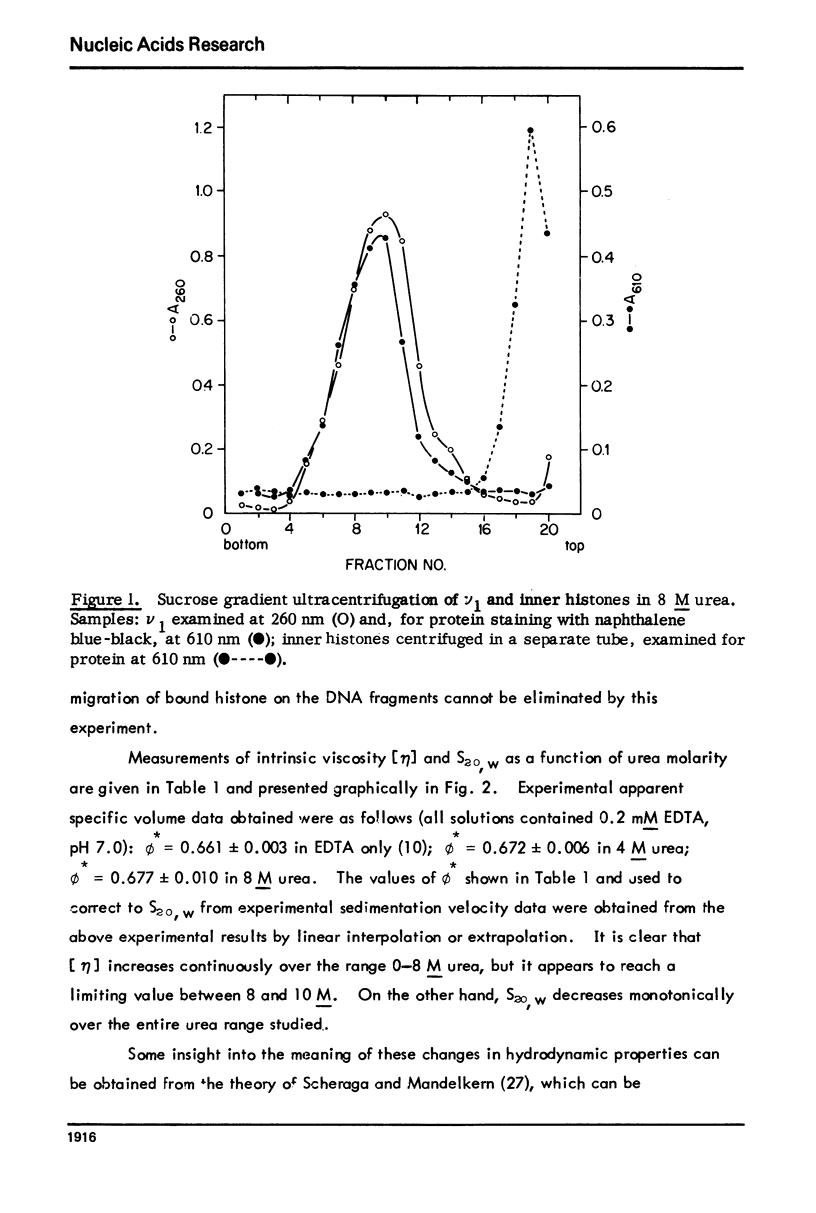
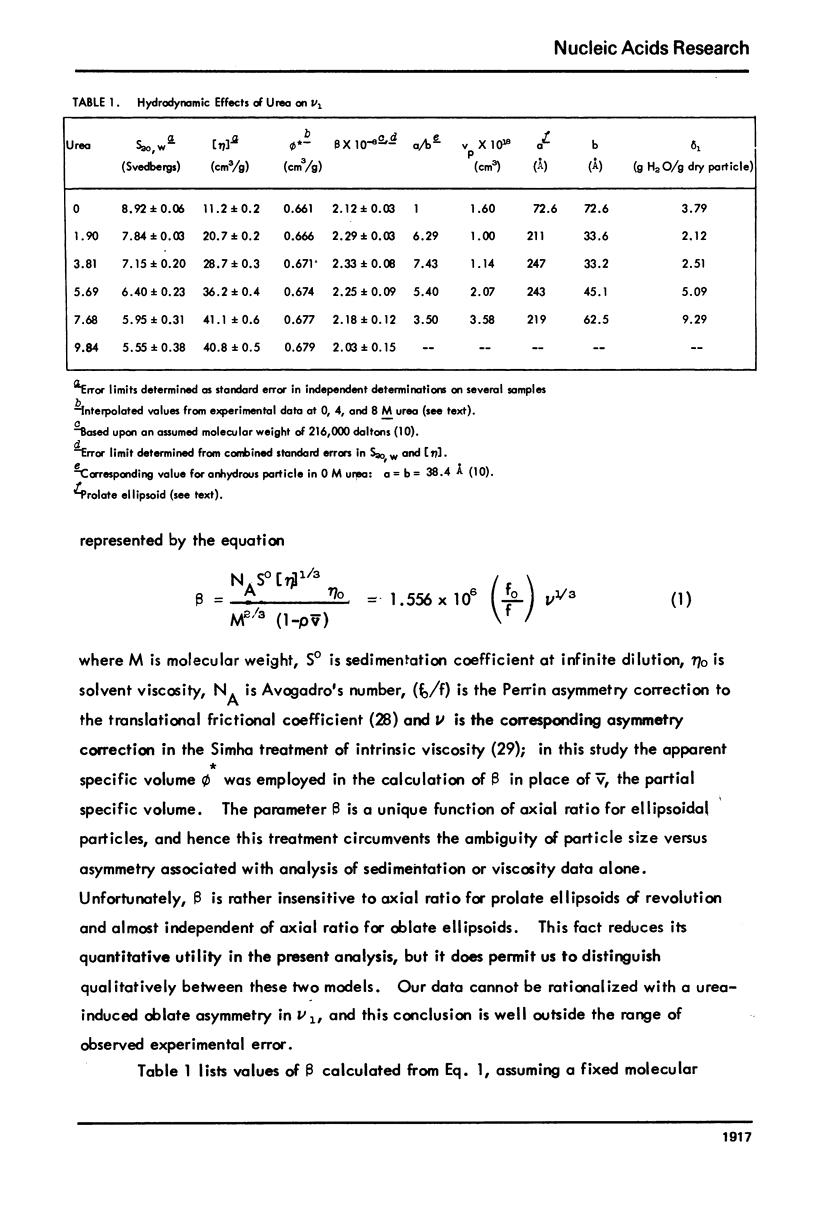
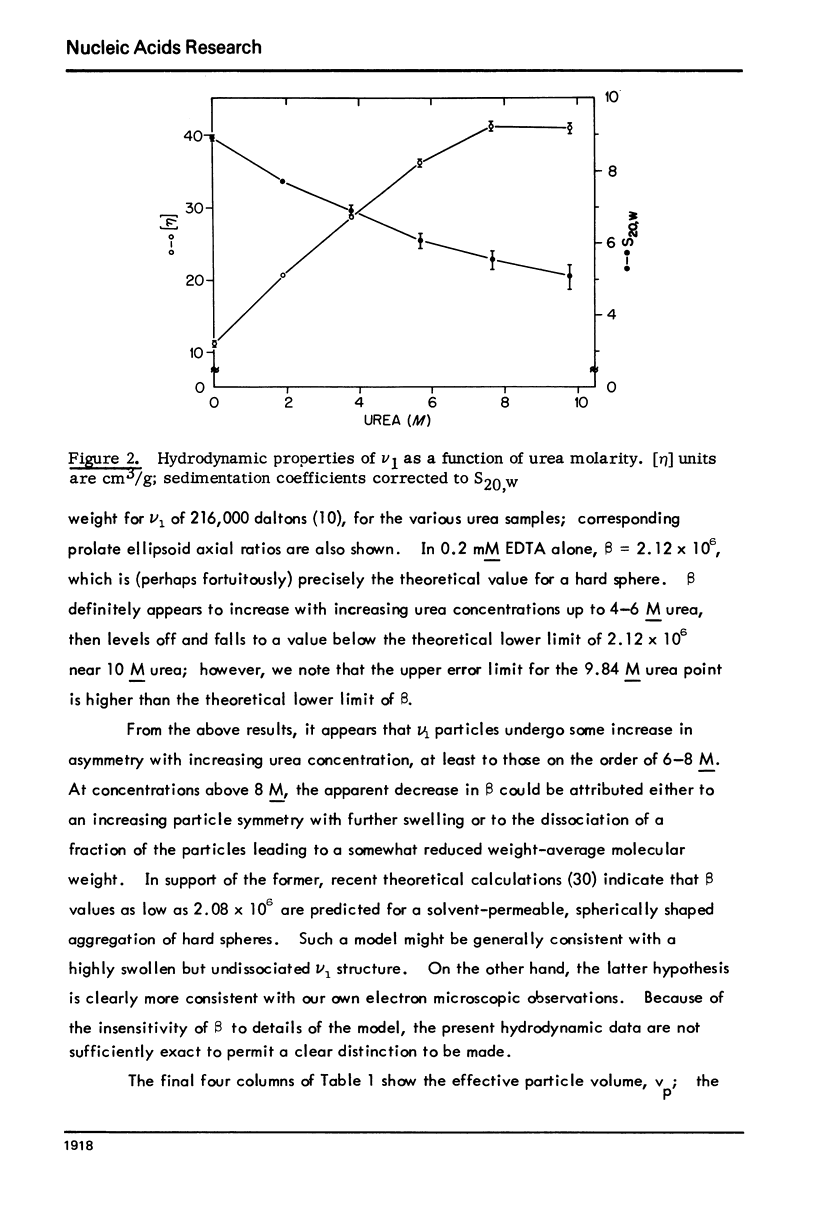
Abstract
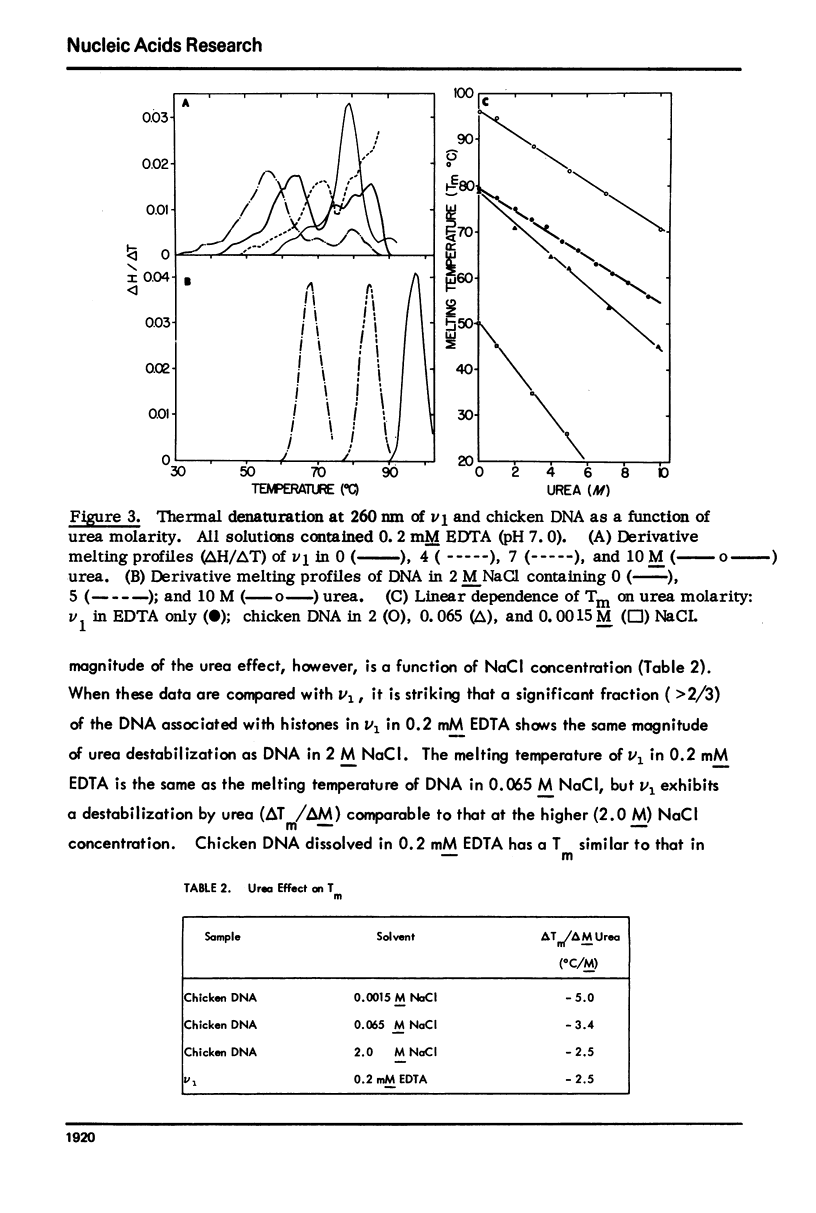
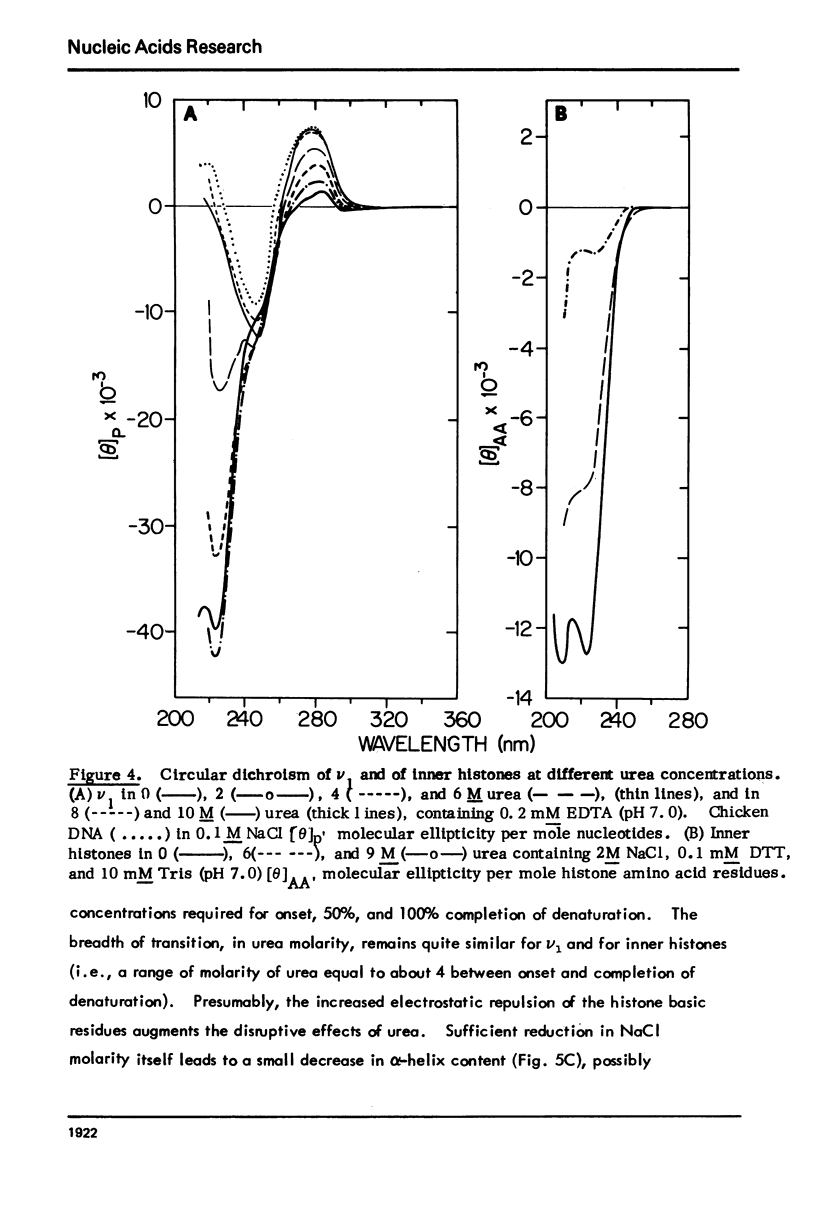
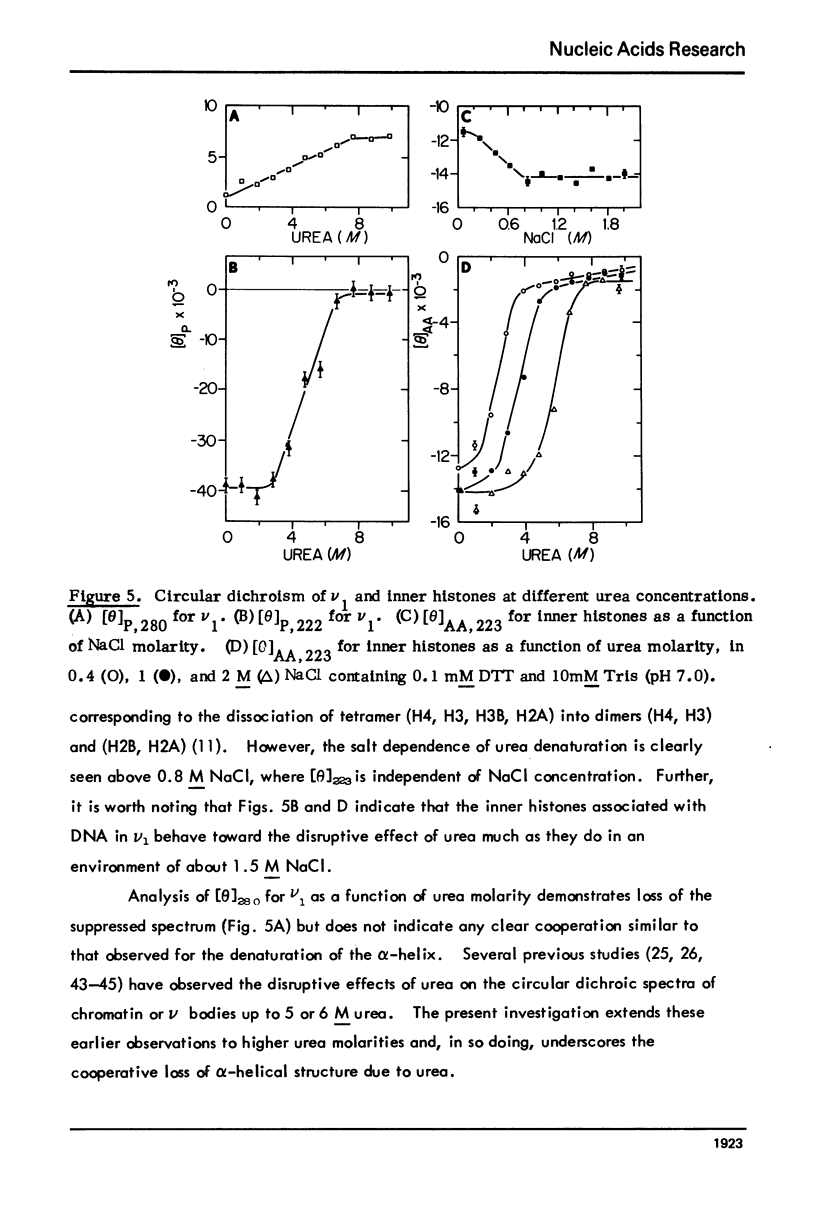
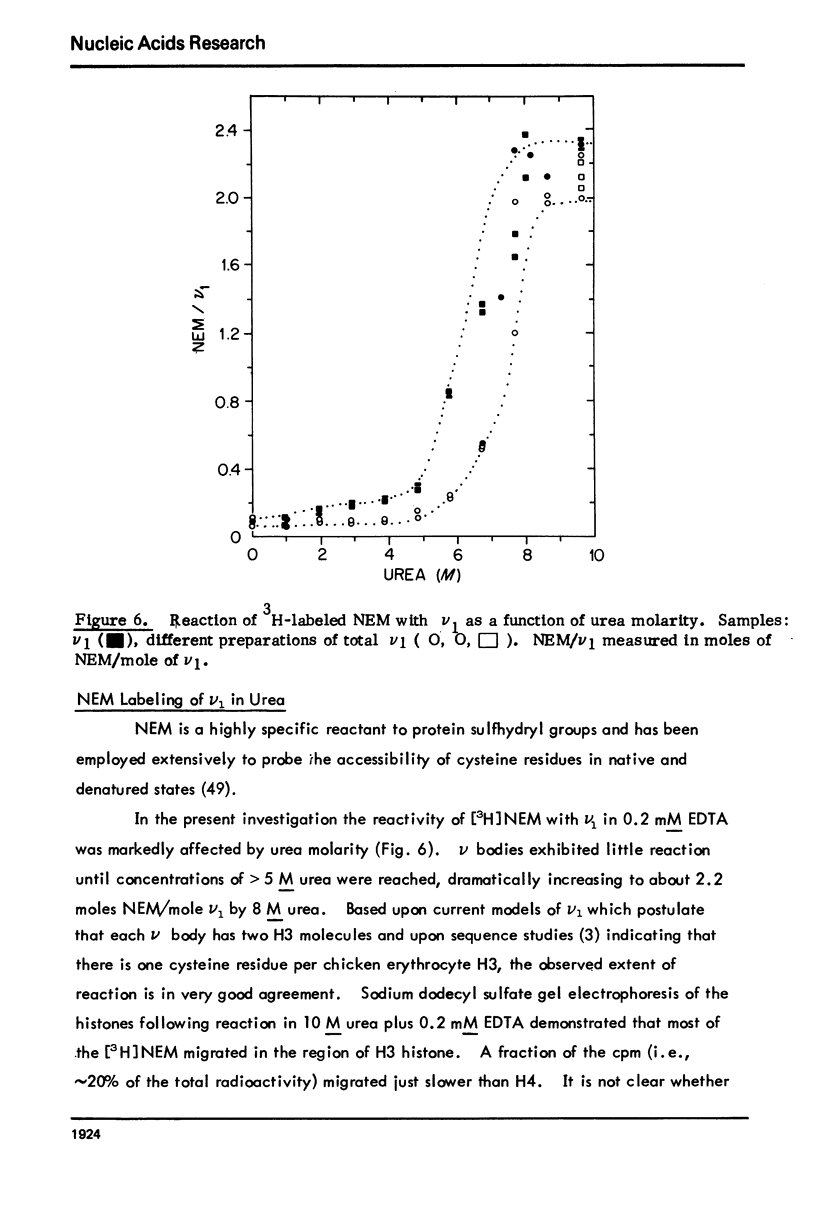
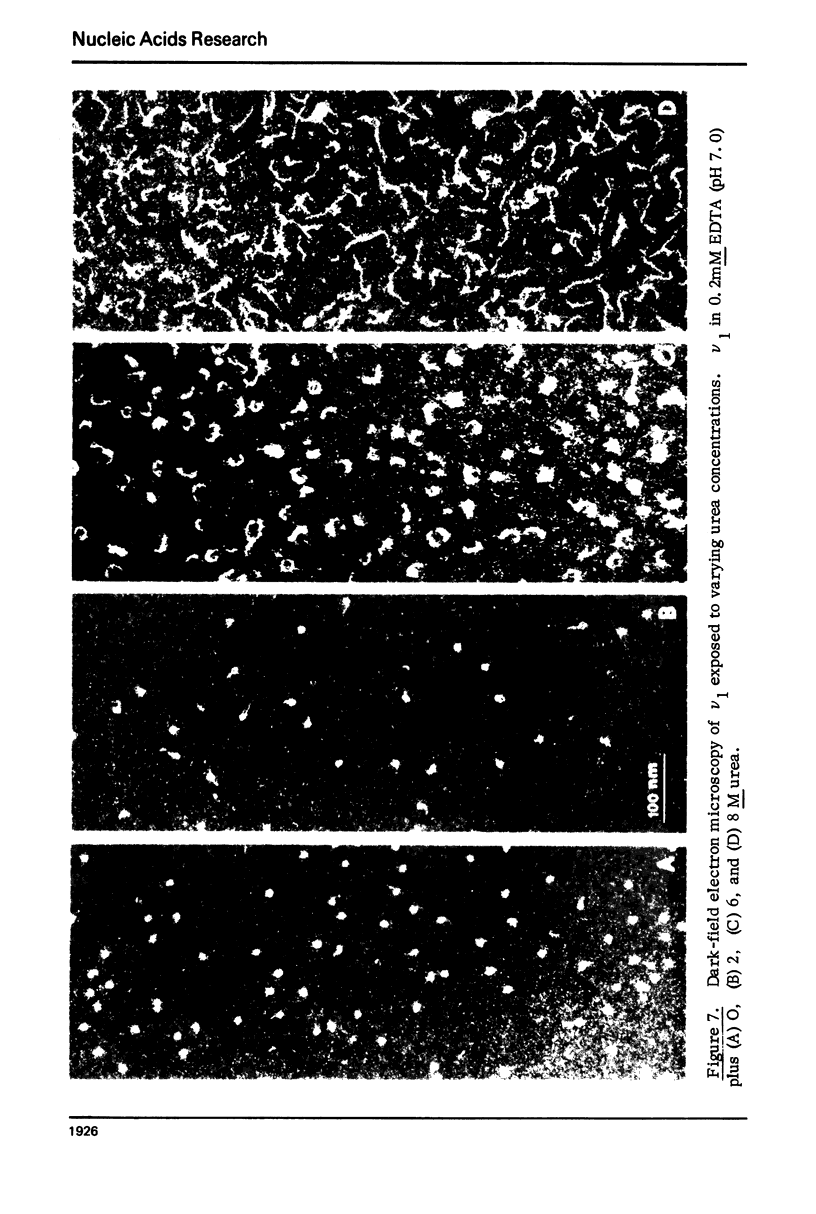
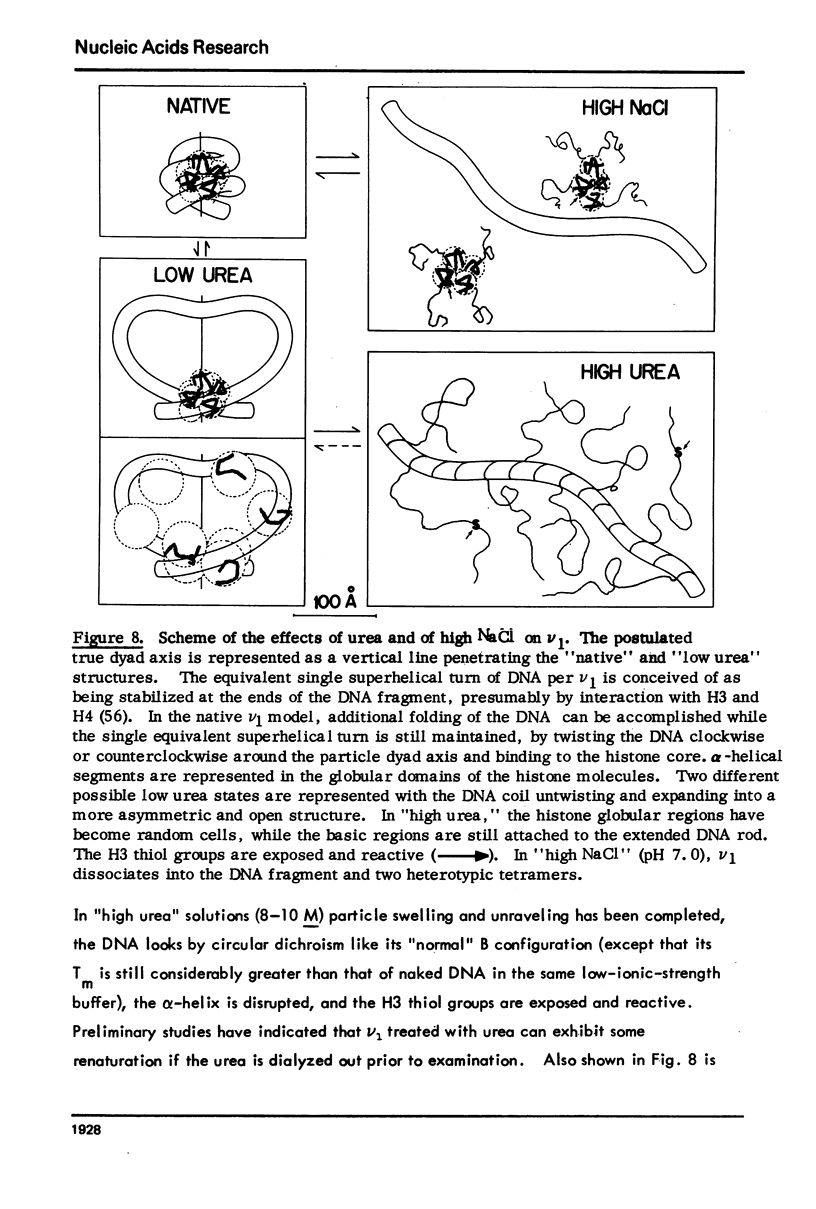
Monomer chromatin nu bodies (nu1) from chicken erythrocyte nuclei were exposed to 0-10 M urea plus 0.2 mM EDTA (PH 7). Alterations in nu1 conformation were examined using hydrodynamic methods (i.e., S, eta, and (formula: see text)), thermal denaturation, circular dichroism, reactivity of histone thiol groups to N-ethyl maleimide, and electron microscopy. The two domains of a nu body (i.e., the DNA-rich shell and the protein-rich core) aeared to respond differently to the destabilizing effects of increasing urea; DNA conformation and stability exhibited noncooperative changes; the