Abstract
Background:
Diarrheal diseases are a major cause of morbidity and mortality in resource-limited countries. Among the bacterial pathogens, diarrheagenic E. coli (DEC) are most frequently implicated in cases of epidemic and endemic diarrhea worldwide. The objective of this study was to determine the prevalence of DEC in stool specimens from patients with acute diarrhea using polymerase chain reaction (PCR).
Materials and Methods:
Escherichia coli stool samples were collected from 115 hospitalized children and adults with acute diarrhea in Mangalore, a coastal city, in southern India. PCR amplification of eae, bfp, stx, ehx genes were used for detection of enteropathogenic (EPEC) and shigatoxigenic E. coli (STEC), lt and st genes were used for enterotoxigenic E. coli (ETEC) and astA gene for enteroaggregative E. coli (EAEC).
Results:
During the 24 month study period, of the 115 stool samples, DEC type was detected in 20 (17.4%) using the PCR method. The most prevalent DEC was atypical EPEC accounting for 12 (10.4%) cases followed by 4 cases of EAEC (3.4%) and 4 of STEC (3.4%). No ETEC strains were isolated from any of the examined stool samples.
Conclusion:
This study suggests that the atypical EPEC are the newly emerging group among DEC stains in Southern India. Further studies are needed to evaluate the epidemiology and virulence properties of atypical EPEC strains.
Keywords: Atypical, diarrhea, enteropathogenic, Escherichia coli, polymerase chain reaction
INTRODUCTION
Diarrheal diseases are the major cause of death in children under 5 years of age in resource-poor countries, resulting in approximately 2.5 million deaths each year worldwide.[1] Among the bacterial pathogens, diarrheagenic E. coli (DEC) are most frequently implicated in cases of epidemic and endemic diarrhea worldwide.[2] However, the detection of DEC strains is difficult since these strains cannot be easily distinguished from the normal fecal flora using conventional phenotypic methods. In vitro assays that detect toxins, adherence, or invasion phenotypes can also detect DEC, but such methods are cumbersome. In recent years, with the introduction of polymerase chain reaction (PCR) in clinical laboratories, it has become possible to detect genes encoding virulence factors in bacterial isolates, allowing the rapid diagnosis of DEC strains.[3] Molecular identification and classification of DEC is established by the presence or absence of one or more specific virulence genes, which are absent in the commensal E coli.[2] DEC strains are classified into 6 distinct pathogenic categories (pathotypes) including enterohemorrhagic E. coli (EHEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and diffusely adherent E. coli (DAEC).[2]
Among the DEC, EPEC is an important cause of pediatric diarrhea, resulting in high morbidity and mortality in resource-poor settings. Based on molecular diagnosis, EPEC strains account for 5-10% of pediatric diarrhea in resource-poor countries. EPEC is subdivided into typical and atypical strains based on the attaching and effacing intimin (eae) gene and plasmid-mediated bundle-forming pilus (bfpA) gene.[4] Typical EPEC strains are eae + bfpA + stx-, whereas atypical EPEC strains carry the eae gene but lack the bfpA and stx gene.[4] For many years, typical EPEC was considered to be a major cause of persistent diarrhea in infants, but recent studies indicate that atypical EPEC are more prevalent than typical EPEC in both resource-rich and resource-poor countries and may be an emerging pathogen. Very few studies have been performed in India to investigate the prevalence and characterization of DEC in patients with diarrhea.[5,6] The objective of this study was to determine the prevalence of DEC in stool specimens from patients with acute diarrhea in Mangalore, India using PCR.
MATERIALS AND METHODS
Study population
Between July 2002 and June 2004, a total of 115 Escherichia coli stool samples were collected from hospitalized patients (95 adults and 20 children) admitted with diarrhea at Kasturba Medical College Hospital, K. S. Hegde Charitable Hospital and Government Wenlock Hospital in Mangalore, a coastal city, in the State of Karnataka, India by random sampling method. Diarrhea was defined by the occurrence of >3 loose stools, liquid or watery or at least 1 bloody stool in a 24 h period. Samples from patients who received antibiotics before admission or during their hospital stay were excluded from the study. The study protocol was reviewed by the ethics committee of K. S. Hegde Medical Academy.
