Cancer chemoprevention uses natural, synthetic, or biological substances to reverse, suppress, or prevent either the initial phase of carcinogenesis or the progression of neoplastic cells to cancer1. It holds promise for overcoming problems associated with the treatment of late-stage cancers. However, the broad application of chemoprevention is compromised currently by limited effectiveness and potential toxicity. In an effort to overcome these challenges, we developed a new chemoprevention approach that specifically targets premalignant tumor cells for apoptosis. This procedure is based on our finding that a deficiency in the adenomatous polyposis coli (APC) gene and subsequent activation of β-catenin lead to the repression of cellular caspase-8 inhibitor cFLIP expression through activation of c-Myc and that 9-cis-retinyl acetate (RAc) independently upregulates tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptors and suppresses decoy receptors. Thus, the combination of TRAIL and RAc induces apoptosis in APC-deficient premalignant cells without affecting normal cells in vitro. In addition, we show that short-term and non-continuous TRAIL and RAc treatment induces apoptosis specifically in intestinal polyps, strongly inhibits tumor growth, and prolongs survival in multiple intestinal neoplasms C57BL/6J-ApcMin/J (ApcMin) mice. With our approach, we further demonstrate that TRAIL and RAc induce significant cell death in human colon polyps, providing a potentially selective approach for colorectal cancer chemoprevention by targeting APC-deficient cells for apoptosis.
TRAIL, or Apo2L, is a membrane-bound TNF family ligand2,3. While TRAIL induces apoptosis in cancer cells, it does not harm normal cells4,5. To test the response of premalignant tumor cells to TRAIL, we isolated primary adenoma cells from intestinal polyps (benign adenomas) of ApcMin mice6-8. We also isolated intestinal epithelial cells from wild-type mice as controls (Supplementary Fig. 1a). TRAIL treatment did not induce any apoptosis in the isolated epithelial cells (Fig. 1a and Supplementary Fig. 1b). Previous reports indicate that certain retinoids synergize TRAIL-induced apoptosis in some cancer cell lines by inducing either TRAIL or its death receptors 9-11. We explored the effect of RAc on TRAIL sensitivity in isolated primary adenoma and normal epithelial cells. Although RAc alone had no effect on the survival of either normal cells from control mice or adenoma cells, it specifically sensitized the adenoma cells to TRAIL-induced cell death (Fig. 1a and Supplementary Fig. 1b). These results show that RAc and TRAIL synergistically induce apoptosis in intestinal adenoma cells derived from ApcMin mice.
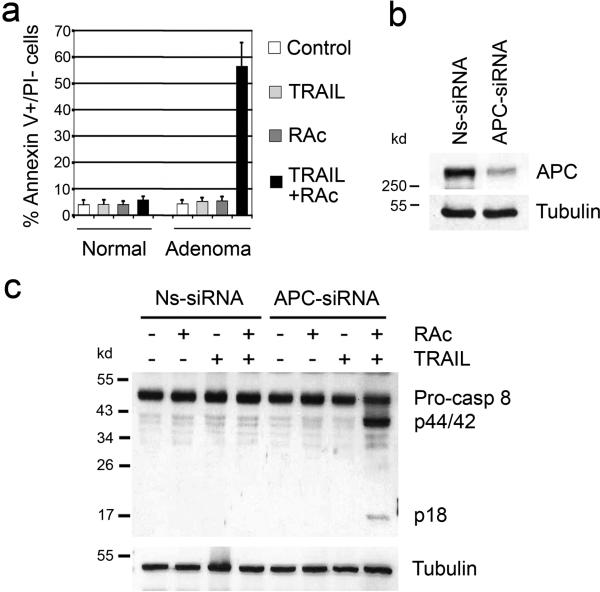
Figure 1. TRAIL and RAc induce apoptosis in APC-deficient cells.
(a) Primary epithelial cells were isolated from normal mouse intestine and polyps from ApcMin mice and treated with TRAIL (50 ng/ml for 24 h), RAc (6.8 ng/ml for 48 h), or both (RAc for 48 h then TRAIL for 24 h). Cells were stained with annexin V-FITC and propidium iodide. Early apoptotic cells (annexin V positive, PI negative) were counted. The data represent results from three independent experiments. Average and standard deviation are shown. (b) Immortalized normal human colon epithelial cells (NCM356) were transfected with either nonspecific Ns-siRNA or APC-siRNA, and APC protein was detected 48 h after transfection by Western blotting. (c) The transfected NCM356 cells were treated and harvested, and both full-length pro-caspase 8 and cleaved forms (p44/42 and p18) were detected by Western blotting.
The major difference between adenoma cells and normal cells is the loss of the wild-type APC allele in adenoma cells12. To test the role of APC in cell sensitivity to TRAIL and RAc, we knocked down APC expression in an immortalized normal human colon epithelial cell line, NCM356, using APC small interfering RNA (APC-siRNA) (Fig. 1b). We observed that knockdown of APC and treatment with TRAIL and RAc resulted in caspase 8 cleavage and an increase in caspase 3/7 activity (Fig. 1c and Supplementary Fig. 1c). Furthermore, caspase 8 inhibitor Z-IETD blocked caspase 3/7 activity (Supplementary Fig. 1c), suggesting that caspase 8 processing resulted in its activation. Similar results were obtained using an immortalized normal human bronchial epithelial cell line (BEAS-2B-BW1799 or BW1799) (Supplementary Fig. 2). These results demonstrate that APC deficiency is the key factor in sensitizing normal cells to apoptosis induced by the combination of TRAIL and RAc.
