Abstract
Sulfotransferases are enzymes that catalyze the transfer of sulfo groups from a donor, for example 3′-phosphoadenosine 5′-phosphosulfate, to an acceptor, for example the amino or hydroxyl groups of a small molecule, xenobiotic, carbohydrate, or peptide. These enzymes are important targets in the design of novel therapeutics for treatment of a variety of diseases. This review examines assays used for this important class of enzyme, paying particular attention to sulfotransferases acting on carbohydrates and peptides and the major challenges associated with their analysis.
Keywords: Sulfotransferases, Sulfonation, Carbohydrates, Enzyme assays, Peptides
Introduction
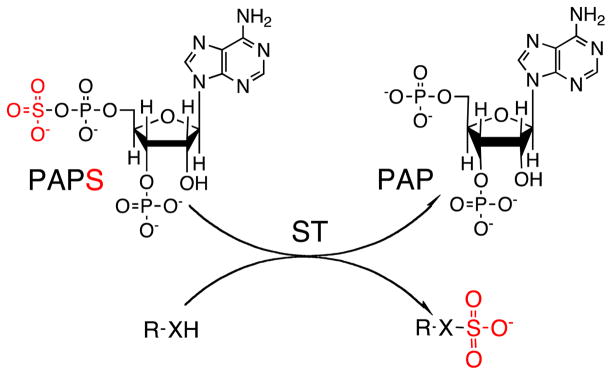
Sulfotransferase (ST) enzymes are responsible for catalyzing the transfer of a sulfo group from a donor molecule, usually 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to an acceptor, for example a sugar, alcohol, phenol, or amine (Scheme 1). STs can be either cytosolic or membrane-associated (Fig. 1). Cytosolic STs are key phase II metabolizing enzymes involved in the clearance of small endogenous and exogenous compounds, for example hormones, bioamines, drugs, and a variety of xenobiotic agents. Most of our understanding of STs comes from the study of these cytosolic enzymes [1–4]. Membrane-associated STs sulfonate larger biomolecules, for example carbohydrates and proteins, and have recently been implicated in many critical biological processes [5]. Structure-based sequence alignments have indicated that the structural fold and the PAPS-binding site are conserved between cytosolic and Golgi STs. In both classes of enzyme, reactions involving the transfer of sulfo groups are believed to proceed by in-line attack of the nucleophile at the sulfate group of PAPS [3, 6].
Scheme 1.
ST-catalyzed transfer of sulfo group (red) from PAPS donor to R–XH acceptor (R = sugar or peptide or small cytosolic molecules. When X = O the ST is an O-STand when X = NH/NR the ST is an N-ST)
Fig. 1.
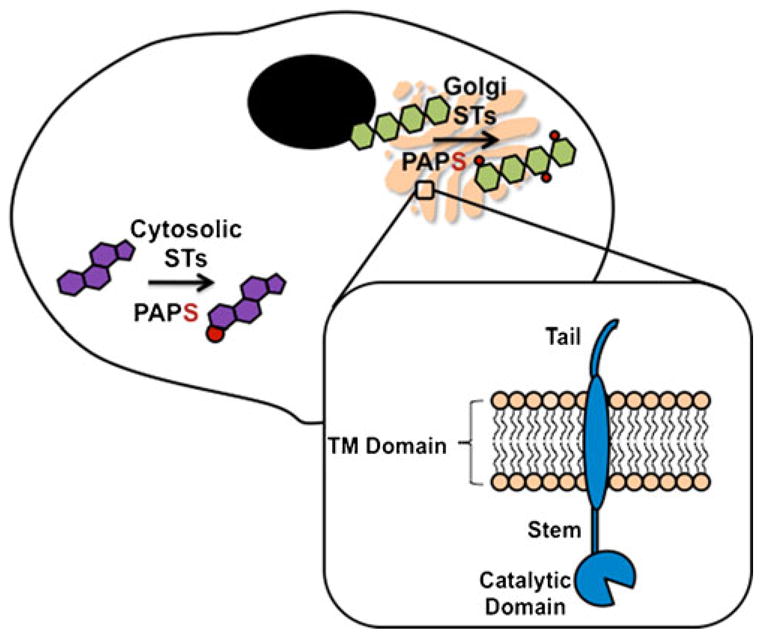
PAPS is biosynthesized in the cytosol of an eukaryotic cell and used there by cytosolic STs to transfer sulfo groups (red) to small endogenous and exogenous compounds, for example steroids. PAPS is also transported to the Golgi where it is used by membrane STs to transfer sulfo groups (red) to hydrophilic molecules, for example carbohydrates. STs in the Golgi consist of a cytosolic tail, transmembrane (TM) domain, and catalytic domain
Sulfo group transfer is an important reaction in the chemical metabolism of drugs, chemical carcinogens, hormones, bile acids, neurotransmitters, peptides, and lipids [7]. Although STs are emerging therapeutic drug targets and may be susceptible to enzyme-specific small molecule inhibitors, this aspect of drug design is currently under-exploited. As the action and mechanisms of these enzymes become better understood and high-throughput screening assays are developed, discovery of ST inhibitors might also afford novel drugs for treatment of cancer, inflammation, and infection, to improve and complement current therapy [8].
Robust enzyme activity assays are crucial to accelerate the progress of new drug development related to STs, because they can provide optimum reaction control at minimum cost of reagents and time. Enzyme assays quantify enzyme performance by monitoring observable signals during conversion of substrate to product. Activity is expressed in terms of “units”, usually defined as the formation of one μmole of product per minute at a specified pH and temperature [9]. A variety of assay methods may be useful for determining the activity of a particular enzyme. Each assay measures reaction velocity and should be reliable, free from false positives or negatives, and easy to perform. Selection of an assay method usually depends on convenience, availability of reagents and apparatus, and assay sensitivity and throughput. Unique challenges in the design and development of ST assays include enzyme type (i.e., cytosolic, Golgi, native, recombinant truncated—catalytic domain, recombinant fusion protein—catalytic domain fused with another protein), purity, stability, and substrate availability.
