor, 2010). Chao diversity indices were calculated using the FastGroupII server http://fastgroup.sdsu.edu/fg_tools.htm). Coverage of libraries was calculated using the diversity tool (http://www.aslo.org/lomethods/free/2004/0114a.html) developed by Kemp and Aller (2004) Distance matrices for 16S rRNA and amoA genes were constructed with UniFrac (http://bmf.colorado.edu/unifrac) (Lozupone et al., 2006). To assess the sources of variation in the 16S rRNA and amoA UniFrac matrices, we carried out permutational multivariate analysis of variance based on 1000 permutations (McArdle, 2001) using the function adonis in vegan package (Oksanen et al., 2008). Spearman's rank (rs) correlations were run to investigate the relationship between environmental parameters and gene abundance. All statistical analyses were run in the R environment (http://www.r-project.org/) with the pvclust package for cluster analysis.
Nucleotide sequence accession numbers
Sequences were deposited in GenBank under the accession numbers HE589603 to HE589764 for the 16S rRNA gene and HE598566 to HE598696 for the amoA gene (see also Supplementary Table S2 for detailed assignation). Gene sequences were deposited following MIENS (Minimal Information about an Environmental Study Standard) and meta-data have the accession numbers ERS054479–ERS054484.
Results
The complete field data set of the vertical changes in temperature, oxygen, DOC, nitrogen compounds and prokaryotes abundance during the annual study can be found in Supplementary Material (Table S1). Water temperatures ranged from 0 (in the slush in winter-spring) to 13 °C (in the summer surface layers). In July–September, the water column was thermally stratified with the thermocline located between 10 and 35 m. During the ice-cover period, the lake was inversely stratified (warmer water downward). Below 35 m the water column was thermally stable at 4 °C all over the year. Oxygen profiles ranged from 170 to 400 μℳ throughout the year. Oxygen saturation was found in most of the samples and the lowest concentration was detected close to the bottom in October and April. Chlorophyll a (Chl a) concentrations ranged from 0.07 to 4.44 μg l−1 and were consistently higher in summer with a deep chlorophyll maximum at 35 m, and a secondary-abundance peak at 10 m. DOC ranged from 12 to 100 μℳ with higher concentrations in surface waters in summer. Concentrations of ammonium, nitrate and nitrite ranged from 0.08 to 4.55, 5.8 to 11.2 and 0.02 to 0.10 μℳ, respectively, nitrate being the predominant form of inorganic nitrogen with maximal concentrations in summer around the thermocline. Ammonium concentrations showed higher concentration in bottom waters than in surface with the highest concentration found at the transition from summer to autumn. We also observed ammonium accumulation in the slush layers. Nitrite concentrations showed two maxima at 20 and 60 m during the ice-free period, and an additional maximum in the slush layers during the ice-covered period. Cell abundance ranged from 3 × 105 to 1 × 106 bacterial cells per ml, and from 2 × 103 to 1 × 105 archaeal cells per ml. These cells abundances were within the same range as those previously reported for Pyrenean lakes (Auguet and Casamayor, 2008).