Inactivation of APC leads to stabilization and nuclear translocation of β-catenin and transcriptional activation of target genes, such as c-Myc and cyclin D113. To investigate the signaling events in the APC pathway required for sensitizing cells to TRAIL and RAc, we transfected either stabilized β-catenin14 or full-length c-Myc into NCM356 cells (Supplementary Fig. 3a and b). Expression of either β-catenin mutant or c-Myc led to TRAIL- and RAc-mediated caspase 8 cleavage and activation (Fig. 2a and Supplementary Fig. 3c). Transfection of cyclin D1 had no effect on TRAIL- and RAc-induced apoptosis in NCM356 and other cells (data not shown). We then determined the requirement of c-Myc for sensitizing cells to TRAIL and RAc. Knockdown of APC led to an increase in c-Myc expression, as expected. Co-transfection of c-Myc-siRNA blocked APC deficiency-mediated c-Myc induction and caspase 8 cleavage/activation in response to TRAIL and RAc (Fig. 2b and Supplementary Fig. 3c). We further confirmed these observations using independent siRNAs (Supplementary Fig. 4). These results indicate that the effect of APC on cell response to TRAIL and RAc is largely dependent on APC/β-catenin-mediated activation of c-Myc.
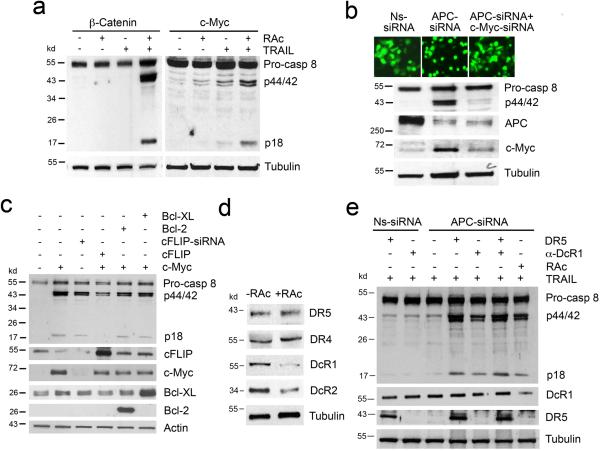
Figure 2. Down regulation of cFLIP by APC/β-catenin-mediated activation of c-Myc and modulation of TRAIL receptor expression by RAc contribute to the activation of TRAIL signaling.
(a) NCM356 cells were transfected with either β-catenin or c-Myc and treated with TRAIL, RAc, or both. Cleavage of caspase 8 was detected by Western blotting. (b) NCM356 cells were transfected with Ns-siRNA, APC-siRNA (GFP tagged), or APC-siRNA and c-Myc-siRNA. The transfected cells were treated with RAc and TRAIL, and photographs were taken. Expression of APC, c-Myc, and caspase 8 was detected by Western blotting. (c) NCM356 cells were transfected with c-Myc, cFLIP-siRNA, c-Myc and cFLIP, c-Myc and Bcl-2, or c-Myc and Bcl-XL. The transfected cells were treated with RAc and TRAIL. Expression of c-Myc, cFLIP, Bcl-2, Bcl-XL, and caspase 8 was detected by Western blotting. (d) NCM356 cells were treated with RAc for 48 h, and expression of DR4, DR5, DcR1, and DcR2 was detected by Western blotting. (e) NCM356 cells were transfected with either APC-siRNA or APC-siRNA and DR5 or were pretreated with an anti-DcR1 antibody after APC-siRNA transfection. Nonspecific Ns-siRNA was used as control. Transfected cells were treated with TRAIL alone or with TRAIL and RAc, as indicated. Cleavage of caspase 8 was detected by Western blotting.
Other studies have shown that c-Myc directly suppresses expression of cFLIP and that this activity correlates with TRAIL sensitivity15. NCM356 cells expressed readily detectable cFLIP and transfection of c-Myc resulted in a significant decrease in cFLIP expression and caspase 8 cleavage (Fig. 2c). While direct suppression of cFLIP expression by cFLIP-siRNA led to caspase 8 cleavage and activation in NCM356 cells treated with TRAIL and RAc, rescue of cFLIP expression by transfection of a cFLIP expression plasmid prevented c-Myc-induced caspase 8 processing and activation in response to TRAIL and RAc (Fig. 2c and Supplementary Fig. 3c). Similar results were obtained in BW1799 cells (Supplementary Fig. 5). In addition to suppressing cFLIP expression, c-Myc could also act on mitochondria to facilitate TRAIL-induced apoptosis16. To test this possibility, we performed rescue experiments using Bcl-2 and Bcl-XL. Overexpression of either Bcl-2 or Bcl-XL did not inhibit c-Myc-mediated caspase 8 processing and activation in response to TRAIL and RAc (Fig. 2c and Supplementary Fig. 3c). These results indicate that APC deficiency sensitizes normal cells to TRAIL and RAc by suppressing cFLIP expression through the activation of β-catenin and c-Myc.