A brief review of ST assays, published over 20 years ago, highlighted an ion-pairing (IP) extraction method for assay of aryl-STs [10]. This method relies on methylene blue and 2-naphthol as substrates, thin-layer chromatography (TLC) for product separation, and radioisotopic detection. Over the past two decades, in-vitro ST assays largely continue to rely on incorporation of radioactive sulfur in PAP35S to determine ST substrate specificity and ST activity. This review briefly describes these and other more recently developed methods that are useful in determining ST activity to enable the better understanding of ST catalysis needed to elucidate substrate-binding mechanisms and for the pharmacological design of potent and specific ST inhibitors that are useful as new drugs.
Radiometric activity assays
Cytosolic ST enzyme activity and substrate specificity have mostly been determined by quantification of the transfer of a PAP35S sulfo group to different substrates followed by separation of the 35S-labeled products by use of gel-filtration chromatography [11, 12], affinity chromatography [13–15], immobilization techniques [16], membranes [17], or high performance liquid chromatography (HPLC) [18, 19]. These procedures can be tedious for routine and detailed kinetic studies of ST enzymes. However, the high sensitivity and speed of these assays cause them to be widely used. Rates of reactions are non-continuously determined by periodically removing samples from the reaction mixture. Radioactivity of either product or residual substrate is then measured by liquid scintillation counting [20]. Such discontinuous methods are tedious and their accuracy can vary substantially, potentially compromising the quality of the kinetic data reported. Thus, most literature studies using a radioassay have not reported kinetic data. Despite these limitations, radiometric assays are still widely adopted for determining enzyme activity because of their universality—the radiolabel is incorporated in the PAP35S substrate common to all STs.
The activities of a variety of Golgi-derived STs have also been assayed radiometrically. These enzymes pose a greater challenge because, unlike cytosolic enzymes, which act on hydrophobic substrates that can be readily separated, with the product, from PAP35S, Golgi-derived STs catalyze the conversion of hydrophilic substrates to an even more hydrophilic product, confounding their separation from PAP35S.
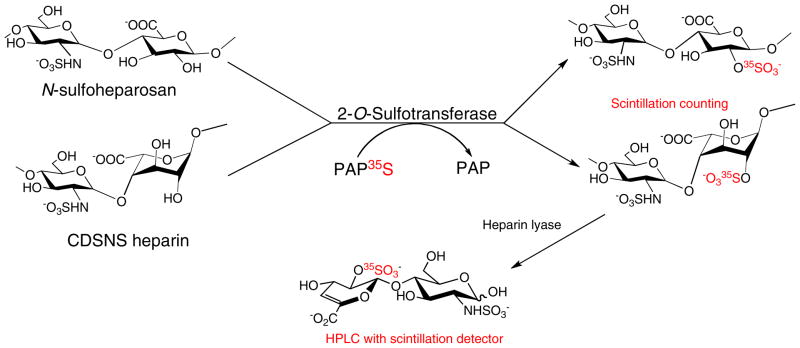
Chondroitin 6-ST (C6ST) and chondroitin 4-ST (C4ST) activity have been determined by using PAP35S with glycosaminoglycan substrates—chondroitin, chondroitin sulfate, and dermatan sulfate. Enzyme conversion was followed by ethanol precipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), or membrane filtration to separate the sulfated reaction products, and subsequent disaccharide analysis using HPLC and scintillation counting to determine the amount of radioactivity incorporated into the polysaccharide [21]. The activity of heparan sulfate 2-O-ST (2-O-ST), which transfers a sulfo group to the 2-O-position of glucuronic or iduronic acid, has also been determined by measuring the incorporation of 35S sulfo groups into N-sulfoheparosan or completely de-sulfonated N-sulfonated heparin (CDSNS) [22] (Scheme 2). The 2-O-35S sulfo polysaccharide was purified by diethylaminoethyl (DEAE) chromatography and assessed by scintillation counting. The position of the sulfo group in the polysaccharide was determined by disaccharide analysis using reversed-phase ion-pairing (RPIP) HPLC.
Scheme 2.
2-O-ST sulfonates N-sulfoheparosan and CDSNS heparin. The polysaccharide product can be quantified and then treated with heparin lyase to afford disaccharides for characterization against disaccharide standards, using HPLC with scintillation detection to establish the position of the added sulfo group
The activity of tyrosylprotein ST (TPST), which catalyzes the transfer of sulfo group from PAPS to the phenolic oxygen of tyrosine residues within highly acidic groups of proteins and polypeptides, was determined by measuring the transfer of 35S sulfo groups to an immobilized peptide substrate by use of liquid scintillation counting [16]. A medium-throughput radiolabel transfer-based assay for the well-characterized N-acetyl glucosamine (GlcNAc)-6-ST NodH from Rhizobium meliloti involved separation of the product from the excess PAP35S substrate, by use of silica gel thin-layer chromatography, and quantification by phosphorimaging. In addition to the activity, the Michaelis–Menten constant (Km) was reported for PAPS, as well as inhibition constants (Ki). This study led to the discovery of the first reported carbohydrate ST inhibitors from a kinase-directed library [23].
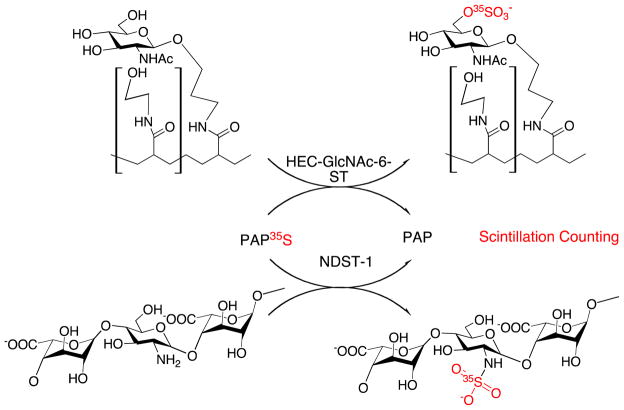
Other substrate–product separation methods have been used in radiometric ST assays. These methods have found application with substrates ranging from small molecules being acted on by cytosolic STs, for example α-naphthol, to large molecules being acted on by Golgi STs, for example proteoglycans, in which sulfated products and donor substrate PAP35S are separated by electrophoresis using SDS-PAGE. Electrophoresis-based separations have been demonstrated with both carbohydrate STs and TPSTs [24]. Dot-blotting radiometric activity assays have been used in high-throughput screening, demonstrating the activity of two carbohydrate STs, heparan sulfate N-deacetylase/N-ST and high-endothelial cell (HEC) GlcNAc-6-O-ST on PAP35S and de-N-sulfonated heparin and N-acetylglucosamine, respectively [17] (Scheme 3). The radiolabeled products were captured on a membrane and then eluted, ready to be quantified, thus avoiding the need for a complicated purification step or repeated washing that is usually required in sample preparation for scintillation counting. Although Km for PAPS and Ki values were reported, Km could not be determined for the polymeric carbohydrate substrates, because of their heterogeneous nature and the presence of multiple sulfation sites on each polymer chain. This method is useful for inhibitor screening and should be useful for high-throughput microplate assays of other carbohydrate STs.