Sample collection, isolation, and identification of E. coli
Stool samples were collected in sterile plastic vials, transported to a microbiology laboratory and initially screened for the presence of leukocytes, red blood cells, ova, and cysts of parasites using conventional microscopy and cultured on the same day to isolate E. coli. Samples were inoculated into selective enrichment medium [Lauryl sulphate tryptose broth (LSTB)] and after overnight incubation, subcultured onto Sorbitol MacConkey's agar and MacConkey's agar (Hi-Media, Mumbai, India). The culture plates were incubated at 37°C for 18-24 h. Lactose and non-lactose fermenting colonies from MacConkey agar and sorbitol fermenting and non-sorbitol fermenting colonies from sorbital agar were selected and subjected to conventional biochemical tests for identification of E. coli as described previously. About 20 biochemically confirmed E. coli colonies from each sample were further characterized by pathotype specific, virulence gene targeted PCR assays.
DNA extraction
DNA from stool samples was extracted using QIAamp DNA stool mini kit (Qiagn, GmbH, Hilden, Germany). The test cultures were grown overnight in 3 ml Luria Bertani broth (Hi-Media, Mumbai) and centrifuged at 10,000 rpm for 10 minutes. The resultant bacterial pellet was washed twice in sterile distilled water, resuspended in 100 ml sterile distilled water, and was heated at 95°C for 15 min in a hot bath. The tubes were then kept in ice. The cell debris was settled by centrifugation at 10,000 rpm for 10 min, and the supernatant was used as template for PCR amplification. All the PCR reactions were performed in a PTC-100 thermocycler (M.J. Research Inc., USA) for 30 cycles. Crude lysates were also prepared from enrichment broths inoculated with stool samples for PCR. Genomic DNA was extracted from bacteria using Cetyl Trimethyl Ammonium Bromide (CTAB) extraction method.[7]
PCR, primers, and products
All PCR primers and molecular reagents were purchased from Bangalore Genei, India.
Each PCR assay was performed in a 30 μl reaction volume containing 3 μl of 10 × PCR buffer (100 mM Tris HCl, 20 mM MgCl2, 500 mM KCl), 200 μM concentrations of each of the 4-deoxyribonucleotide triphosphates (dNTPs), 0.5 μg of each primer, and 1U Taq DNA polymerase (Bangalore Genei, India). The DNA samples carrying the relevant virulence gene (s) served as positive controls in each reaction. Sterile distilled water served as negative control.
Detection of virulence genes associated with DEC
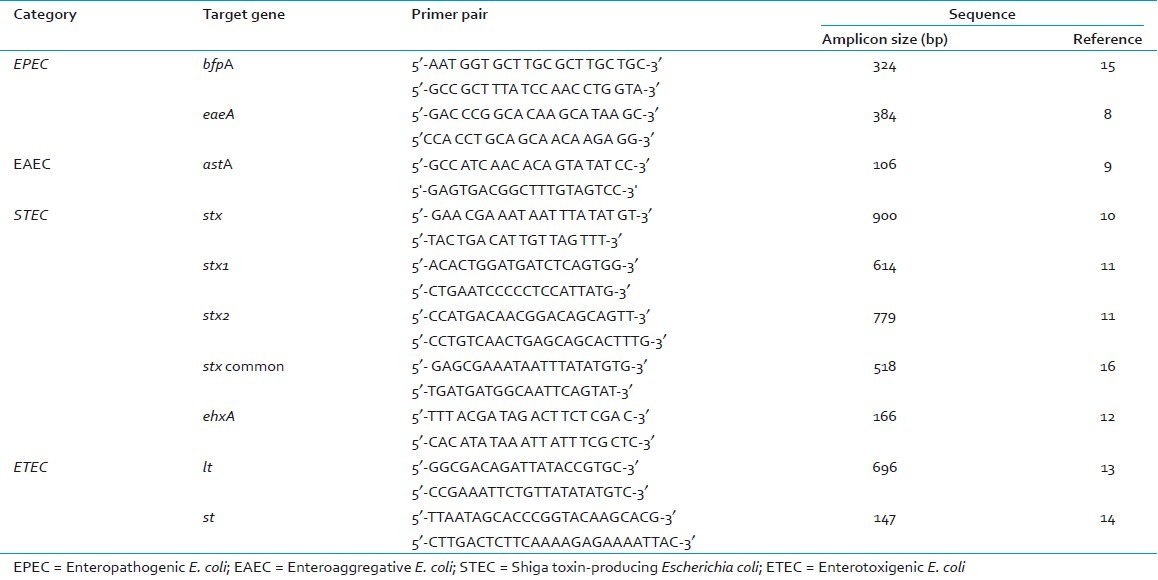