Previous reports have shown that certain retinoids sensitize some cancer cells to TRAIL-induced apoptosis by upregulating either TRAIL or TRAIL receptors DR4 and DR5 9,10. To test the effect of RAc on normal cells in our setting, we examined the expression of TRAIL receptors in NCM356, BW1779, and another immortalized normal human colon epithelial cell line CRL1831 cells after RAc treatment. Although RAc did not induce TRAIL expression (data not shown), it induced DR4 or DR5 expression in these cells (Fig. 2d and Supplementary Fig. 6a). Interestingly, RAc strongly suppressed DcR1 expression in these cells (Fig. 2d and Supplementary Fig. 6a). In addition, the DcR2 expression was inhibited by RAc in NCM356 cells (Fig. 2d). We further showed that RAc caused similar changes in mRNA levels in NCM356 cells (Supplementary Fig. 6b), suggesting that RAc modulates TRAIL receptor expression at the levels of mRNA.
Overexpression of DR5 sensitized APC-knockdown cells to TRAIL-induced caspase 8 cleavage and activation (Fig. 2e and Supplementary Fig. 6c). Pretreatment of APC-knockdown cells with a DcR1-neutralizing antibody to block TRAIL-DcR1 interaction also partially induced caspase 8 cleavage and activation after TRAIL treatment, and this effect was further enhanced in DR5-transfected cells (Fig. 2e and Supplementary Fig. 6c). Similar results were obtained in BW1799 cells (Supplementary Fig. 6d and e). These data suggest that both induction of DR5 and suppression of DcR1 contribute to the effect of RAc in sensitizing APC-deficient cells to apoptosis.
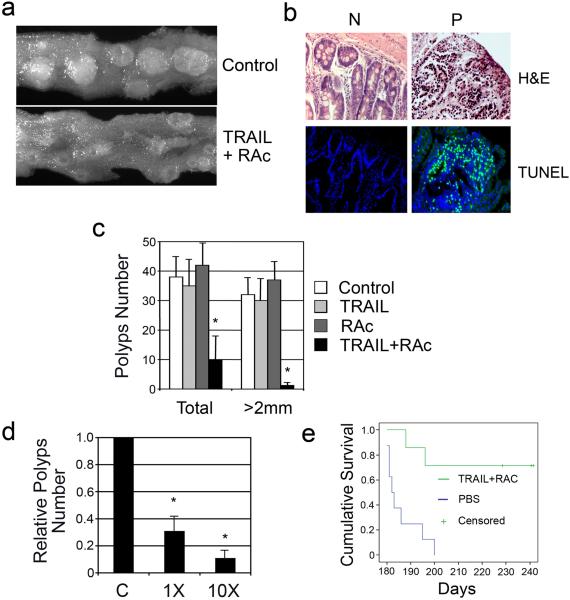
We next tested the effect of TRAIL and RAc in ApcMin mice in vivo. We first treated the 3-month-old ApcMin mice with TRAIL and low-dose RAc for 15 treatment cycles over six weeks. We observed that TRAIL and RAc significantly inhibited the growth of intestinal polyps as compared to controls (Fig. 3a). Histological analysis of tumors and adjacent normal tissues in treated mice showed that many cells in tumors, but not in normal tissues, contain condensed nuclei, suggesting that cells in the tumors are dying (Fig. 3b). This observation was confirmed by terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling (TUNEL) analysis (Fig. 3b). When the total numbers and sizes of the polyps were compared 1 month after the six week treatment period, the treated mice displayed an average 4- to 5-fold decrease in the total number of polyps relative to the mice treated with PBS control (Fig. 3c). The reduction in large polyps (≥2 mm) was much more pronounced (to about 1/30 of control) (Fig. 3c). These data demonstrate that the combination of TRAIL and RAc induces apoptosis specifically in Apc-defective polyps and effectively suppresses the tumor growth.
Figure 3. Treatment with TRAIL and RAc induces cell death in the polyps, inhibits tumor growth, and promote survival in ApcMin mice.
(a) The ApcMin mice were treated with either control phosphate buffered saline (PBS) or TRAIL and RAc for 15 treatment cycles. Intestinal polyps were examined 1 month after the treatment and photographs were taken. (b) The polyp samples and adjacent sections were stained by H&E and TUNEL stain. Surrounding normal tissue was used as the control. Representative photographs are shown. (c) Intestinal polyps in the entire intestinal track were examined and counted. Each treatment group contained 5-7 mice. Average and standard deviations are shown. An asterisk indicates P<0.005. (d). The ApcMin mice were treated with either 1X or 10X doses of TRAIL and RAc for two cycles within a week. The intestinal polyps in the entire intestinal track were examined and counted. Each treatment group contained 5-7 mice. Average and standard deviation are shown. An asterisk indicates P<0.005. (e) Kaplan-Meier survival analysis of ApcMin mice. ApcMin mice at 4 month were treated with either TRAIL (3 mg/kg) and RAc (68 μg/kg) or control PBS every 3 weeks for 5 times. The endpoint was reached when mice were either moribund or at day 243. Mean survival time was 212.8 ± 5.9 in TRAIL and RAc group (7 mice) and 186 ± 2.6 in the PBS treated group (8 mice) with log-rank test p≤0.001. All statistical analyses were performed using SPSS software (version 16.0).