Scheme 3.
HEC-GlcNAc-6-ST and NDST-1 act on N-acetylglucosamine and de-N-sulfonated heparin, respectively. The 35S-labeled product with O-sulfo and N-sulfo groups can easily be separated from PAP35S and quantified by scintillation counting
Although radiometric ST assays are so prevalent, they remain expensive with PAP35S costing ~$1,000/100 μCi, and 35S has a half-life of only 87.1 days. Disposal of spent radioisotopes and regulatory compliance are also added costs associated with these assays.
Photometric activity assays
Photometric activity assays can be used for continuous rapid kinetic determinations for high-throughput screening of potential substrates and inhibitors. Enzyme kinetic data, for example Km and Vmax are often easily determined by spectrophotometric methods by taking advantage of reactants that afford a new chromophore or undergo a detectable shift in absorption at a characteristic wavelength during a reaction. Most of the assays reported monitor the production of p-nitrophenol (PNP) [25–29], a few assays measure 2-naphthol sulfation rates [30], and one assay describes the photometric measurement of cysteine formed from sulfite [31].
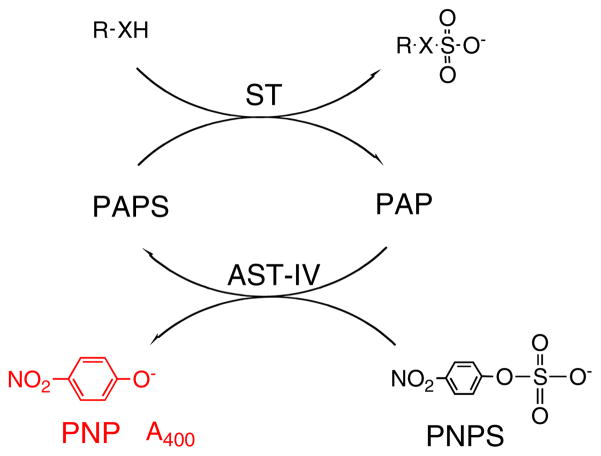
One novel ST-coupled assay system involves the well-studied enzyme, aryl sulfotransferase IV (AST-IV), which transfers a sulfo group from PAPS to aryl alcohols [27]. In this assay, PAPS is continuously regenerated in ST-catalyzed reactions by using p-nitrophenyl sulfate (PNPS) as a sulfo group donor, facilitating colorimetric monitoring of the PNP product at 400 nm (Scheme 4). The linear double-reciprocal plot generated gave apparent Km and Vmax of several carbohydrate substrates and PAP in the reverse-physiological reaction similar to published values from the physiological reaction. This assay could potentially be applied to rapid kinetic determinations for carbohydrate and protein STs, enabling high-throughput screening for potential ST substrates and inhibitors. Such an assay might be useful in the biomedical screening of blood samples and other tissues for specific ST activity or the concentration of ST substrates.
Scheme 4.
PAPS regeneration system. AST-IV catalyzes the transfer of the sulfo group from the less expensive donor, PNPS. The sulfo group is then transferred to the substrate of interest by the O-ST enzyme. The PNP formed in the coupled reaction absorbs at 400 nm
Another photometric method, called electrophoresis-mediated microanalysis (EMMA), based on capillary electrophoresis with ultraviolet detection, has been used to assay the activity of a phenol ST (SULT1A1) with PNP as a substrate. This assay uses initial rate kinetics to afford a Km for PNP that is consistent with previously reported values [28]. EMMA uses electrophoresis to separate the inhibitor PAP from PAPS before injection of enzyme and substrate inside the capillary. Photometric assays such as these are convenient, fast, and simple, only requiring a substrate that is cleaved into a colorimetric co-product. This assay is limited by small path lengths and requires concentrations above the nanomolar range. However, compared with other spectrophotometric assays, the EMMA method is rapid, automatable, and requires only nanoliter volumes of potentially expensive reagents.
Fluorimetric activity assays
Assays based on fluorescence are continuous, more sensitive than photometric assays, and have sensitivity comparable with that of end-point radioisotope assays [32]. High sensitivity also enables the use of low substrate concentrations and requires small amounts of enzyme. Many assays take advantage of compatible fluorescent substrates, for example 4-methylumbelliferyl sulfate (MUS) [32, 33], 2-naphthyl substrate [34], or pyrene 1-sulfate [35]. Others attach fluorescent moieties to substrates of interest, for example oligosaccharides [36] or a variety of acids [37].
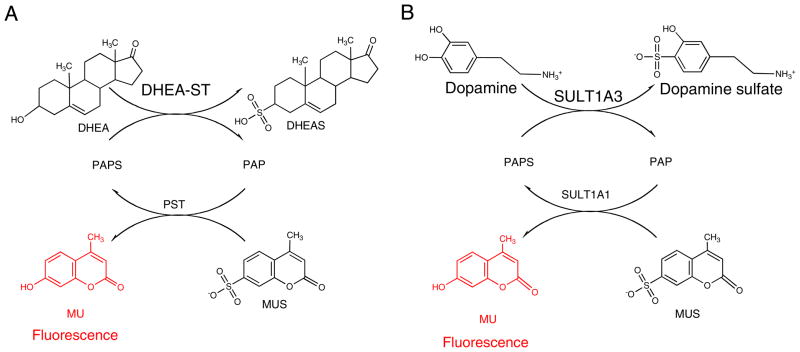
A fluorimetry-based activity assay has been described that utilizes the regeneration of PAPS from PAP by the auxiliary enzyme phenol ST with MUS as the sulfo donor (Scheme 5A) [32]. The reaction was coupled with the alcohol ST, human dehydroepiandrosterone (hDHEA–ST), and the 4-methylumbelliferone (MU) product served as a fluorescent indicator of enzyme turnover to monitor the alcohol ST activity. The Vmax, Km, and Ki values for MUS and dehydroepiandrosterone (DHEA) were determined from initial rate kinetics of the 450 nm emission on 360 nm excitation. This suitable excitation wavelength was determined to prevent interference from the absorbance of MUS. The observed fluorescence intensity was significantly dependent on the pH value but less dependent on temperature. The change of fluorescence intensity of 4-methylumbelliferone was sufficiently sensitive to measure the activity of nanogram or picomole amounts of enzyme, comparable with a previously reported radiometric assay. This method has the potential for the development into a high-throughput assay for measuring the AST activity of biological samples using a microplate reader. The activity of other enzymes, associated with either sulfonation (STs) or desulfonation (sulfatases) might also be determined by use of this approach.