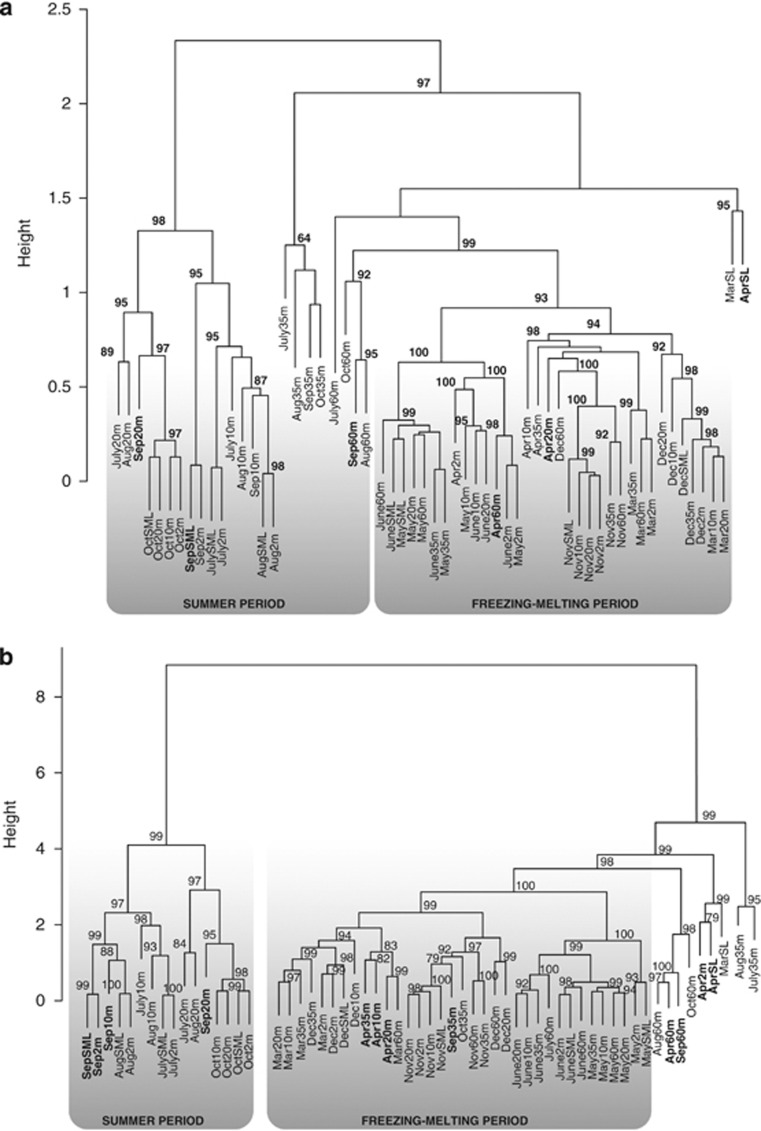
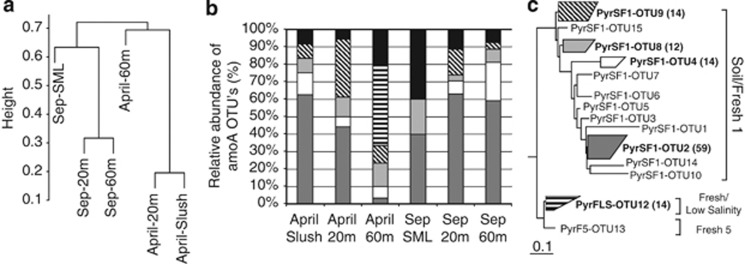
The DGGE analysis of the archaeal 16S rRNA gene produced a reproducible genetic fingerprint for each sample in different runs. The number of DGGE bands for the whole set of samples ranged between 5 and 12 per sample. Similarity and permutational multivariate analysis of variance analyses made on the manually assigned fingerprint showed ‘depth'as a significant source of variation (R2=0.31, P<0.01), particularly during the summer period (Figure 1a). Here, we identified two different clusters of samples according to the archaeal community composition, (i) surface samples up to 20 m, and (ii) samples from 20 m to the bottom of the lake. Samples also clustered according to the temporal variations (R2=0.57, P<0.001) into two main data sets, one for the freezing–melting period, and another for summer as previously reported in other Pyrenean lakes (Auguet et al., 2011). In fact, the spatio-temporal variations in archaeal diversity mirrored the most significant sources of variation in environmental parameters as determined by a hierarchical clustering and permutational multivariate analysis of variance analysis of the whole environmental data set (Figure 1b) (‘depth' R2=0.22, P<0.01; ‘month' R2=0.65, P<0.001). On the basis of these results, we selected samples representative of each period (that is, April and September) and each depth for more detailed genetic analyses by qPCR and cloning of both 16S rRNA and amoA genes.
Figure 1.
(a) Spatio-temporal clustering according to the similarity matrix generated from archaeal 16S rRNA gene DGGE fingerprints and UPGMA clustering method. Code indicates month.depth. Six selected samples (bold labeling) were further studied by cloning and qPCR analysis. (b) Samples clustering according to changes in environmental parameters (chlorophyll a, DOC, nutrients, oxygen concentrations, cells abundance and temperature) based on a hierarchical ascendant classification analysis.
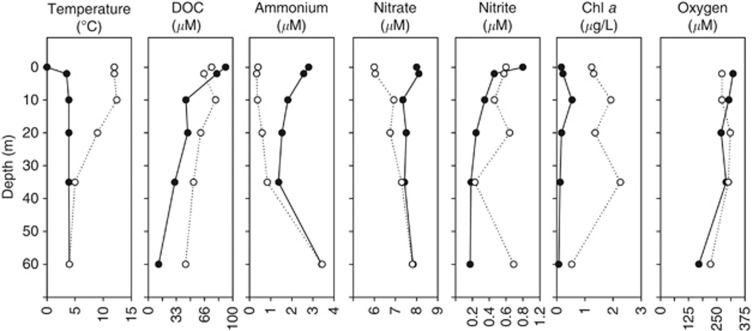
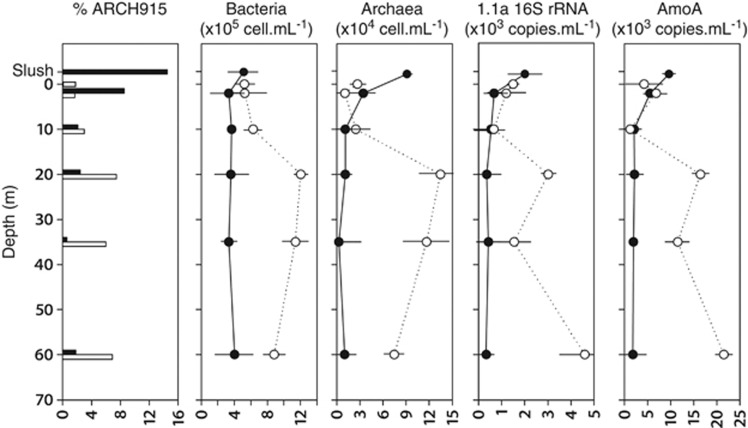
In April, the lake was thermally mixed with a constant water column temperature of 4 °C below the ice cover. DOC, nitrite, ammonium, Chl a and oxygen concentrations gradually decreased with depth although ammonium concentration remarkably increased from 35 to 60 m (Figure 2). The cell abundances (CARD-FISH counts) and quantitative distribution of specific archaeal genes showed little variations with depth, ranging from 3.3 × 102 to 6.7 × 10216S rRNA gene copies ml−1 and from 1.8 × 103 to 5.6 × 103 amoA gene copies ml−1 (Figure 3, black labels for April). Interestingly, the slush layer showed higher nutrients concentrations (particularly enriched in nitrite) and cell abundances than the water column below (Figure 3). This was particularly true for archaea. In the slush, we observed the highest proportion of positive CARD-FISH hybridized archaeal cells (14.5% of total DAPI counts), and the highest concentration of MG 1.1a+SAGMGC−1 16S rRNA (4.3 times the average value in the water column) and