Biochemically, accumulation of soluble β-catenin and induction of c-Myc expression were seen in the polyps of ApcMin mice but not in normal tissue (Supplementary Fig. 7a). cFLIP protein was readily detectable in normal tissue, while it was significantly reduced in polyps (Supplementary Fig. 7b). Consistent with our in vitro observations, RAc induced DR5 expression and repressed DcR1 in both the polyps in ApcMin mice.
Since TRAIL and RAc together target APC-deficient cells for apoptosis, this combination has the potential to achieve chemoprevention in short-term therapy. To test this, we applied two short consecutive TRAIL and RAc treatment cycles to ApcMin within a week and analyzed the intestinal tumors 2 weeks after treatment. After only two treatments, we observed a 69% reduction in the number of polyps when 3 mg/kg of TRAIL and 68 μg/kg of RAc were injected into ApcMin mice (Fig. 3d). The tumor numbers were further reduced to 10% of the control when 10-fold higher doses of TRAIL and RAc were used (Fig. 3d). These data indicate that short-term treatment with TRAIL and RAc is capable for inhibiting growth of premalignant cells in ApcMin mice. To test the effect of TRAIL and RAc treatment on the survival of ApcMin mice, we applied 5 non-consecutive TRAIL and RAc treatments to ApcMin mice within 4 months (average 1 treatment every 3 weeks). All mice in the control group treated with PBS died within 7 months; however, five of the seven treated mice survived beyond 8 months (P≤0.001) (Fig. 3e) demonstrating that non-continuous TRAIL and RAc treatment yields a long-term survival benefit for ApcMin mice. While it is likely that the two early deaths in the treatment group resulted from a late start in treatment, it cannot be ruled out that intrinsic resistance, heterogeneity, genetic or epi-genetic changes of premalignant cells can influence the effect of TRAIL treatment. Further study will provide a more in-depth assessment.
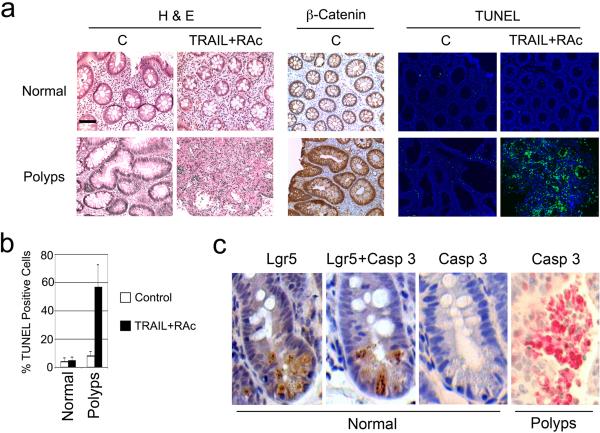
As a first step to demonstrate the efficacy and relevance of TRAIL and RAc treatment in humans, we treated biopsy samples of both normal and tumor regions from consenting familial adenomatous polyposis (FAP) patients under organ culture conditions as reported 17,18. A cross section of normal colon tissue showed a round and defined structure of individual villus consisting of a monolayer of colon epithelial cells and membrane β-catenin staining pattern, while dysplastic colon polyps showed much larger villi, bigger nuclei, an irregular structure consisting of multiple epithelial layers, and enhanced nuclear β-catenin staining (Fig. 4a). Treatment of normal tissue slices with TRAIL and RAc did not induce significant cell death, while the same treatment in the polyp samples resulted in significant cell death, as detected by TUNEL assay (Fig. 4a). Quantification of TUNEL-positive cells showed that TRAIL and RAc killed an average of 57% of the cells in the human colon polyps (Fig. 4b). These results verify that TRAIL and RAc are effective against human polyps from FAP patients in vitro and that our approach holds promise for the chemoprevention of colon cancer in humans.
Figure 4. Effect of TRAIL and RAc on human colon polyps and ISCs, and synthetic lethal interaction of TRAIL, RAc and APC.
(a) Tissue slices from both the normal region and colon polyps from FAP patients were treated with RAc (6.8 ng/ml) for 48 h and then with TRAIL (100 ng/ml) for an additional 24 h (TRAIL+RAc). The samples treated with vehicle were used as controls (C). H&E staining, β-catenin staining, and TUNEL assay (green) were performed. Nuclei were revealed by DAPI staining (blue). Scale bar equals 100 μm. Representative photographs are shown. (b) TUNEL-positive cells (green) were counted against the number of nuclei in multiple fields under a microscope. Average and standard deviations are shown. The data are derived from samples of 4 patients. (c) ApcMin mice were treated with 5 non-continuous cycles of TRAIL and RAc. Both the normal tissue and polyps of the intestine were serially sectioned and stained with either Lgr5 (brown), or activated caspase 3 (red), or both. Multiple serial sections were analyzed and representative photographs are shown.