Scheme 5.
Determination of ST activity by increase in MU fluorescence. A DHEA-ST and B SULT1A3 activity by regeneration of PAPS with auxiliary SULT1A1 enzyme reducing the fluorescence of MU
A similar coupled real-time fluorimetric enzyme assay was developed for monoamine-preferring phenol ST (SULT1A3). SULT1A1 was used to regenerate PAPS, again using MUS as a sulfo group donor (Scheme 5B). MU was continuously monitored as SULT1A3-catalyzed sulfo group transfer to dopamine with PAPS [33] to determine activity, Vmax, Km, and Ki values of SULT1A3. These values were consistent with values previously determined by use of radiometric assay procedures at comparable sensitivity [38].
A fluorescence-based HPLC assay for determination of human estrogen ST (SULT1E1) inhibition was developed using 1-hydroxypyrene to investigate the inhibitory effect of endocrine-disrupting compounds (EDC) [35]. SULT1E1 is involved in the regulation of 17β-estradiol responsiveness and is believed to protect peripheral tissues from excessive estrogenic effects. 1-Hydroxypyrene was selected as the substrate because of its fluorescent properties, the fluorescent properties of its metabolite, pyrene 1-sulfate, and because it is noncarcinogenic unlike the metabolites of benzo [a]pyrene, which had been used in previous studies. A gradient HPLC separation of 1-hydroxypyrene and pyrene 1-sulfate on a reversed-phase C18 column was developed and optimized The formation of pyrene 1-sulfate was quantified fluorescently, with detection limits of 0.1 pmol for pyrene 1-sulfate and 1-hydroxypyrene. The double reciprocal plot of the kinetic data afforded Km and Vmax values for 1-hydroxypyrene similar to those previously reported. A library of 19 compounds with known estrogenic properties was tested at a single concentration, and IC50 values were determined for the ten strongest SULT1E1-inhibiting compounds. Negative controls representing 100 % inhibition and positive controls representing 0 % inhibition were used to demonstrate assay reliability. The method was highly reproducible for SULT1E1 activity screening and inhibition studies. This assay offers opportunities to investigate human tissue samples in the presence of EDCs, and can be used to establish causative predictions between levels of EDCs and health problems associated with SULT1E1.
In summary, although fluorescence-based assays are convenient and extremely sensitive, they can be difficult to design and develop, particularly when the substrates and/or products are not fluorescent.
Mass spectrometric activity assays
Unfortunately, many ST substrates and reaction products do not contain a chromophore or fluorophore necessary to perform spectrophotometric and/or spectrofluorimetric assays, and in some cases radioisotope use is undesirable. Assays based on mass spectroscopy are useful in these cases. Modern soft-ionization methods enable sensitive analysis of sulfated compounds without fragmentation of the fragile O-sulfo and N-sulfo linkages and can provide unambiguous differentiation of substrate from product. Most of the reported mass spectrometric assays have utilized electrospray ionization–mass spectrometry (ESI–MS) [39–42], some have combined MS analysis with liquid chromatography (LC–MS) [43] and one has used the hydrogen–deuterium exchange upon epimerization procedure with liquid chromatography–mass spectrometry (DEEP–LC–MS) [44].
An activity assay based on ESI–MS using an ion-trap mass spectrometer was developed and applied to a bacterial carbohydrate ST, NodST, in which the enzyme catalyzes the transfer of a sulfo group from PAPS to chitobiose, yielding PAP and 6-O-sulfochitobiose [39]. The total analysis time was reportedly comparable with that of a standard spectrophotometric assay. The activity and Km for PAPS and chitobiose, the Vmax and Ki for PAP, and the mode of inhibition were all determined. The Km value for PAPS was consistent with literature values obtained by TLC assay, validating the ESI–MS assay as a reliable and accurate method for determining the kinetic data for NodST. Unlike the earlier radiometric method using TLC, ESI–MS was also capable of obtaining the Km value for chitobiose. The substrates and products of many ST-catalyzed reactions, including those catalyzed by NodST, do not have the chromophores needed for spectrophotometric methods. Strategies relying on the synthesis of artificial chromogenic or fluorogenic substrates are time-consuming to develop and are not always feasible. In contrast, ESI–MS-based assays can usually be used to assay any ST as long as an internal standard with a structure and ionization efficiency similar to those of either product or substrate is available. An additional advantage is that substrate and product concentrations can often be simultaneously analyzed during the course of the reaction, making the assay an efficient and accurate method for determining enzyme kinetic data. It is noteworthy that substrate concentration is in excess at the beginning of the reaction and is depleted at reaction completion so the accuracy of the measurement of substrate and product is not identical throughout the reaction. LC–MS will continue to be used to investigate the catalytic mechanism of NodST and to identify ST inhibitors. In the future it is likely that ESI–MS based assays will be adapted to study many other mammalian carbohydrate STs of therapeutic interest.