archaeal amoA (times 3.5) genes (Figure 3, right panels). In September the lake was thermally stratified and three differentiated water masses were observed along the vertical axis, that is, epilimnion (0–10 m), metalimnion (thermocline, 10–40 m) and hypolimnion (40 m bottom) (Figure 2). The epilimnion showed the lowest proportion of (i) positively hybridized archaeal cells, (ii) cell abundances and (iii) specific archaeal genes (Figure 3, white labels for September). The microbial parameters markedly increased in the metalimnion with the archaeal abundance peak found at 20 m matching the peak of nitrite concentration (Figure 2, Supplementary Table S1). The hypolimnion was enriched in different nitrogen compounds, and the abundances for MG 1.1a+SAGMGC−1 16S rRNA and archaeal amoA genes showed a second abundance peak at the bottom of the lake (Figure 3). Overall, only nitrite significantly explained the vertical variability of specific archaeal gene abundances (P<0.05) after multiple linear regression analysis. In addition, the abundances of the 16S rRNA and amoA genes were significantly correlated (rs=0.88, P<0.01, n=12, slope=4.8).
Figure 2.
Vertical profiles of temperature, DOC, Nitrogen species, Chl a and oxygen, along the water column of Lake Redon in April (black symbols) and September (white symbols). The slush sample was sampled in April and the SML (0 m) in September.
Figure 3.
Vertical profiles of (i) the percentage of archaea (% ARCH915) versus total cell counts, (ii) the abundance of bacterial and archaeal cells as determined by CARD-FISH and (iii) the concentrations of MG 1.1a+SAGMGC-1 16S rRNA and archaeal amoA gene copies as determined by qPCR, along the water column of Lake Redon in April (black symbols) and September (white symbols). The slush sample was sampled in April and the SML (0 m) in September. Standard deviations <10% applies for all the data.
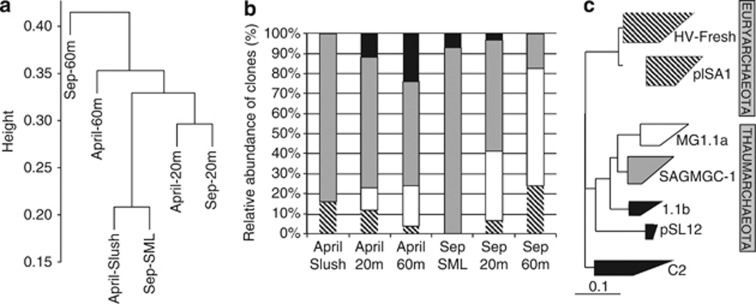
As a whole, the qPCR results indicated three potential ammonia oxidation hotspots in the lake, (i) on the top (slush) in winter–spring, (ii) at the thermocline and (iii) at the bottom in summer–autumn. A similar vertical distribution between MG 1.1a+SAGMGC−1 16S rRNA and archaeal amoA genes was also observed. In order to unveil the phylogenetic composition for archaeal 16S rRNA and amoA genes, and the vertical changes in identity, six samples from three selected depths (top, 20 m, and 60 m) were cloned for April and September. Most of the 16S rRNA gene sequences matched Thaumarchaeota (close to 90%) and only a few Euryarchaeota were detected (Figure 4). Thaumarchaeota were spread in the clusters MG 1.1a, SAGMAGC−1, pSL12 (mostly found at the bottom waters during the ice-cover period), the soil group 1.1b (found at the SML), and a few sequences from the crenarchaeotal group C2. Overall, the members of SAGMAGC-1 and MG 1.1a accounted for most archaeal 16S rRNA gene sequences (>80%). The euryarchaeotal sequences detected, essentially belonged to the widespread clades HV-Fresh (also named LDS, for Lake Dagow sediment) and PlSA1 (also named RC V, for Rice Cluster V) (Auguet et al., 2010; Barberán et al., 2011).
Figure 4.