A major concern in chemoprevention is potential toxicity associated with treatment. We have shown that TRAIL and RAc target the APC-β-catenin-c-Myc signaling pathway for apoptosis. Since activation of β-catenin is involved in stem cell self-renewal and maintenance in adult tissues19, TRAIL and RAc could possibly interact in a detrimental way with stem cells. In order to investigate this, we first tested the effect of TRAIL and RAc on intestinal stem cells (ISCs) in ApcMin mice. Using a putative ISC marker leucine-rich-repeat-containing G-protein-coupled receptor (Lgr5)20, we noted that most of the Lgr5 positive cells in the normal sections were located at the bottom of the crypt as reported20 (Fig. 4c). Staining of active caspase 3 revealed extensive staining in tumor sections similar to the findings shown in Figure 3b (Fig. 4c). More importantly, no staining was detected in serial sections from the normal region, including Lgr5+ cells in the crypt regions where the ISC resides (Fig. 4c). We also studied the effect of long-term TRAIL and RAc treatment on tissue-resident stem cells isolated from pararenal adipose tissue (mASCs) of ApcMin mice21 (Supplementary Fig. 8). We found no detectable effect on morphology (data not shown), neural differentiation (data not shown), proliferation and apoptosis, senescence, osteogenesis, and adipogenesis both in vitro and in vivo (Supplementary Fig 9 and 10). Taken together, these results suggest that TRAIL and RAc exert no negative effect on stem cells (ISCs and mASCs) in mice.
In summary, we discovered a synergistic interaction among TRAIL, RAc, and tumor suppressor APC, which results in the specific induction of apoptosis in APC-deficient cells by TRAIL and RAc (Supplementary Fig. 11). Induction of apoptosis represents a most potent cellular mechanism against cancer, and selectively eliminating premalignant tumor cells by TRAIL and RAc is an effective method of chemoprevention. More importantly, we demonstrate that the chemopreventive effect of TRAIL and RAc can be achieved by short-term intermittent and non-continuous treatment cycles. Thereby, the potential side effects and costs often associated with long-term treatment could be minimized and controlled.
Methods Summary
Recombinant soluble human TRAIL was prepared according to a published method22. For TRAIL and RAc treatment, the cells were first treated with RAc (6.8 ng/ml) for 48 h, and then TRAIL (50ng/ml or 500ng/ml) was added. Cell viability was determined using Annexin V-FITC Apoptosis Detection Kit (Sigma). Caspase 3/7 activity was measured using Apo-ONE Homogeneous Caspase 3/7 assay kit (Promega). Transfection and Western blotting were performed as described previously23. The apoptotic cells in tumor samples were detected by TUNEL assay using an apoptosis detection kit (R&D Systems) or caspase 3 staining using an anti-cleaved caspase 3 antibody (Cell Signaling). ApcMin mice were purchased from the Jackson Laboratory. For treatment, ApcMin mice at 3 months of age were first given RAc via i.p. injection at 68 μg/kg. After 48 h, TRAIL was injected i.p. at 3 mg/kg. After 24 h, RAc was injected again, followed by TRAIL injection. The procedure was repeated for 6 weeks resulting in a total of 15 treatment cycles. Whole intestines were collected immediately after euthanasia and opened longitudinally so that polyps could be counted and measured. Tissues were processed by either formalin fixation or by being frozen in liquid nitrogen for tissue protein extraction, H&E staining, immunohistochemical analysis, and TUNEL assay. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of The University of Texas M. D. Anderson Cancer Center. Fresh human tissue samples were obtained from consenting FAP patients according to an Institutional Review Board-approved protocol.
Methods
Cell lines and transfection
Primary mouse intestinal epithelial cells and adenoma cells were isolated from either normal C57/B6 mice or ApcMin mice as reported24. Briefly, the small intestine or polyps were cut into 2- to 3-mm segments. The tissues were transferred to a 15-ml tube, washed at least 10 times in 10 ml of fresh Hank's buffered salt solution (HBSS) with vigorous shaking, and diced into <1-mm3 pieces using a sharp scalpel blade. The tissue pieces were transferred to a 15-ml tube with 10 ml Dulbecco's modified Eagle's medium (DMEM) containing 100 U/ml penicillin and 100 μg/ml streptomycin. The mixtures were allowed to settle under gravity for 1 min and all but a small amount at the bottom was carefully removed. This procedure was repeated 5 times. The mixtures were then washed with DMEM 3 times and the pellet was resuspended in the epithelial cell medium containing equal volumes of DMEM and Ham's F-12 medium with the following additives: 5 μg/ml insulin, 10 ng/ml epidermal growth factor, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.2% D-glucose, and 2% fetal bovine serum (FBS). CRL1831 cells were obtained from ATCC. NCM356 cells were obtained from INCELL (San Antonio, TX) and grown in A52 medium supplemented with 30 μg/ml bovine pituitary extract, 8 μg/ml vitamin C, 2 mM glutamine and 1 nM dexamethasone. A52 was custom made without folic acid, which was added at concentrations of 25 nM, 50 nM, 75 nM and 150 nM before use. BW1799 cells were kindly provided by Dr. Reuben Lotan of The University of Texas M. D. Anderson Cancer Center and maintained in keratinocyte-SFM medium (Gibco).