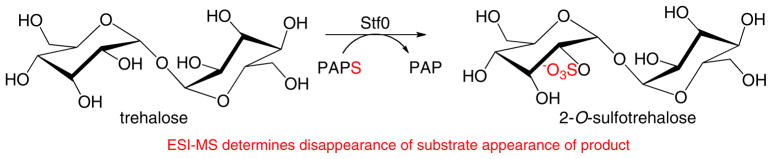
ESI–MS assays have been applied to microbial STs, for example in the assay of Mycobacterial carbohydrate ST (Stf0) [40]. This enzyme catalyzes sulfo group transfer from PAPS to trehalose to form 2-O-sulfotrehalose (Scheme 6). The product was quantified relative to internal standard by use of single-point normalization factors. Initial rate kinetics were studied. Analysis of both saturation and double-reciprocal plots using nonlinear fitting to the Michaelis–Menten equation led to the determination of the catalytic and product inhibition mechanisms, Km, and turnover number (kcat) values for trehalose and PAPS. The kinetic constants for trehalose were in excellent agreement with those previously obtained by use of a TLC assay. The results for PAPS were the first reported values and the Km value of PAPS is similar to those measured for other STs.
Scheme 6.
Transfer of sulfo group from PAPS to trehalose by Stf0
Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry has also been used to detect noncovalent complexes. This was also the first study to provide detailed mechanistic data for Stf0. The power of mass spectrometry is that in addition to providing an assay of enzymatic activity, it also can provide an understanding of the structure, mechanism, and function of biologically important STs.
An LC–ESI–MS-based assay of tyrosylprotein ST-1 and ST-2 that relies on peptides as substrates was developed to determine kinetic data for the purified individual isozymes [43]. A 1:1 mixture of both isozymes was used to address the possibility that the two isozymes work in synergy within the Golgi. This HPLC–ESI–MS assay, unlike previous radioisotopic analysis, was able to differentiate between the formation of monosulfated and disulfated products by using RP-LC coupled directly to a linear ion-trap mass spectrometer equipped with an ESI source. The optimum concentrations of TPST-2, TPST-1, and the 1:1 TPST-1–TPST-2 mixture in the assay reactions were determined to be in the micromolar range. Initial velocity kinetics of each reaction were fitted to the Michaelis–Menten equation to obtain Km, Vmax, and kcat for PAPS and other substrates. The accuracy and precision of the method for quantification were internally validated by running replicates of monosulfated and disulfated standards and the average matrix spike recoveries were 103±7 % and 101±12 % for the monosulfated and disulfated products, respectively. This assay uniquely determines differences in the kinetic constants of sulfation reactions in peptides with multiple tyrosine sulfation sites, providing critical information on enzyme specificity. It can also be used to test potential inhibitors of these isozymes, and to help determine the catalytic mechanism of TPST-1 and TPST-2. Development of this method has proved to be an essential step in investigating the kinetic data of sequential tyrosine sulfation of chemokine receptors by TPSTs and in determining their catalytic mechanism. LC–ESI–MS analysis should also be applicable to the study of other chemokine receptor substrates, to test potential inhibitors, and to determine the mechanisms of tyrosylprotein ST isozymes.
Because radiolabeled materials are not required in the ESI–MS assay, and because the analysis time can be comparable with that of standard UV techniques, mass spectrometry-based assays are expected to become competitive with, and in some cases more convenient than, traditional methods. Mass spectrometric assays have a distinct advantage over radiometric assays because they can differentiate between mono, di, and polysulfonation, common in many natural products, providing critical information on ST specificity.
Conclusions and perspectives
This review has focused on comparing recent advances in radiometric, photometric, fluorimetric, and mass spectrometric ST assay methods (Table 1). Other promising but less popular techniques for measuring ST activity include chemoluminescence [45] and HPLC [46].
Table 1.
ST enzymes and assay methods
| Enzyme | Substrate/acceptor | Detected product | Assay method | Km |
|---|---|---|---|---|
| Human cytosol-localized | ||||
| Aryl sulfotransferase IV (AST-IV) | Aryl alcohol | O-Sulfated ester, PAPS* | P* | 26.4 μmol L−1 |
| N-deacetylase/N-sulfotransferase (NDST-1) | -GlcAβ1-4GlcNα1-4- | -GlcAβ1-4GlcNSα1-4-, PAPS* | R* | 1.0 μmol L−1 |
| Phenol sulfotransferase (SULT1A1) | Phenol, PAP | Sulfated phenol, PAPS*, 4-methylumbelliferone (MU) | P*, F | 0.84 μmol L−1 |
| Dehydroepiandrosterone sulfotransferase (DHEA-ST: SULT2A1) | DHEA | DHEAS, PAPS* | F* | 4.7 μmol L−1 |
| Monoamine-preferring phenol ST (SULT-1A3) | Dopamine, catecholamine, phenol | Dopamine sulfate, sulfated phenol, PAPS* | F* | 6.8 μmol L−1 |
| Estrogen ST (SULT-1E1) | Estrogen, 1-hydroxypyrene | Estrogen sulfate, pyrene-1-sulfate*, PAPS | F* | 6.4 nmol L−1 |
| Human Golgi-localized | ||||
| Chondroitin 4-sulfotransferase (C4ST) | GlcAβ1-3GalNAcβ1-4 | GlcAβ1-3GalNAc(4S)β1-4 | R | n.r. |
| Chondroitin 6-sulfotransferase (C6ST) | NeuAcα1-3 Galβ1-4GlcNAc- | NeuAcα1-3 Gal (6S)β1-4GlcNAc- | R | n.r. |
| NeuAcα1-3 Galβ1-4GlcNAc(6S) | NeuAcα1-3 Gal(6S)β1-4GlcNAc(6S) | R | n.r. | |
| Heparan sulfate 2-sulfotransferase (HS2ST) | -IdoAα1-4GlcNSα1-4 | -IdoA(2S)-GlcNS_α1-4 | R | n.r. |
| Tyrosylprotein sulfotransferase (TPST), TPST-1, TPST-2 | Tyrosine within highly acidic motif of polypeptide/protein | Tyrosine-sulfated protein | R, M* | 21 mmol L−1 |
| High endothelial cell-GlcNAc6-O-ST (HEC-GlcNAc6ST: LSST) | 6-OH of GlcNAc | 6-Sulfo sialyl Lewis X, PAPS* | R* | 3.3 μmol L−1 |
| Bacteria | ||||
| N-acetyl glucosamine (GlcNAc)-6-sulfotransferase (GlcNAc-6-O-ST: NodST) | Chitobiose | 6-O-Sulfochitobiose, PAPS* | R*, M* | 4.3 μmol L−1, 6.7 μmol L−1 |
| Mycobacterial carbohydrate sulfotransferase (StfO) | Trehalose | Trehalose-2-sulfate | M* | 15 mmol L−1 |
R, radiometric assay; P, photometric assay; F, fluorimetric assay, M, mass spectrometric assay; n.r., not reported
Km values reported for substrate and assay methods are indicated with an asterisk
Radiolabeling is still the recommended method for assaying ST enzyme activity. Although these assays are extremely sensitive, their hazard levels, accuracy, and non-continuous nature keep them from being an ideal method. Because of the ease of application, radiometric methods are often not optimized and only adopted for quick use. Radiolabeled assays will certainly continue to be used, but will require improvement and proper adjustment for each enzyme system.