The following virulence associated genes of DEC were targets for PCR amplification: attaching and effacing factor (eaeA), shiga-like toxins (stx, stx1, stx2), heat labile toxin (lt), heat stable toxin (st), bundle forming pilus (bfpA), enteroaggregative stable toxin (astA), and hemolysin (ehxA). A total of 2000 E. coli biochemically confirmed isolates from samples (1000 isolates from direct stool samples and 1000 isolates from enrichment broths) were tested by PCR for presence of various virulence genes associated with DEC. PCR was performed individually using each primer pair as previously described[8–15] [Table 1]. PCR products were separated on a 2% agarose gel, stained with ethidium bromide and photographed using a gel documentation system. E. coli reference strain EDL 933 was used as positive control in all PCR reactions for EPEC and STEC. E coli ATCC 25922 was used as negative controls in each batch of reactions.
Table 1.
Oligonucleotide primer pairs used in the study for detection of diarrheagenic E.coli
Cloning and sequencing of astA PCR product
AstA gene from the strain ST-103 isolated from our study was amplified using primers East1F and East1R.[14] The PCR product was purified using PCR purification kit (Millipore, USA). The purified PCR product was used for cloning and sequencing using TOPO – TA cloning kit (Invitrogen, USA).
RESULTS
Prevalence of diarrheagenic E. coli
Of the 115 stool samples obtained from patients with acute diarrhea during the 24-month study period, DEC type was detected in 20 (17.4%) using the PCR method [Table 2]. The most prevalent DEC was EPEC accounting for 12 (10.4%) cases followed by 4 cases of EAEC (3.4%) and 4 cases of Shiga toxin-producing Escherichia coli [STEC] (3.4%) [Table 3]. No ETEC strains were isolated from any of the examined stool samples. Irrespective of whether a sample is positive for pathogenic E. coli or not by direct PCR, isolates of E. coli were collected from all the samples before and after enrichment. Thus, a total of 2000 E. coli isolates were obtained from all the samples, 1000 each from the samples before and after enrichment in LSTB [Table 2]. The isolates were biochemically identified as E. coli and then subjected to PCR for virulence genes associated with different pathogenic groups of E. coli. Of these, 105 and 143 isolates of E. coli, from direct and enrichment samples respectively, were identified as pathogenic types based on the presence of the virulence genes [Table 2].
Table 2.
Prevalence of pathogenic E. coli in stool samples determined by PCR
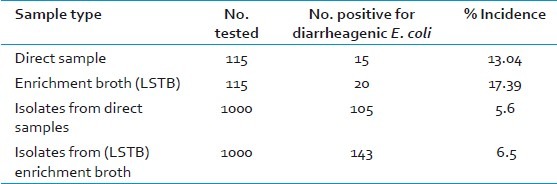
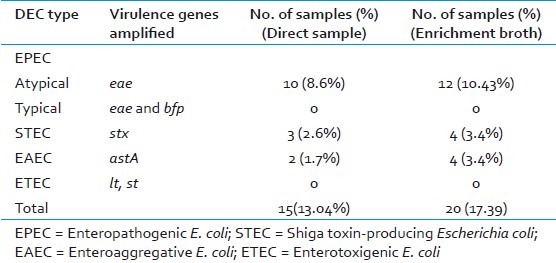
Table 3.
Prevalence of diarrheagenic E. coli (DEC) pathotypes in participants with acute diarrhea (n = 115)
Detection of virulence genes and characterization of DEC
PCR amplification of eae, bfp, stx, ehx genes were used for detection of EPEC and STEC, lt and st genes were used for ETEC and astA gene for EAEC [Table 3]. All EPEC isolates were positive for eae gene but negative for bfp and stx gene and were classified as atypical EPEC genotype (eae+, bfp-, stx-). A total of 12 (10.4%) stool samples (and 81 isolates from these samples) were positive for eae gene, but negative for bfp and stx gene [Figure 1].