Antibodies and other materials
Anti-full length and anti-cleaved caspase 8, anti-cleaved caspase 3, anti-cFLIP, anti-DR5, anti-Bcl-2, and anti-Bcl-XL antibodies were purchased from Cell Signaling Technology. Anti-c-Myc and anti-cytokeratin-20 (CK-20) antibodies were purchased from Santa Cruz Biotechnology Inc. Anti-APC was obtained from Calbiochem. Anti-β-actin and anti-α-tubulin were purchased from Sigma. Anti-DcR1 and anti-DcR2 antibodies were purchased from Imgenex. Anti-DR4 antibody was obtained from Upstate. Anti-β-catenin antibody was purchased from BD Transduction Laboratories. Fluorescein isothiocyanate-conjugated anti-mouse CD14 and CD31 and phycoerythrin-conjugated anti-mouse CD44 and CD90 antibodies were purchased from BD Pharmingen. Anti-Lgr5 antibody was obtained from Abcam. Recombinant soluble TRAIL was prepared according to published results22. RAc was purchased from Sigma.
Plasmids and vector-based siRNA construction
The c-Myc, β-catenin, Bcl-2, and Bcl-XL expression plasmids were described previously14,23,25. DR5 expression plasmid was kindly provided by Dr. Wafik El-Deiry of the University of Pennsylvania School of Medicine. The full-length cFLIP cDNA was generated by reverse-transcription polymerase chain reaction of total RNA from HeLa cells. All siRNAs except APC-siRNA(2) and c-Myc-siRNA(2) were generated in pSUPER as described elsewhere23. The target sequence of human APC is 5′-ggaagtattgaagatgaag-3′, of human cFLIP 5′-agaggtaagctgtctgtcg-3′, and of human c-Myc 5′-ttcaagaga-3′. The APC-siRNA(2) and c-Myc-siRNA(2) were purchased from Open Biosystems with target sequences of 5′-gctgtgaaattcacagtaata-3′ and 5′-ccgagaacagttgaaacacaaa-3′, respectively.
Quantitative PCR
Total RNA was extracted using Trizol reagent according to the manufacturer's instructions (Invitrogen). The cDNA was prepared using SuperScript cDNA Synthesis Kit (Invitrogen). Quantitative PCR was performed using the SYBR Green Quantitative PCR kit from Thermo Scientific in a Bio-Rad C1000TM Thermal Cycler following the manufacturer's protocol. Amplification was carried out in a total volume of 20 μL for 40 cycles of 15 seconds at 95°C, 20 seconds at 60°C, and 30 seconds at 72°C. Samples were run in triplicate and their relative expression was determined by normalizing expression of each target GAPDH. These were then compared with the normalized expression in Control untreated sample to calculate a fold change value. Primer sequences were as follows: human GAPDH 5'-TGCACCACCAACTGCTTAGC-3' and 5'-GGCATGGACTGTGGTCATGAG-3'; human DR4 5'-TGTACGCCCTGGAGTGACAT-3' and 5'-CACCAACAGCAACGGAACAA-3'; human DR5 5'-CACTCACTGGAATGACCTCCTTT-3' and 5'-GTGCAGGGACTTAGCTCCACTT-3'; human DcR1 5'-CCCTAAAGTTCGTCGTCGTCAT-3' and 5'-GGGCAGTGGTGGCAGAGTA-3'; human DcR2 5'-ACAGAGGCGCAGCCTCAA-3' and 5'-ACGGGTTACAGGCTCCAGTATATT-3',
Ex vivo organ cultures
Paired tumor and adjacent normal samples from consenting patients (male and female) undergoing colonoscopy at the MD Anderson Cancer Center Gastrointestinal Oncology and Digestive Diseases endoscopy and surgical unit were collected, transferred to research lab in cold media, and processed within 1 h in a sterile tissue culture hood. The culture condition is modified based on previous reports17,18. Briefly, the culture medium consists of a 1:1 mixture Dulbecco's modified Eagles's medium and Ham's F12 with 5% fetal calf serum (FCS), 10mM HEPES, 0.5% DMSO, 0.5ug/ml Hydrocortisone, 1% MEM vitamins solution, 5μg/ml insulin, 5μg/ml transferin, 5ng/ml selenium, 100 IU/ml penicillin, 100 μg/ml streptomycin, 15 μg/ml gentamicin and 15 μg/ml Ciproxin. The tissue is washed five times in culture media. The tissue is placed in a sterile glass dish where all necrotic and visible connective tissue is removed. The tissue is sliced gently into very thin pieces using a blade and placed in a 6-well tissue culture dish with one ml of growth media. The dish is placed in a 37°C, 5% CO2 incubator for 24 h. It is inspected visually for contamination and the media is pulled off and replaced after every 2 days. After 1 day in culture, the tissue slices were subject to various treatment protocols.