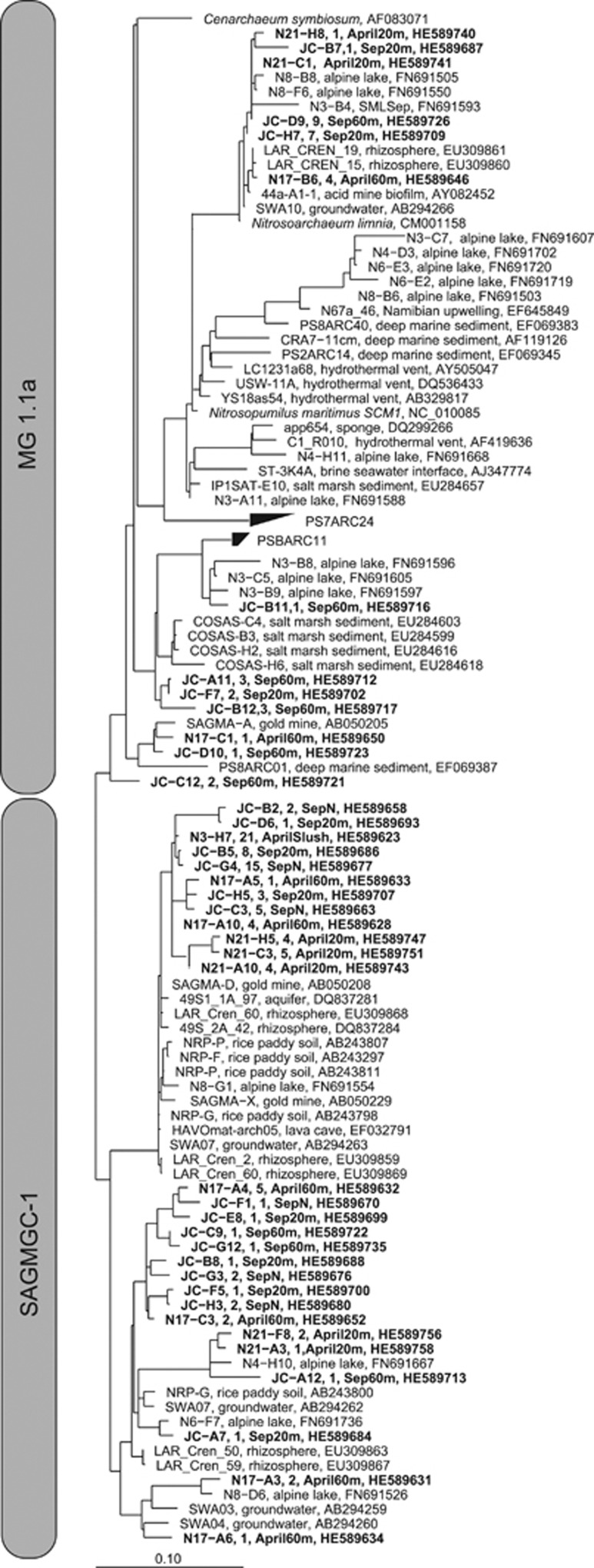
Different analyses carried out with sequence data from the six 16S rRNA clone libraries of Lake Redon in April and September. (a) Hierarchical clustering analysis (UPGMA algorithm with Jackknife supporting values, 100 replicates). Distances between clusters expressed in UniFrac units (distance 0 means identical environments, distance 1 means two environments containing mutually exclusive lineages). (b) Archaeal community structure (relative abundances). Thaumarchaeota Group 1.1a (white label) and SAGMAGC-1 (gray label) are the dominant groups. Other minor thaumarchaeotal/crenarchaeotal groups detected are shown together (black label), as well as the euryarchaeotal groups (shaded label). (c) Collapsed maximum parsimony tree showing the phylogenetic position of the archaeal groups detected. See more details in Figure 5. Scale bar, 10% estimated divergence.
Interestingly, the archaeal assemblages from the two most peculiar samples of this study (that is, winter slush and summer SML) were largely dominated (c. 85–90% of the sequences) by SAGMAGC-1 thaumarchaeotal sequences rooted with the MG 1.1a (89% identity between both the clades) (Figure 4). SAGMAGC-1 was also detected as the predominant clade along the whole water column for most of the samples. Remarkably, sequences from the MG 1.1a were not detected in the slush and SML layers, and its contribution was larger in the summer-stratified hypolimnion. Most of the MG 1.1a sequences were closely related (>97% identity in the 16 S rRNA gene sequence) to Nitrosoarchaeum limnia (Figure 5, upper clade), a novel AOA recently enriched from low-salinity sediments (Blainey et al., 2011). SAGMGC−1 sequences, in turn, were closely related (between 99 and 94% identity) to the AOA Nitrosotalea devanaterra (Lehtovirta-Morley et al., 2011) and to environmental thaumarchaeotal sequences previously found in lakes, aquifers, rhizosphere and rice–paddy soil (Figure 5, lower clade).
Figure 5.
Maximum-parsimony tree of the 16S rRNA gene sequences from the Marine 1.1a and SAGMAGC-1 groups obtained along the water column of Lake Redon (in bold label) in April and September. GenBank accession numbers are shown for each sequence. Scale bar, 10% estimated divergence.