core protein structure revealed cooperative destabilization between 4 and 7 M urea. Companion studies on the conformation of the inner histone "heterotypic tetramer" also revealed cooperative destabilization with increasing urea concentration.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansevin A. T., Hnilica L. S., Spelsberg T. C., Kehm S. L. Structure studies on chromatin and nucleohistones. Thermal denaturation profiles recorded in the presence of urea. Biochemistry. 1971 Dec 7;10(25):4793–4803. doi: 10.1021/bi00801a030. [DOI] [PubMed] [Google Scholar]
- Bartley J. A., Chalkley R. The viscosity of nucleohistone in urea. Biochim Biophys Acta. 1968 Jun 26;160(2):224–228. doi: 10.1016/0005-2795(68)90090-1. [DOI] [PubMed] [Google Scholar]
- Bekhor I., Bonner J., Dahmus G. K. Hybridization of chromosomal RNA to native DNA. Proc Natl Acad Sci U S A. 1969 Jan;62(1):271–277. doi: 10.1073/pnas.62.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffa L., Saccomani G., Tamburro A. M., Scatturin A., Vidali G. Chromosomal nucleoproteins: CD studies on reconstituted nucleohistones from avian erythrocytes. Int J Protein Res. 1971;3(6):357–363. doi: 10.1111/j.1399-3011.1971.tb01731.x. [DOI] [PubMed] [Google Scholar]
- Bramhall S., Noack N., Wu M., Loewenberg J. R. A simple colorimetric method for determination of protein. Anal Biochem. 1969 Oct 1;31(1):146–148. doi: 10.1016/0003-2697(69)90251-6. [DOI] [PubMed] [Google Scholar]
- CASASSA E. F., EISENBERG H. THERMODYNAMIC ANALYSIS OF MULTICOMPONENT SOLUTIONS. Adv Protein Chem. 1964;19:287–395. doi: 10.1016/s0065-3233(08)60191-6. [DOI] [PubMed] [Google Scholar]
- Carlson R. D., Olins A. L., Olins D. E. Urea denaturation of chromatin periodic structure. Biochemistry. 1975 Jul 15;14(14):3122–3125. doi: 10.1021/bi00685a013. [DOI] [PubMed] [Google Scholar]