Isolation and culture of adipose tissue-derived stem cells
Subcutaneous adipose tissue was obtained from three mice (one wild-type and two ApcMin) or ApcMin mice that had received 5 cycles of TRAIL and RAc treatment within 4 months following our institutional guidelines. Cells were isolated from adipose tissue as described previously 26,27. Adipose tissue was minced and incubated for 90 min at 37°C on a shaker with Liberase Blendzyme 3 (Roche) at a concentration of 4 U/g fat tissue in phosphate-buffered saline (PBS). The digested tissue was sequentially filtered through 100-μm and 40-μm filters (Fisher Scientific) and centrifuged at 450 g for 5 min. The supernatant containing adipocytes and debris was discarded; the pelleted cells were washed twice with HBSS (Cellgro) and finally resuspended in stem cell growth medium (SCGM), which contained alpha-modification of Eagle's medium (αMEM, Cellgro), 20% FBS (Atlanta Biologicals), 2 mM glutamine (Cellgro), and 100 U/ml penicillin with 100 μg/ml streptomycin (Cellgro). Cells adhering to plastic were designated mice adipose tissue-derived stem cells (mASCs) and grown in Nunclon culture flasks (Nunc) at 37°C in a humidified atmosphere containing 5% CO2, followed by daily washes to remove red blood cells and non-attached cells. After the mASCs became confluent (passage 0), cells were digested and seeded at a density of 3,000 cells/cm2 (passage 1).
Assays of mASCs
For in vitro treatment assays, mASCs were cultured in the presence TRAIL (50ng/ml), or RAc (6.8 ng/ml), or both for two months (about 60 doublings). For effect of TRAIL and RAc treatment in vivo, mASCs were harvested from ApcMin mice that had received 5 cycles of treatments with TRAIL (3 mg/kg) and RAc (68 μg/kg) in 4 months. Cell proliferation was determined by counting live cells. Apoptosis was measured using Annexin V staining (BD Biosciences) using a FACSCalibur flow cytometer (Becton Dickinson) and Cell Quest software (Becton Dickinson). Cell senescence was determined by β-galactosidase activity at pH 6 using the Senescent Cells Staining Kit (Cell Signaling) according to the provided protocol. Osteogenic differentiation of mASCs was induced in osteogenic medium containing high-glucose DMEM supplemented with 10% FBS, 0.1 μM dexamethasone, 200 μM L-ascorbic acid, and 10 mM β-glycerol phosphate (Sigma). The medium was changed every 3 days for 2 weeks. In the control experiment, mASCs were cultured in control medium containing high-glucose DMEM plus 10% FBS for 2 weeks. To assess mineralization, calcium deposits in cultures were stained with Alizarin Red S (Sigma). The osteogenic efficiency was then determined using Osteogenesis Quantitation Kit (Chemicon) following the manufacture's instructions. The adipogenic ability of mASCs was induced in adipogenic medium for up to 2 weeks as described previously26,27. The adipogenic medium contained low-glucose DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin (Cellgro), 100 μM L-ascorbic acid, 1 μM dexamethasone, 0.5 mM 1-methyl-3-isobutylxanthine, 100 μM indomethacin, and 10 μg/ml insulin (Sigma). mASCs of the control group were cultured in low-glucose DMEM plus 10% FBS (control media). The medium was changed every 3 days. Adipogenesis of cells was assessed by incubating cells with Oil Red O solution (Sigma) to stain neutral lipids in the cytoplasm. The adipogenic efficiency of mASCs was then determined using Adipogenesis Quantitation Kit (Chemicon) according to the protocol provided by the manufacturer.
Immunohistochemistry and Immunofluorescence staining
Tissues in paraffin block were sectioned at 4-μM in thickness. After deparaffinization, Antigen retrieval was performed using citrate buffer (Vector lab, Burlingame, CA) heated in a pressure cooker for 25 min and then cooled to room temperature. Blocking of endogenous peroxides was accomplished by incubating sections in 3% hydrogen peroxide (Sigma) for 5 min. Antibodies were incubated with sections overnight at 4°C. Immunostaining was performed by using DAB (3,3'-diaminobenzidine) or AEC (3-Amino-9-ethylcarbazole) (Dako) according to the manufacturer's instructions. Sections were counterstained with hematoxylin for 1 min, rinsed in water and cover slipped permanently for light microscopy.
The mASCs cultured on glass cover slips were washed 3 times with PBS and fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature. Cells were then washed 3 times with PBS alone or PBS containing 0.3% Triton X-100 (Sigma) and blocked by treatment with 10% donkey serum for 30 min at room temperature. Cells were then incubated with primary antibodies fluorescein isothiocyanate-conjugated anti-mouse CD14 and CD31 or phycoerythrin-conjugated anti-mouse CD44 and CD90 in a humidified chamber for 1 h at 37°C. Thereafter, 4',6-diamidino-2-phenylindole (DAPI) was used to stain the cell nuclei. The images of cells were taken under a fluorescence microscope.
Statistical analysis
The two-tailed Student's t-test or one-way analysis of variance (ANOVA) was used to compare differences between groups. Values with P<0.05 were considered statistically significant. Survival analyses were performed using SPSS software (Version 16.0).
Acknowledgements
We thank Haiyun Yang for technical support, Dr. Reuben Lotan of M. D. Anderson Cancer Center for the BW1799 cells, and Dr. Wafik El-Deiry at University of Pennsylvania School of Medicine for DR5 expression plasmid; Dr. Mark Chambers, Mr. Jeremy Yate, Ms. LaToya Ingram, Ms. Debbie Chow, and Ms. Guadalupe Reyes for their involvement in IRB approval and human tissue acquisition. We also thank Jody Vykoukal, Ph.D., Ana Elena Kadala JD, and Datasha Malone for their help in editing the manuscript. This work was supported by NIH grant AI063063 and Institutional Funds from M. D. Anderson Cancer Center (to XW) and in part by Alliance of Cardiovascular Researchers grant 543102 (to E. Alt).