Photometric and fluorimetric assays are generally less hazardous, less costly, high-throughput, and enable accurate and continuous analysis. Unfortunately, these assays cannot always be applied, as many substrates and/or products of interest do not contain chromophores or fluorophores. The preparation of chromogenic or fluorogenic substrates remains a challenge of design and synthesis for the chemist.
Mass spectrometric assays are an excellent option for substrates and/or products which are neither chromogenic nor fluorogenic. These assays are highly accurate with low hazards, but they require costly equipment and are discontinuous. Because mass spectrometry is increasingly becoming accessible in operation and maintenance, its use to assay enzymes and to screen inhibitors in pharmaceutical research is gradually increasing [47]. These assays are especially interesting for carbohydrate ST application because they enable measurement of kinetic constants for polymeric substrates with multiple reactive sites. Moreover these carbohydrate STs are attracting increasing interest for applications in inflammation and cancer and as novel therapeutic targets [48].
Each assay method described in this review has benefits and limitations that should be taken into account when selecting the optimum method for a particular ST (Table 2). In the future, less expensive, safer, and more sensitive assays will be required for obtaining better insight into the activity and substrate-binding properties of STs. Defined substrates, carefully designed for individual STs, especially those that enable label-free detection, should also provide more insight. Further method development of ST assays is critical, because advances in assay development for a single ST can often be applied broadly to many STs for a variety of reactions and end applications. STs increasingly have a variety of important applications and improved assays are clearly required to determine their activity and take full advantage of their potential.
Table 2.
Comparison of ST assay methods
| Assay Method | Benefits | Limitations |
|---|---|---|
| Radiometric | Highly sensitive, fast, specific | Hazardous, expensive, discontinuous |
| Photometric | Fast, inexpensive, continuous, high-throughput capable, kinetic constants easily measurable, requires few sample-processing steps, accurate | Requires chromogenic substrate, relatively less sensitive |
| Fluorimetric | Highly sensitive, fast, continuous, inexpensive, high-throughput capable, kinetic constants easily measurable, specific, requires few sample-processing steps, accurate | Requires fluorogenic substrate |
| Mass spectrometric | Highly specific, highly sensitive, highly accurate | Requires expensive equipment, discontinuous, requires specialized skills |
Acknowledgments
The authors are grateful to the National Institutes of Health for supporting this research with grants GM38060, HL094463 and HL096972. Priscilla Paul is grateful for an NIH-funded predoctoral fellowship.
Abbreviations
- ADP
Adenosine diphosphate
- APS
Adenosine 5′-phosphosulfate
- AST-IV
Aryl sulfotransferase type IV
- ATP
Adenosine triphosphate
- CDSNS
Completely de-sulfonated N-sulfonated heparin
- DEAE
Diethylaminoethyl
- EDC
Endocrine-disrupting compound
- EMMA
Electrophoresis-mediated microanalysis
- ESI–MS
Electrospray ionization–mass spectrometry
- GlcNAc
N-Acetylglucosamine
- hDHEA-ST
Human dehydroepiandrosterone sulfotransferase
- HEC
High endothelial cell
- HPLC
High-performance liquid chromatography
- IP
Ion-pairing
- kcat
Turnover number
- Ki
Inhibition constant
- Km
Michaelis–Menten constant
- MU
Methylumbelliferone
- NDST-1
N-Deacetylase N-sulfotransferase type 1
- NodST
Bacterial carbohydrate sulfotransferase
- N-ST
N-Sulfotransferase
- NPAPS
3′-Phosphoadenosine 5′-phosphosulfate
- O-ST
O-Sulfotransferase
- PNP
p-Nitrophenyl
- PNPS
p-Nitrophenylsulfate
- PPi
Pyrophosphate
- RP
Reversed phase
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- ST
Sulfotransferase
- STF0
Mycobacterial carbohydrate sulfotransferase
- SULT-1A1
Phenyl sulfotransferase type 1A1
- SULT-1A3
Phenyl sulfotransferase type 1A3
- SULT1E1
Estrogen sulfotransferase type 1E1
- TLC
Thin-layer chromatography
- TM
Transmembrane
- Vmax
Maximum velocity
Contributor Information
Priscilla Paul, Department of Chemical and Biological Engineering, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA.
Jiraporn Suwan, Department of Chemistry and Chemical Biology, Rensselaer Polytechnic Institute, Troy, NY 12180, USA.
Jian Liu, Division of Medicinal Chemistry and National Products, College of Pharmacy, University of North Carolina, Chapel Hill, NC 27599, USA.