For the archaeal amoA gene, 130 clones were sequenced (Table 1), which ended in 17 unique operational taxonomic units (OTUs, 5% cutoff ranging from 50 to 94% nucleotide identity). The recovered amoA genes were shared between 60 and 88% nucleotide identity with Nitrosopumilus maritimus. Interestingly, we noticed that, within a single lake sample, we could find OTUs diverging up to 38% in nucleotide sequence. Five OTUs represented 80±20% of the sequences, that is, OTUs numbers 2, 4, 8, 9 and 12 (Figure 6, panels b and c), and up to 98% of the amoA gene sequences fell into two previously reported freshwater clusters obtained from Lake Llebreta, a shallow alpine lake belonging to the Limnological Observatory of the Pyrenees (Supplementary Figure. S1). Most of the sequences matched the soil/fresh-1 group (cluster A, in Auguet et al., 2011). In turn, OTU−12 (very abundant at the bottom of the lake in winter–spring, and responsible for splitting this sample from the remaining April samples, Figure 6a) fell into the fresh/low-salinity group (cluster C), which has 95% identity with the amoA sequence of N. limnia. Probably, OTU−12 belongs to those Archaea from the 1.1a group which were closer to N. limnia. Overall, community similarity analysis of the 16S rRNA and amoA gene libraries showed consistent vertical differences in the composition of the archaeal sequences (P<0.05, permutational multivariate analysis of variance test). Despite the fact that relative abundances in clone libraries cannot be translated to real abundances, and truly quantitative methods are needed to substantiate and further explore these findings, the set of samples were treated in exactly the same way and, therefore, any bias should apply the same and the comparison would still be valid. In addition, the quite limited number of clones analyzed per sample (Table 1) is in agreement with the high coverage values found (>77% coverage on average for the 16S rRNA gene, and 82% coverage on average for the amoA gene, Table 1), and the low number of DGGE bands found for each sample (5–12), indicating a low archaeal richness.
Figure 6.
Different analyses carried out with sequence data from the six amoA clone libraries of Lake Redon in April and September as in Figure 4. (a) Hierarchical clustering analysis. (b) AOA community structurere presented by relative abundances of the five most represented OTUs (i.e., OTUs 2, 4, 8, 9 and 12, see labels and number of sequences within each group in panel c) whereas the remaining OTUs are condensed (black bars). (c) Collapsed maximum likelihood tree inferred by RaxML and based on 579 nucleotides positions. See more details in Supplementary Figure. S1. Scale bar, 10% estimated divergence.
Discussion
Recently described phylogenetic lineages within the domain Archaea are significant components in freshwater plankton (Auguet and Casamayor, 2008; Barberán et al., 2011). Archaeal 16S rRNA and amoA genes had been previously reported to be abundant and genetically diverse in surface waters of alpine lakes (Auguet and Casamayor, 2008; Auguet et al., 2011). To understand better, the ecology of these microorganisms, we investigated the relative abundance, distribution and phylogenetic composition along the vertical gradient in a deep oligotrophic alpine lake. The recurrent dominance of AOA lineages and the strong correlations detected between abundances of MG 1.1a+SAGMGC-1 16S rRNA and archaeal amoA genes, and nitrite concentrations both in surface waters (Auguet et al., 2011), and along the vertical gradient (present study) suggest a pivotal role of archaea in the N biogeochemistry of oligotrophic high-altitude lakes. In agreement, we did not found either ammonia-oxidizing bacteria (AOB) after specifically looking for the bacterial amoA gene, or β- and γ-proteobacterial nitrifiers when looking at the bacterial 16S rRNA gene clone libraries (Hervàs and Casamayor, 2009). This result was expected as ammonium concentrations in the water are mostly below the AOB affinity threshold (>1 μM NH4+, Bollmann et al., 2002). Therefore, ultra-oligotrophic waters from mountain areas may constitute an ideal habitat for AOA as the low ammonium concentrations in situ would select for AOA because of their higher affinity to ammonium (Km130 nM NH4+, Martens-Habbena et al., 2009) as compared with AOB.
Obviously, caution is needed before a direct link between amoA gene abundance and nitrification activity can be established in absence of bulk nitrification measurements. In some cases it has been reported that the archaeal amoA gene could be an evolutionary relict that is not functional anymore (Mussmann et al., 2011). Nonetheless, in Lake Redon the match found between the vertical abundance of nitrite and peaks in the abundance of both amoA and 16 S rRNA genes strongly suggests potential active ammonium oxidation zones related to archaea. Intriguingly, abundances of the 16S rRNA and the amoA genes differ markedly. Studies in marine environments reported an archaeal amoA to 16 S ratio of 0.5:1 and lower (De Corte et al., 2009). In Lake Redon ratios of 5:1–8:1 were observed, suggesting that either not all 16S genes of amoA gene containing organisms were detected with the qPCR assay or that the archaea present in the lake have a higher copy number of amoA genes per genome.