- Chang C., Li H. J. Urea perturbation and the reversibility of nucleohistone conformation. Nucleic Acids Res. 1974 Aug;1(8):945–958. doi: 10.1093/nar/1.8.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Fric I., Sponar J. Circular dichroism of native and reconstituted nucleohistones. Biopolymers. 1971;10(9):1525–1531. doi: 10.1002/bip.360100908. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwin B. I., Smith S. G., Peebles P. T. Two active forms of RD-114 virus DNA polymerase in infected cells. Cell. 1975 Sep;6(1):45–52. doi: 10.1016/0092-8674(75)90072-0. [DOI] [PubMed] [Google Scholar]
- Gilli S. J., Thompson D. S. A rotating cartesian-diver viscometer. Proc Natl Acad Sci U S A. 1967 Mar;57(3):562–566. doi: 10.1073/pnas.57.3.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Murphy R. F., Bonner J. Structure of transcriptionally active chromatin. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4404–4408. doi: 10.1073/pnas.72.11.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon S., Johnson R. S., Wolf B., Chan A. Mixed conformations of deoxyribonucleic acid in chromatin: a preliminary report. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3263–3267. doi: 10.1073/pnas.69.11.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington R. E. Flow birefringence of T2 bacteriophage DNA. Biopolymers. 1968;6(1):105–116. doi: 10.1002/bip.1968.360060109. [DOI] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. The effect of urea on staphylococcal nuclease digestion of chromatin. Biochem Biophys Res Commun. 1975 Dec 15;67(4):1391–1400. doi: 10.1016/0006-291x(75)90181-3. [DOI] [PubMed] [Google Scholar]
- Johnson R. S., Chan A., Hanlon S. Mixed conformations of deoxyribonucleic acid in intact chromatin isolated by various preparative methods. Biochemistry. 1972 Nov 7;11(23):4347–4358. doi: 10.1021/bi00773a023. [DOI] [PubMed] [Google Scholar]
- Kawahara K., Tanford C. Viscosity and density of aqueous solutions of urea and guanidine hydrochloride. J Biol Chem. 1966 Jul 10;241(13):3228–3232. [PubMed] [Google Scholar]