Jonathan S. Dordick, Department of Chemical and Biological Engineering, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA. Department of Biology, Rensselaer Polytechnic Institute, Troy, NY 12180, USA. Department of Biomedical Engineering, Rensselaer Polytechnic Institute, 110 8th Street, Biotechnology Center, 4005, Troy, NY 12180, USA
Robert J. Linhardt, Email: linhar@rpi.edu, Department of Chemical and Biological Engineering, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA. Department of Chemistry and Chemical Biology, Rensselaer Polytechnic Institute, Troy, NY 12180, USA. Department of Biology, Rensselaer Polytechnic Institute, Troy, NY 12180, USA. Department of Biomedical Engineering, Rensselaer Polytechnic Institute, 110 8th Street, Biotechnology Center, 4005, Troy, NY 12180, USA
References
- 1.Yoshinari K, Petrotchenko EV, Pedersen LC, Negishi M. Crystal structure-based studies of cytosolic sulfotransferase. J Biochem Mol Toxicol. 2001;15:67–75. doi: 10.1002/jbt.1. [DOI] [PubMed] [Google Scholar]
- 2.Falany C. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 3.Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC. Structure and function of sulfotransferases. Arch Biochem Biophys. 2001;390:149–157. doi: 10.1006/abbi.2001.2368. [DOI] [PubMed] [Google Scholar]
- 4.Jancova P, Anzenbacher P, Anzenbacherova E. Phase II drug metabolizing enzymes, Biomedical Papers of the Medical Faculty of the University Palacký, Olomouc. Czechoslovakia. 2010;154:103–116. doi: 10.5507/bp.2010.017. [DOI] [PubMed] [Google Scholar]
- 5.Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem. 2004;43:3526–3548. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- 6.Rath VL, Verdugo D, Hemmerich S. Sulfotransferase structural biology and inhibitor discovery. Drug Discov Today. 2004;9:1003–1011. doi: 10.1016/S1359-6446(04)03273-8. [DOI] [PubMed] [Google Scholar]
- 7.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 8.Hemmerich S, Verdugo D, Rath VL. Strategies for drug discovery by targeting sulfation pathways. Drug Discov Today. 2004;9:967–975. doi: 10.1016/S1359-6446(04)03261-1. [DOI] [PubMed] [Google Scholar]
- 9.Shuler ML, Kargi F. Bioprocess engineering: basic concepts. 2. Prentice Hall; 2001. [Google Scholar]
- 10.Ramaswamy SG, Jakoby WB. Sulfotransferase assays. Elsevier; 1987. [DOI] [PubMed] [Google Scholar]
- 11.Jansson L, Höök M, Wasteson A, Lindahl U. Biosynthesis of heparin. Solubilization and partial characterization of N- and O-sulphotransferases. Biochem J. 1975;149:49–55. doi: 10.1042/bj1490049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang H-G, Evers MR, Xia G, Baenziger JU, Schachner M. Molecular cloning and characterization of chondroitin-4-O-sulfo-transferase-3. A novel member of the HNK-1 family of sulfotransferases. J Biol Chem. 2002;277:34766–34772. doi: 10.1074/jbc.M204907200. [DOI] [PubMed] [Google Scholar]
- 13.Aikawa J, Grobe K, Tsujimoto M, Esko JD. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. Structure and activity of the fourth member, NDST4. J Biol Chem. 2001;276:5876–5882. doi: 10.1074/jbc.M009606200. [DOI] [PubMed] [Google Scholar]
- 14.Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J Biol Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- 15.Burkart MD, Izumi M, Chapman E, Lin C-H, Wong C-H. Regeneration of PAPS for the enzymatic synthesis of sulfated oligosaccharides. J Org Chem. 2000;65:5565–5574. doi: 10.1021/jo000266o. [DOI] [PubMed] [Google Scholar]
- 16.Ouyang Y-B, Lane W, Moore K. Tyrosylprotein sulfotransferase: purification and molecular cloning of an enzyme that catalyzes tyrosine O-sulfation, a common posttranslational modification of eukaryotic proteins. Proc Nat Acad Sci USA. 1998;95:2896–2901. doi: 10.1073/pnas.95.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verdugo D, Bertozzi C. A 96-well dot-blot assay for carbohydrate sulfotransferases. Anal Biochem. 2002;307:330–336. doi: 10.1016/s0003-2697(02)00060-x. [DOI] [PubMed] [Google Scholar]
- 18.Sim MK, Hsu TP. Sensitive assays for the determination of monoamine oxidase and phenol sulphotransferase activity in small tissue samples. J Pharmacol Meth. 1990;24:157–163. doi: 10.1016/0160-5402(90)90026-h. [DOI] [PubMed] [Google Scholar]
- 19.Klein M, Papenbrock J. Kinetics and substrate specificities of desulfoglucosinolate sulfotransferases in Arabidopsis thaliana. Physiol Plant. 2009;135:140–149. doi: 10.1111/j.1399-3054.2008.01182.x. [DOI] [PubMed] [Google Scholar]
- 20.Eisenthal D. Enzyme assays: a practical approach. Oxford University Press; New York: 2002. [Google Scholar]
- 21.Habuchi O, Matsui Y, Kotoya Y, Aoyama Y, Yasuda Y, Noda M. Purification of chondroitin 6-sulfotransferase secreted from cultured chick embryo chondrocytes. J Biol Chem. 1993;268:21968–21974. [PubMed] [Google Scholar]
- 22.Bethea HN, Xu D, Liu J, Pedersen LC. Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis. Proc Nat Acad Sci USA. 2008;105:18724–18729. doi: 10.1073/pnas.0806975105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armstrong A, Potley Chang, Nierengarten Cook, Bowman Bishop, Gray Shokat, Schultz Bertozzi. Discovery of Carbohydrate sulfotransferase Inhibitors from a Kinase-Directed Library. Angew Chem International Edition. 2000:39. doi: 10.1002/(sici)1521-3773(20000403)39:7<1303::aid-anie1303>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Ethen C, Larson S, Prather B. A versatile polyacrylamide gel electrophoresis based sulfotransferase assay. BMC Biotechnol. 2010;10:11. doi: 10.1186/1472-6750-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudlacek P, Anderson R, Liebentritt D, Johnson G, Huerter C. Human skin and platelet minoxidil sulfotransferase activities: biochemical properties, correlations and contribution of thermolabile phenol sulfotransferase. J Pharmacol Exp Ther. 1995;273:582–590. [PubMed] [Google Scholar]