Figure 1.
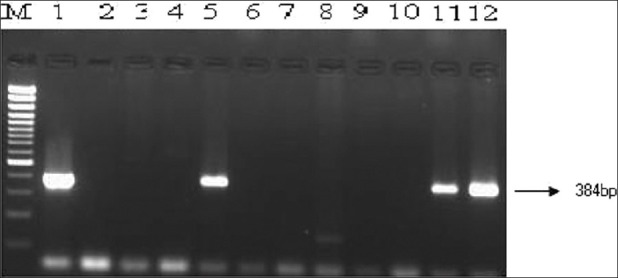
Direct detection of eaeA gene of enteropathogenic and enterohemorrhagic E. coli in stool samples by PCR. Lane M, 100 bp DNA ladder (Genetix); Lane 1, Reference strain EDL 933; Lane 2, Negative control strain; Lanes 3-12, Stool 91 to Stool 100; Lanes 5, 11 and 12, Stool samples positive for eae genes
STEC strains differ genotypically from atypical EPEC by their possession of stx genes. In this study, 2 sets of PCR with primers common to stx1 and stx2 genes[10] were used to detect strains carrying variants of stx genes. A total of 4 (3.4%) stool samples (and 22 isolates from these samples) were positive for stx genes. The primers described by Yamasaki et al[16] detected the pathogen in 2 samples, while the primers described by Lin et al[10] detected the pathogen in 1 sample; stx2 gene was detected in 1 sample.
A total of 4 samples (and 40 isolates from these samples) were astA positive. This gene encodes enteroaggregative E. coli Stable Toxin (EAST1), which is a homologue of ETEC ST.[2] The PCR product obtained from one of the strain (St-103) was cloned. The derived sequence under AF411067 corresponded to Aggregative Adhesion Fimbria (AAF) Type III-encoding operon in EAEC. The PCR amplified region from ST103 aligned with AF411067 from nt 5398 to nt 5502 with a similarity of 95%. The region 5398 to 5514 of AF411067 corresponded to astA gene, thus indicating that the strain ST103 possesses the astA gene of EAEC.
DISCUSSION
This study was undertaken to investigate the prevalence of DEC in diarrheal patients by use of PCR in adult and pediatric patients with acute diarrhea in Mangalore, a coastal city, in South India . In this study, DEC was isolated in 17.4% of fecal samples from participants with diarrhea. The prevalence and other epidemiologic characteristics of DEC as etiologic agents of diarrhea vary globally from region to region, and even between and within countries in the identical geographic location.
In this study, atypical EPEC strains were found to be the dominant DEC and may be emerging pathogens. The role of typical EPEC strains causing acute and chronic watery childhood diarrhea in developing countries is well-known. However, data from India is limited.[5,6] In one study from Kashmir, EPEC was detected in 7.6% of fecal samples from patients with diarrhea.[6]
A significant association of typical EPEC strains (eae+ bfpA+ stx-) with diarrhea was reported previously from developing countries.[2] In recent years, several studies have shown that atypical EPEC strains (eae+, bfpA- and stx-) are more prevalent than typical EPEC strains in developed countries as well as in resource-limited countries including Mexico, Nicaragua, Vietnam, and Mozambique. In contrast to typical EPEC, atypical EPEC strains appear to be common in domestic animals,[4] and have been reported in food-borne and water-borne outbreaks.[17] The role of atypical EPEC in childhood diarrhea is still unclear, and future studies are warranted to investigate the epidemiology and pathogenesis of atypical EPEC strains.
STEC strains differ geno and phenotypically from atypical EPEC by their possession of stx genes.[2] In the present study, we isolated a very low proportion of STEC strains. These data is consistent with the low prevalence of STEC infection in India.[5,6] There may be several variants of stx gene in E. coli. While 2 major groups, stx1 and stx2 are recognized, within each of the 2 toxin types, several variants have been identified including stx1c, stx1d, stx2c, stx2d, and stx2activatable, stx2e, stx2f and stx2g. It is prudent to use several alternative primers to detect STEC due to variations in the genes. Most STEC disease has been described in the United States, Europe, Australia, South America, and parts of Asia. Although STEC infections are very rare in India,[5,6,18] it has been detected in meat and fish samples indicating that this pathogen may pose a public health threat in the near future.