- Mandel R., Fasman G. D. Chromatin and nucleosome structure. Nucleic Acids Res. 1976 Aug;3(8):1839–1855. doi: 10.1093/nar/3.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Olins D. E., Olins A. L., Von Hippel P. H. Model nucleoprotein complexes: studies on the interaction of cationic homopolypeptides with DNA. J Mol Biol. 1967 Mar 14;24(2):157–176. doi: 10.1016/0022-2836(67)90324-5. [DOI] [PubMed] [Google Scholar]
- Ramm E. I., Vorob'ev V. I., Birshtein T. M., Bolotina I. A., Volkenshtein M. V. Circular dichroism of DNA and histones in the free state and in deoxyribonucleoprotein. Eur J Biochem. 1972 Feb 15;25(2):245–253. doi: 10.1111/j.1432-1033.1972.tb01690.x. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- Senior M. B., Olins D. E. Effect of formaldehyde on the circular dichroism of chicken erythrocyte chromatin. Biochemistry. 1975 Jul 29;14(15):3332–3337. doi: 10.1021/bi00686a007. [DOI] [PubMed] [Google Scholar]
- Shih T. Y., Fasman G. D. Conformation of deoxyribonucleic acid in chromatin: a circular dichroism study. J Mol Biol. 1970 Aug 28;52(1):125–129. doi: 10.1016/0022-2836(70)90182-8. [DOI] [PubMed] [Google Scholar]
- Shih T. Y., Lake R. S. Studies on the structure of metaphase and interphase chromatin of Chinese hamster cells by circular dichroism and thermal denaturation. Biochemistry. 1972 Dec 5;11(25):4811–4817. doi: 10.1021/bi00775a026. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Histones H3 and H4 interact with the ends of nucleosome DNA. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4400–4404. doi: 10.1073/pnas.73.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Sober H. A. Circular dichroism of calf liver nucleohistone. Biochemistry. 1970 Aug 4;9(16):3103–3109. doi: 10.1021/bi00818a001. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Preparation and physical characterization of a homogeneous population of monomeric nucleosomes from HeLa cells. Nucleic Acids Res. 1976 Sep;3(9):2255–2266. doi: 10.1093/nar/3.9.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm F. X., de Murcia G. M., Champagne M. H., Daune M. P. Conformational changes of histones and DNA during the thermal denaturation of nucleoprotein. Eur J Biochem. 1974 Jun 15;45(2):431–443. doi: 10.1111/j.1432-1033.1974.tb03567.x. [DOI] [PubMed] [Google Scholar]
- Williams R. E., Lurquin P. F., Seligy V. L. Circular dichroism of avian-erythrocyte chromatin and ethidium bromide bound to chromatin. Eur J Biochem. 1972 Sep 25;29(3):426–432. doi: 10.1111/j.1432-1033.1972.tb02005.x. [DOI] [PubMed] [Google Scholar]
- Yaneva M., Dessev G. Persistence of the ten-nucleotide repeat in chromatin unfolded in urea, as revealed by digestion with deoxyribonuclease i. Nucleic Acids Res. 1976 Jul;3(7):1761–1767. doi: 10.1093/nar/3.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]