- 26.Sundaram R, Szumlanski C, Otterness D, van Loon J, Weinshilboum R. Human intestinal phenol sulfotransferase: assay conditions, activity levels and partial purification of the thermolabile form. Drug Metab Dispos. 1989;17:255–264. [PubMed] [Google Scholar]
- 27.Burkart MD, Wong CH. A continuous assay for the spectrophotometric analysis of sulfotransferases using aryl sulfotransferase IV. Anal Biochem. 1999;274:131–137. doi: 10.1006/abio.1999.4264. [DOI] [PubMed] [Google Scholar]
- 28.Novakova H, Dyck V, Glatz VS. Study of enzyme kinetics of phenol sulfotransferase by electrophoretically mediated microanalysis. J Chromatogr. 2004;1032:319–326. doi: 10.1016/j.chroma.2003.11.082. [DOI] [PubMed] [Google Scholar]
- 29.Lin ES, Yang YS. Colorimetric determination of the purity of 3′-phospho adenosine 5′-phosphosulfate and natural abundance of 3′-phospho adenosine 5′-phosphate at picomole quantities. Anal Biochem. 1998;264:111–117. doi: 10.1006/abio.1998.2800. [DOI] [PubMed] [Google Scholar]
- 30.Frame LT, Ozawa S, Nowell SA, Chou HC, DeLongchamp RR, Doerge DR, Lang NP, Kadlubar FF. A simple colorimetric assay for phenotyping the major human thermostable phenol sulfotransferase (SULT1A1) using platelet cytosols. Drug Metab Dispos. 2000;28:1063–1068. [PubMed] [Google Scholar]
- 31.Ara T, Sekiya J. Non-radioactive adenosine 5′-phosphosulfate sulfotransferase assay by coupling with sulfite reductase and O-acetylserine(thiol)lyase. Biosci Biotechnol Biochem. 1997;61:621–624. doi: 10.1271/bbb.61.621. [DOI] [PubMed] [Google Scholar]
- 32.Chen W-T, Liu M-C, Yang Y-S. Fluorimetric assay for alcohol sulfotransferase. Anal Biochem. 2005;339:54–60. doi: 10.1016/j.ab.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Lu LY, Hsu YC, Yang YS. Spectrofluorimetric assay for monoamine-preferring phenol sulfotransferase (SULT1A3) Anal Biochem. 2010;404:241–243. doi: 10.1016/j.ab.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Arand M, Robertson LW, Oesch F. A fluorimetric assay for quantitating phenol sulfotransferase activities in homogenates of cells and tissues. Anal Biochem. 1987;163:546–551. doi: 10.1016/0003-2697(87)90261-2. [DOI] [PubMed] [Google Scholar]
- 35.Reinen J, Vriese E, Glatt H, Vermeulen NPE. Development and validation of a fluorescence HPLC-based screening assay for inhibition of human estrogen sulfotransferase. Anal Biochem. 2006;357:85–92. doi: 10.1016/j.ab.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Wilson JJ, Brodbelt JS. Ultraviolet photodissociation at 355 nm of fluorescently labeled oligosaccharides. Anal Chem. 2008;80:5186–5196. doi: 10.1021/ac800315k. [DOI] [PubMed] [Google Scholar]
- 37.Best MD, Brik A, Chapman E, Lee LV, Cheng W-C, Wong C-H. Rapid discovery of potent sulfotransferase inhibitors by diversity-oriented reaction in microplates followed by in situ screening. ChemBioChem. 2004;5:811–819. doi: 10.1002/cbic.200300841. [DOI] [PubMed] [Google Scholar]
- 38.Pai T, Ohkimoto K, Sakakibara Y, Suiko M, Sugahara T, Liu M. Manganese stimulation and stereospecificity of the Dopa (3,4-dihydroxyphenylalanine)/tyrosine-sulfating activity of human monoamine-form phenol sulfotransferase. Kinetic studies of the mechanism using wild-type and mutant enzymes. J Biol Chem. 2002;277:43813–43820. doi: 10.1074/jbc.M200785200. [DOI] [PubMed] [Google Scholar]
- 39.Pi N, Armstrong JI, Bertozzi CR, Leary JA. Kinetic analysis of NodST sulfotransferase using an electrospray ionization mass spectrometry assay. Biochemistry. 2002;41:13283–13288. doi: 10.1021/bi020457g. [DOI] [PubMed] [Google Scholar]
- 40.Pi N, Hoang MB, Gao H, Mougous JD, Bertozzi CR, Leary JA. Kinetic measurements and mechanism determination of Stf0 sulfotransferase using mass spectrometry. Anal Biochem. 2005;341:94–104. doi: 10.1016/j.ab.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Pi N, Leary JA. Determination of enzyme/substrate specificity constants using a multiple substrate ESI–MS assay. J Am Soc Mass Spectrom. 2004;15:233–243. doi: 10.1016/j.jasms.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Ge X, Sirich TL, Beyer MK, Desaire H, Leary JA. A strategy for the determination of enzyme kinetics using electrospray ionization with an ion trap mass spectrometer. Anal Chem. 2001;73:5078–5082. doi: 10.1021/ac0105890. [DOI] [PubMed] [Google Scholar]
- 43.Danan LM, Yu Z, Hoffhines AJ, Moore KL, Leary JA. Mass spectrometric kinetic analysis of human tyrosylprotein sulfotransferase-1 and -2. J Am Soc Mass Spectrom. 2008;19:1459–1466. doi: 10.1016/j.jasms.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babu P, Victor XV, Nelsen E, Nguyen TKN, Raman K, Kuberan B. Hydrogen/deuterium exchange-LC–MS approach to characterize the action of heparan sulfate C5-epimerase. Anal Bioanal Chem. 2011;401:237–244. doi: 10.1007/s00216-011-5087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hempel N, Negishi M, McManus ME. Human SULT1A genes: cloning and activity assays of the SULT1A promoters. Meth Enzymol. 2005;400:147–165. doi: 10.1016/S0076-6879(05)00009-1. [DOI] [PubMed] [Google Scholar]
- 46.Kanno N, Nagahisa E, Sato M, Sato Y. Nonradioactive assay for adenosine 5′-phosphosulfate sulfotransferase using reversed-phase ion-pair high-performance liquid chromatography. Biochem Mol Biol Int. 1993;29:47–55. [PubMed] [Google Scholar]
- 47.Greis KD. Mass spectrometry for enzyme assays and inhibitor screening: an emerging application in pharmaceutical research. Mass Spectrom Rev. 2007;26:324–339. doi: 10.1002/mas.20127. [DOI] [PubMed] [Google Scholar]
- 48.Hemmerich S. Carbohydrate sulfotransferases: novel therapeutic targets for inflammation, viral infection and cancer. Drug Discov Today. 2001;6:27–35. doi: 10.1016/s1359-6446(00)01581-6. [DOI] [PubMed] [Google Scholar]