Interestingly, we unveiled ammonium oxidation zones changing vertically with seasons. In summer–autumn, nitrification at the bottom of the lake could be related to the ammonium produced in the sediment by microbial decomposition of organic material from early summer phytoplankton blooms (Catalan et al., 2006), and the oxygen-minimum zone observed. In winter–spring, microbial activities and biomasses are by far larger in the slush layers generated by episodic events of snow deposition, flooding by lake water, melting, freezing and atmospheric depositions (that is rain or snow) (Felip et al., 1995) than in the water column. Atmospheric depositions are the main source of reactive nitrogen in remote mountain catchments (Catalan et al., 1994), particularly ammonium with concentrations in the rain up to 30fold higher than mean values measured in the lake water (Auguet et al., 2011). High influence of airborne dust nutrients has also been reported (Hervàs et al., 2009; Reche et al., 2009) that may fuel AOA nitrification activity. These results reflect the wide set of ecological and thermal conditions where AOA are distributed, ranging from hot springs (Zhang et al., 2008) to the ice layers reported here. Intriguingly, the presence of a potential nitrification zone at 20 m is more difficult to understand because both photoinhibition (Merbt et al., 2012) and preference for a dark environment (Church et al., 2010; Merbt et al., 2011) has been recently reported for some types of AOA.
Another interesting finding of this work is the high relative abundance and high frequency of appearance of the SAGMGC−1 Thaumarchaeota, a sister clade to the marine 1.1 a clade, originally discovered in South Africa gold mines (Takai et al., 2001). This result suggests a significant environmental role for the SAGMGC−1 clade in alpine lakes. Previous reports showed that the members of the SAGMGC−1 clade to be mainly found in freshwater systems, for example, alpine lakes (Auguet and Casamayor, 2008; Auguet et al., 2011) groundwater (Shimizu et al., 2007), aquifers (López-Archilla et al., 2007) and terrestrial habitats such as rhizospheres (Herrmann et al., 2008) and rice–paddy soils (Sakai et al., 2007). Whereas members of the MG 1.1a have been largely implicated in nitrification (Konneke et al., 2005; Hallam et al., 2006; Blainey et al., 2011), for the SAGMGC−1 clade only a very recent study indicates that a cultured strain from an acid soil, Nitrosotalea devanaterra, codes for the amoA gene (Lehtovirta-Morley et al., 2011). Thus, several lines of evidence may support ammonium oxidation potential in freshwater SAGMGC−1. First, SAGMGC-1 belongs to the Thaumarchaeota phylum in which all the cultivated or enriched members contain the amoA gene, and more precisely are closely related to the AOA N. devanaterra. Second, we noticed substantial abundance of amoA gene in a sample (that is, the slush), where the only thaumarchaeal group observed by 16S rRNA gene sequencing was SAGMGC−1. And third, the identity of the amoA gene found in the slush sample was associated with the previously reported soil/fresh 1− amoA cluster A (Auguet et al., 2011).The amoA cluster A is mainly formed by sequences from freshwater origin not yet affiliated to any 16S rRNA-defined population and interestingly fits with the Nitrosotalea subcluster 1 (Pester et al., 2012). Unfortunately, these are indirect findings yet and further work and interdisciplinary approaches are needed to probe that the freshwater SAGMGC−1 members are AOA.
Field studies may provide still helpful insights into the ecology of uncultured SAGMGC−1. In a previous work carried out in 10 alpine lakes, we unveiled the neuston as a selective environment where members of SAGMGC−1 are naturally enriched (Auguet and Casamayor, 2008), in agreement with the result found here. Remarkably, in an annual survey in three connected shallow alpine lakes made in parallel to the present work, we found surface archaeal assemblages dominated by the MG1.1a all around the year (Auguet et al., 2011), although SAGMGC−1 was also detected. In the deep Lake Redon, the two clades segregated inversely along the water column with the relative abundance of SAGMGC−1 vertically decreasing whereas that of MG 1.1a increased. Although caution is needed to translate relative abundances of clones in the clone library to real cell abundance distributions, data suggest that the vertical gradient is as a key factor to understand the ecology of both the groups. In fact, the community similarity analysis indicated archaeal assemblage composition to be significantly related to depth. There are, however, many physical, chemical and biological variables that change with depth, and high-altitude oligotrophic lakes are especially affected by high irradiances and UV penetration, with varying effects associated with altitude (Catalan et al., 2006). It has been reported that parameters such as DOC and oxygen concentrations are potential environmental factor shaping the ecological niches of AOA (Erguder et al., 2009). The recently reported photoinhibition in AOA (Merbt et al., 2012) may provide additional hypothesis to explore differential tolerances to irradiance among clades, as diverse marine planktonic AOA ecotypes (that is, shallow and deep) (Francis et al., 2005; Hallam et al., 2006; Mincer et al., 2007; Beman et al., 2008) may be related to the differential resistance and adaptations to light.The vertical distribution of the MG 1.1a observed in Lake Redon was most closely related to that found for this clade in oceans (that is, below the euphotic zone) and may be associated to a lower tolerance to UV radiations and high light intensities. Conversely, the presence of SAGMGC−1 in surface waters and particularly at the air–water interface suggests a better adaptation to solar irradiance. Nevertheless, different metabolic adaptations and environmental distribution may be contained in the different clusters observed within each clade that would require finer phylogenetic resolution. Altogether, both altitudinal and vertical gradients in high mountain lake districts provide an excellent framework to further explore the ecology and biology of the idiosyncratic AOA populations.