In this study, EAEC accounted for 3.4% of cases. EAEC are a heterogeneous emerging pathogen affecting all ages, prevalent in resource-rich and resource-limited settings, and are associated with acute and persistent watery diarrhea in children, travelers and in individuals infected with HIV/AIDS. Low prevalence of EAEC has been reported from Brazil,[19] wherein EPEC was the most prevalent (10%) followed by ETEC (7.5%) and EAEC (4%). In contrast, higher prevalence of EAEC have been reported from Tanzania (33) and several other regions in India, including Kolkata (9%), northern India (12.3% in acute diarrhea and 34.5% in persistent diarrhea) and Manipal (22%).[20]
In our study, 4 fecal samples were positive for strains harboring the EAST1 gene, which is a homologue of ETEC ST. Several reports also support the role of EAST1 in diarrhea. EAST1 gene is also commonly found in other DEC pathotypes, such as EPEC, ETEC, and EHEC. We determined the partial nucleotide sequence of astA, which showed 95% similarity with the astA sequence in the GenBank. This strongly suggests the involvement of EAEC in diarrhea in this region, and more studies are needed to determine the etiology and the virulence genes associated with this pathogen.
In this study, no ETEC strains were detected. ETEC is a ubiquitous pathogen, commonly transmitted via contaminated food and water, and is a major cause of traveler's diarrhea and infantile diarrhea in developing countries. Previous reports from India have reported varied findings ranging from 0.92% in Kashmir[6] to ~ 12% in Calcutta.
In this study, detection of DEC was done by PCR amplification of virulence genes directly from stool samples and also from lysates obtained after enrichment of stool in LSTB. We found that PCR performed on stool enrichment broth lysates showed a better detection rate (17%) compared with the results of PCR using direct stool samples (13%). This could be due to two reasons. First, although viable DEC may be present in low numbers in some samples, the strains may not be cultured, and therefore, an enrichment step may be necessary for their detection. The presence of inhibitors in the stool specimens is another possibility. Therefore, extraction of DNA after enrichment of the samples may have resulted in better detection rates of DEC.
The advent of PCR technology has greatly simplified the detection of DEC associated with diarrhea. Real-time multiplex PCR is a sensitive, specific, and inexpensive method for detection of DEC. Rapid availability of results helps clinicians make timely appropriate therapeutic decisions since some pathogens warrant antibiotic therapy while certain DEC strains (e.g., STEC), antibiotics are not recommended due to increased risk of hemolytic uremic syndrome (HUS). However, in India and in other resource-limited countries, molecular methods to detect DEC are not available in routine clinical laboratories.
Our study has several limitations. First, there was no control group. Second, certain strains like enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and diffusely adherent E. coli (DAEC) was not tested. Despite these limitations, this study suggests that atypical EPEC is involved in a significant proportion among the diarrheal cases in Mangalore and may be an emerging pathogen. Further studies are needed to investigate the epidemiology and virulence properties of atypical EPEC strains.
ACKNOWLEDGEMENTS
This work was supported by research grants from the Department of Biotechnology, Government of India. We thank the Dean and the administrative and clinical staff of Kasturba Medical College Hospital, K. S. Hegde Charitable Hospital, Nitte University, and Wenlock Hospital for their gracious support.
Footnotes
Source of Support: This work was supported by research grants from the Department of Biotechnology, Government of India and K.S.Hegde Medical Academy, Nitte University Deralakatte.
Conflict of Interest: None declared.