References
- 1.Sporn MB, Dunlop NM, Newton DL, Smith JM. Prevention of chemical carcinogenesis by vitamin A and its synthetic analogs (retinoids). Fed Proc. 1976;35(6):1332–1338. [PubMed] [Google Scholar]
- 2.Pitti RM, et al. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271(22):12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 3.Wiley SR, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3(6):673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 4.Koschny R, Walczak H, Ganten TM. The promise of TRAIL-potential and risks of a novel anticancer therapy. J Mol Med. 2007;85(9):923–935. doi: 10.1007/s00109-007-0194-1. [DOI] [PubMed] [Google Scholar]
- 5.Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov. 2008;7(12):1001–1012. doi: 10.1038/nrd2637. [DOI] [PubMed] [Google Scholar]
- 6.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247(4940):322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 7.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256(5057):668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 8.Fodde R, et al. A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors. Proc Natl Acad Sci U S A. 1994;91(19):8969–8973. doi: 10.1073/pnas.91.19.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altucci L, et al. Retinoic acid-induced apoptosis in leukemia cells is mediated by paracrine action of tumor-selective death ligand TRAIL. Nat Med. 2001;7(6):680–686. doi: 10.1038/89050. [DOI] [PubMed] [Google Scholar]
- 10.Sun SY, Yue P, Hong WK, Lotan R. Augmentation of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis by the synthetic retinoid 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid (CD437) through up-regulation of TRAIL receptors in human lung cancer cells. Cancer Res. 2000;60(24):7149–7155. [PubMed] [Google Scholar]
- 11.Jin F, et al. Activation of nuclear factor-kappaB contributes to induction of death receptors and apoptosis by the synthetic retinoid CD437 in DU145 human prostate cancer cells. Cancer Res. 2005;65(14):6354–6363. doi: 10.1158/0008-5472.CAN-04-4061. [DOI] [PubMed] [Google Scholar]
- 12.Yang K, et al. A mouse model of human familial adenomatous polyposis. J Exp Zool. 1997;277(3):245–254. [PubMed] [Google Scholar]
- 13.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17(1):45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Xia X, et al. Loss of presenilin 1 is associated with enhanced beta-catenin signaling and skin tumorigenesis. Proc Natl Acad Sci U S A. 2001;98(19):10863–10868. doi: 10.1073/pnas.191284198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricci MS, et al. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol Cell Biol. 2004;24(19):8541–8555. doi: 10.1128/MCB.24.19.8541-8555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieminen AI, Partanen JI, Hau A, Klefstrom J. c-Myc primed mitochondria determine cellular sensitivity to TRAIL-induced apoptosis. EMBO Journal. 2007;26(4):1055–1067. doi: 10.1038/sj.emboj.7601551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willson JK, Bittner GN, Oberley TD, Meisner LF, Weese JL. Cell culture of human colon adenomas and carcinomas. Cancer Res. 1987;47(10):2704–2713. [PubMed] [Google Scholar]
- 18.Hasson E, et al. Solid tissues can be manipulated ex vivo and used as vehicles for gene therapy. J Gene Med. 2005;7(7):926–935. doi: 10.1002/jgm.740. [DOI] [PubMed] [Google Scholar]
- 19.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18(5):523–527. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 20.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 21.Bai X, Sadat S, Gehmert S, Alt E, Song YH. VEGF receptor Flk-1 plays an important role in c-kit expression in adipose tissue derived stem cells. FEBS Lett. 2007;581(24):4681–4684. doi: 10.1016/j.febslet.2007.08.063. [DOI] [PubMed] [Google Scholar]
- 22.Ren X, Xu Z, Myers JN, Wu X. Bypass NFkappaB-Mediated Survival Pathways by TRAIL and Smac. Cancer Biol Ther. 2007;6(7) doi: 10.4161/cbt.6.7.4206. [DOI] [PubMed] [Google Scholar]
- 23.Deng Y, Ren X, Yang L, Lin Y, Wu X. A JNK-dependent pathway is required for TNFalpha-induced apoptosis. Cell. 2003;115(1):61–70. doi: 10.1016/s0092-8674(03)00757-8. [DOI] [PubMed] [Google Scholar]
- 24.Evans GS, Flint N, Somers AS, Eyden B, Potten CS. The development of a method for the preparation of rat intestinal epithelial cell primary cultures. J Cell Sci. 1992;101(Pt 1):219–231. doi: 10.1242/jcs.101.1.219. [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci U S A. 1994;91(9):3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai X, et al. Electrophysiological properties of human adipose tissue-derived stem cells. Am J Physiol Cell Physiol. 2007;293(5):C1539–1550. doi: 10.1152/ajpcell.00089.2007. [DOI] [PubMed] [Google Scholar]
- 27.Gimble J, Guilak F. Adipose-derived adult stem cells: isolation, characterization, and differentiation potential. Cytotherapy. 2003;5(5):362–369. doi: 10.1080/14653240310003026. [DOI] [PubMed] [Google Scholar]