Acknowledgments
We are thankful to the Centre de Recerca d'Alta Muntanya (CRAM), Universitat de Barcelona, Vielha for laboratory facilities. C Gutiérrez-Provecho, A Barberán, M Bacardit and L Alonso-Sáez are acknowledged for field and laboratory assistance and J Catalan for scientific feedback and support. We are also thankful for the constructive comments provided by the reviewers and the editor. This research was supported by grants CRENYC CGL2006-12058 and PIRENA CGL2009-13318 to EOC, and CONSOLIDER grant GRACCIE CSD2007-00067 from the Spanish Office of Science and Innovation (MICINN). JCA benefits from a Juan de la Cierva postdoctoral fellow (MICINN).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Agogué H, Brink M, Dinasquet J, Herndl GJ. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–792. doi: 10.1038/nature07535. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Auguet JC, Barberán A, Casamayor EO. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Auguet JC, Casamayor EO. A hotspot for cold crenarchaeota in the neuston of high mountain lakes. Environ Microbiol. 2008;10:1080–1086. doi: 10.1111/j.1462-2920.2007.01498.x. [DOI] [PubMed] [Google Scholar]
- Auguet JC, Nomokonova N, Camarero L, Casamayor EO. Seasonal changes of freshwater ammonia-oxidizing archaeal assemblages and nitrogen species in oligotrophic alpine lakes. Appl Environ Microbiol. 2011;77:1937–1945. doi: 10.1128/AEM.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Casamayor EO. Global phylogenetic community structure and beta-diversity patterns of surface bacterioplankton metacommunities. Aquat Microb Ecol. 2010;59:1–10. [Google Scholar]
- Barberán A, Fernandez-Guerra A, Auguet J-C, Galand PE, Casamayor EO. Phylogenetic ecology of widespread uncultured clades of the Kingdom Euryarchaeota. Mol Ecol. 2011;20:1988–1996. doi: 10.1111/j.1365-294X.2011.05057.x. [DOI] [PubMed] [Google Scholar]
- Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollmann A, Bar-Gilissen MJ, Laanbroek HJ. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl Environ Microbiol. 2002;68:4751–4757. doi: 10.1128/AEM.68.10.4751-4757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan J, Camarero L, Gacia E, Ballesteros E, Felip M. Nitrogen in the Pyrenean lakes (Spain) Hydrobiologia. 1994;274:17–27. [Google Scholar]
- Catalan J, Camarero L, Felip M, Pla S, Ventura M, Buchaca T, et al. High mountain lakes: extreme habitats and witnesses of environmental changes. Limnetica. 2006;25:551–584. [Google Scholar]
- Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Corte D, Yokokawa T, Varela MM, Agogue H, Herndl GJ. Spatial distribution of bacteria and Archaea and amoA gene copy numbers throughout the water column of the eastern Mediterranean Sea. ISMEJ. 2009;3:147–158. doi: 10.1038/ismej.2008.94. [DOI] [PubMed] [Google Scholar]
- Demergasso C, Escudero L, Casamayor EO, Chong G, Balagué V, Pedrós-Alió C. Novelty and spatio-temporal heterogeneity in the bacterial diversity of hypersaline Lake Tebenquiche (Salar de Atacama) Extremophiles. 2008;12:491–504. doi: 10.1007/s00792-008-0153-y. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble P, Haddad PR. Indirect photometric detection of anions in capillary electrophoresis. J Chromatogr. 1999;A834:189–212. [Google Scholar]
- Erguder T.H, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing Archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- Felip M, Sattler B, Psenner R, Catalan J. Highly active microbial communities in the ice and snow cover of high mountain lakes. Appl Environ Microbiol. 1995;61:2394–2401. doi: 10.1128/aem.61.6.2394-2401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrera I, Massana R, Casamayor EO, Balague V, Sanchez O, Pedros-Alio C, et al. High-diversity biofilm for the oxidation of sulfide-containing effluents. Appl Microbiol Biotechnol. 2004;64:726–734. doi: 10.1007/s00253-004-1582-x. [DOI] [PubMed] [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing Archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Lovejoy C, Vincent WF. Remarkably diverse and contrasting archaeal communities in a large arctic river and the coastal Arctic Ocean. Aquat Microb Ecol. 2006;44:115–126. [Google Scholar]
- Galand PE, Gutierrez-Provecho C, Massana R, Gasol JM, Casamayor EO. Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay Microbial Observatory) Limnol Oceanogr. 2010;55:2117–2125. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alingment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:520–536. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfort L, Kim JH, Coolen MJL, Abbas B, Schouten S, Herndl GJ, et al. Diversity of Archaea and detection of crenarchaeotal amoA genes in the rivers Rhine and Tet. Aquat Microb Ecol. 2009;55:189–201. [Google Scholar]
- Herrmann M, Saunders AM, Schramm A. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte. Littorellauniflora. Appl Environ Microbiol. 2008;74:3279–3283. doi: 10.1128/AEM.02802-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervàs A, Casamayor EO. High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol Ecol. 2009;67:219–228. doi: 10.1111/j.1574-6941.2008.00617.x. [DOI] [PubMed] [Google Scholar]