REFERENCES
- 1.Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO Child Health Epidemiology Reference Group.WHO estimates of the causes of death in children. Lancet. 2005;365:1147–52. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley LT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff C, Luthy J, Altwegg M, Baggi F. Rapid detection of DEC by real-time PCR. J Microbiol Methods. 2005;61:335–41. doi: 10.1016/j.mimet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–13. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danashree B, Mallya PS. Detection of shiga-toxigenic Escherichia coli (STEC) diarrhoeagenic stool and meat samples in Mangalore, India. Indian J Med Res. 2008;128:271–7. [PubMed] [Google Scholar]
- 6.Wani SA, Nabi A, Fayaz I, Ahmad I, Nishikawa Y, Qureshi K, et al. Investigation of diarrhoeic faecal samples for enterotoxigenic Shiga toxin-producing and typical or atypical enteropathogenic Escherichia coli in Kashmir, India. FEMS Microbiol Lett. 2006;261:238–44. doi: 10.1111/j.1574-6968.2006.00354.x. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidaman JG, Smith JA, et al. New York: John Wiley & Sons; 1995. Short protocols in molecular biology; pp. 2–11. [Google Scholar]
- 8.Paton A W, Paton J C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfb0111 and rfb0157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yatsuyanagi J, Saito S, Sato H, Miyajima Y, Amano K, Enomoto K. Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreak. J Clin Microbiol. 2002;40:294–7. doi: 10.1128/JCM.40.1.294-297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Z, Kurazono H, Yamasakiand S, Takeda Y. Detection of various variant verotoxin genes in Escherichia coli by polymerase chain reaction. Microbiol Immunol. 1993;37:543–8. doi: 10.1111/j.1348-0421.1993.tb01675.x. [DOI] [PubMed] [Google Scholar]
- 11.Fagan PK, Hornitzky MA, Bettelheim KA, Djordjevic SP. Detection of Shiga-Like Toxin (stx1 and stx2), Intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) Hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl Environ Microbiol. 1999;65:868–72. doi: 10.1128/aem.65.2.868-872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fratamico PM, Sackitey SK, Wiedmann M, Deng MY. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–91. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultsz C, Pool GJ, van Ketel R, de Wever B, Speelman P, Dankert J. Detection of enterotoxigenic Escherichia coli in stool samples by using non-radioactively labeled oligonucleotide DNA probes and PCR. J Clin Microbiol. 1994;32:2393–7. doi: 10.1128/jcm.32.10.2393-2397.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsvik O, Strockbine NA. PCR detection of heat.stable, heat-labile, and Shiga-like toxin genes in Escherichia coli. In: Persing DH, Smith TF, Tenover FC, White TJ, editors. Diagnostic molecular microbiology: Principles and applications. Rochester, NY: Mayo Foundation; 1993. pp. 271–6. [Google Scholar]
- 15.Gunzburg ST, Tornieporth NG, Riley LW. Identification of enteropathogenic Escherichia coli by PCR- Based detection of the bundle forming pilus gene. J Clin Microbiol. 1995;33:1375–7. doi: 10.1128/jcm.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamasaki S, Lin Z, Shirai H, Terai A, Oku Y, Ito H, et al. Typing of verotoxins by DNA colony hybridization with poly- and oligo- nucleotide probes, a bead-enzyme-linked immunosorbent assay, and polymerase chain reaction. Microbial Immunol. 1996;40:345–52. doi: 10.1111/j.1348-0421.1996.tb01078.x. [DOI] [PubMed] [Google Scholar]
- 17.Baba A, Ebuchi S, Uryu K, Hiwaki H, Ogata K, Washimi E, et al. An outbreak of water borne gastroenteritis caused by diarrheagenic Escherichia coli possessing eae gene. Jpn J Infect Dis. 2006;59:59–60. [PubMed] [Google Scholar]
- 18.Sehgal R, Kumar Y, Kumar S. Prevalence and geographical distribution of Escherichia coli 0157:H7 in India: A 10-year survey. Trans R Soc Trop Med Hyg. 2008;102:380–3. doi: 10.1016/j.trstmh.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Franzolin MR, Alves RC, Keller R, Gomes TA, Beutin L, Barreto ML, et al. Prevalence of diarrheagenic Escherichia coli in children with diarrhea in Salvador, Bahia, Brazil. Mem Inst Oswaldo Cruz. 2005;100:359–63. doi: 10.1590/s0074-02762005000400004. [DOI] [PubMed] [Google Scholar]
- 20.Raju B, Ballal M. Multidrug resistant enteroaggregative Escherichia coli diarrhea in rural southern Indian population. Scand J Infect Dis. 2009;41:105–8. doi: 10.1080/00365540802641856. [DOI] [PubMed] [Google Scholar]