- Hervàs A, Camarero L, Reche I, Casamayor EO. Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ. Microbiol. 2009;11:1612–1623. doi: 10.1111/j.1462-2920.2009.01926.x. [DOI] [PubMed] [Google Scholar]
- Kemp PF, Aller JY. Estimating prokaryotic diversity: when are 16S rDNA libraries large enough. Limnol Oceanogr-Meth. 2004;2:114–125. [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Nat. Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D, et al. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci USA. 2009;106:4752–4757. doi: 10.1073/pnas.0812444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifiant acid soil. Proc Natl Acad Sci USA. 2011;108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Llirós M, Casamayor EO, Borrego CM. High archaeal richness in the water column of a freshwater sulfurous karstic lake along an interannual study. FEMS Microbiol. Ecol. 2008;66:331–342. doi: 10.1111/j.1574-6941.2008.00583.x. [DOI] [PubMed] [Google Scholar]
- Llirós M, Gich F, Plasencia A, Auguet JC, Darchambeau F, Casamayor EO, et al. Vertical distribution of ammonia oxidizing crenarchaeota and methanogens in the epipelagic waters of Lake Kivu. Appl Environ Microbiol. 2010;76:6853–6863. doi: 10.1128/AEM.02864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Marès T, Auguet JC, Casamayor EO. Winter to Spring changes in the slush bacterial community composition of a high mountain lake (Lake Redon, Pyrenees) Environ Microbiol Rep. 2012;4:50–56. doi: 10.1111/j.1758-2229.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- López-Archilla AI, Moreira D, Velasco S, López-García P. Archaeal and bacterial community composition of a pristine coastal aquifer in Doñana National Park, Spain. Aquat Microb Ecol. 2007;47:123–139. [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucl Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- McArdle B. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- Merbt S, Auguet JC, Casamayor EO, Martí E. Biofilm recovery in a wastewater treatment plant-influenced stream and spatial segregation of ammonia-oxidizing microbial populations. Limnol Oceanogr. 2011;56:1054–1064. [Google Scholar]
- Merbt S, Stahl DA, Casamayor EO, Marti E, Nicol GW, Prosser JI. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett. 2012;327:41–46. doi: 10.1111/j.1574-6968.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- Mincer TJ, Church MJ, Taylor LT, Preston C, Kar DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- Mussmann M, Brito I, Pitcher A, Sinninghe Damsté JS, Hatzenpichler R, Richter A, et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'hara B, Simpson GL, Stevens MHH.2008Vegan: community Ecology Package. version 111–14. http://vegan.r-forge.r-project.org .
- Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, et al. AmoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol. 2012;14:525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot J, Galand PE, Lovejoy C, Vincent WF. Vertical structure of archaeal communities and the distribution of ammonia monooxygenase A gene variants in two meromictic High Arctic lakes. Environ Microbiol. 2009;11:687–699. doi: 10.1111/j.1462-2920.2008.01846.x. [DOI] [PubMed] [Google Scholar]
- Reche I, Ortega-Retuerta E, Romera O, Pulido-Villena E, Morales-Baquero R, Casamayor EO. Effect of Saharan dust inputs on bacterial activity and community composition in Mediterranean lakes and reservoirs. Limnol Oceanogr. 2009;54:869–879. [Google Scholar]
- Reed D.W, Smith JM, Francis CA, Fujita Y. Responses of ammonia-oxidizing bacterial and archaeal populations to organic nitrogen amendments in low-nutrient groundwater. Appl Environ Microbiol. 2009;76:2517–2523. doi: 10.1128/AEM.02436-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S, Imachi H, Sekiguchi Y, Ohashi A, Harada H, Kamagata Y. Isolation of key methanogens for global methane emission from rice paddy fields: a novel isolate affiliated with the clone cluster rice cluster I. Appl Environ Microbiol. 2007;3:4326–4331. doi: 10.1128/AEM.03008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Akiyama M, Naganuma T, Fujioka M, Nako M, Ishijima Y. Molecular characterization of microbial communities in deep coalseam groundwater of northern Japan. Geobiology. 2007;5:423–433. [Google Scholar]
- Solórzano L. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol Oceanogr. 1969;14:799–801. [Google Scholar]
- Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK. Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol. 2001;67:5750–5760. doi: 10.1128/AEM.67.21.5750-5760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivate mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- van der Wielen P, Voost S, van der Kooij D. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl Environ Microbiol. 2009;75:4687–4695. doi: 10.1128/AEM.00387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Ye Q, Huang Z, Li W, Chen J, Song Z, et al. Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol. 2008;74:6417–6426. doi: 10.1128/AEM.00843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.