Abstract
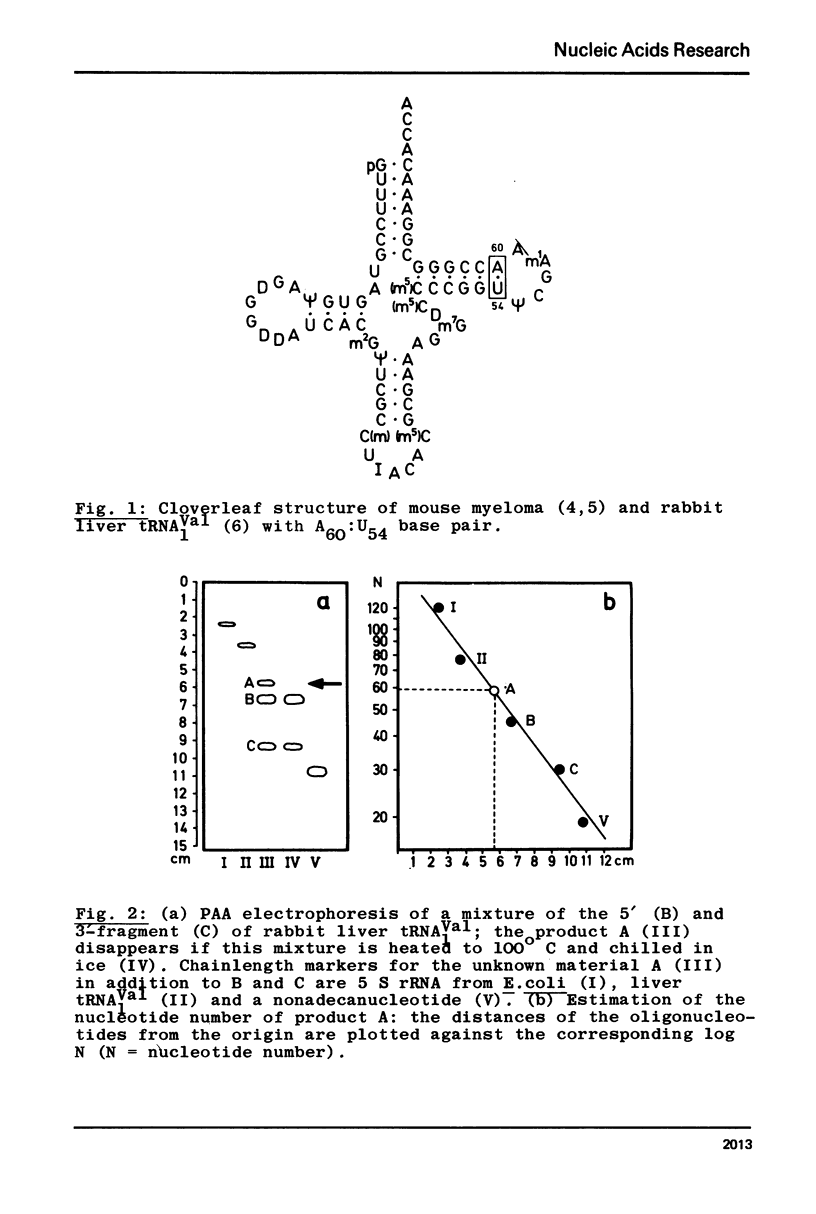
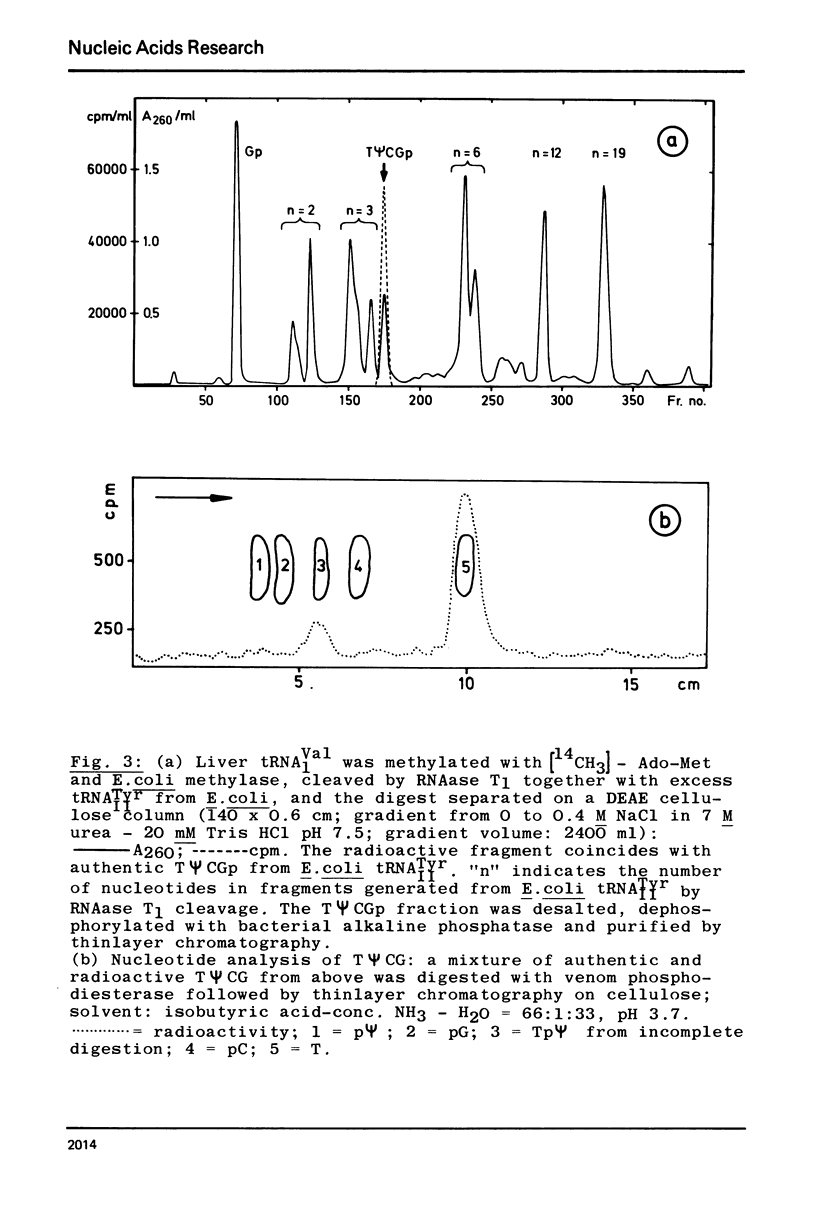
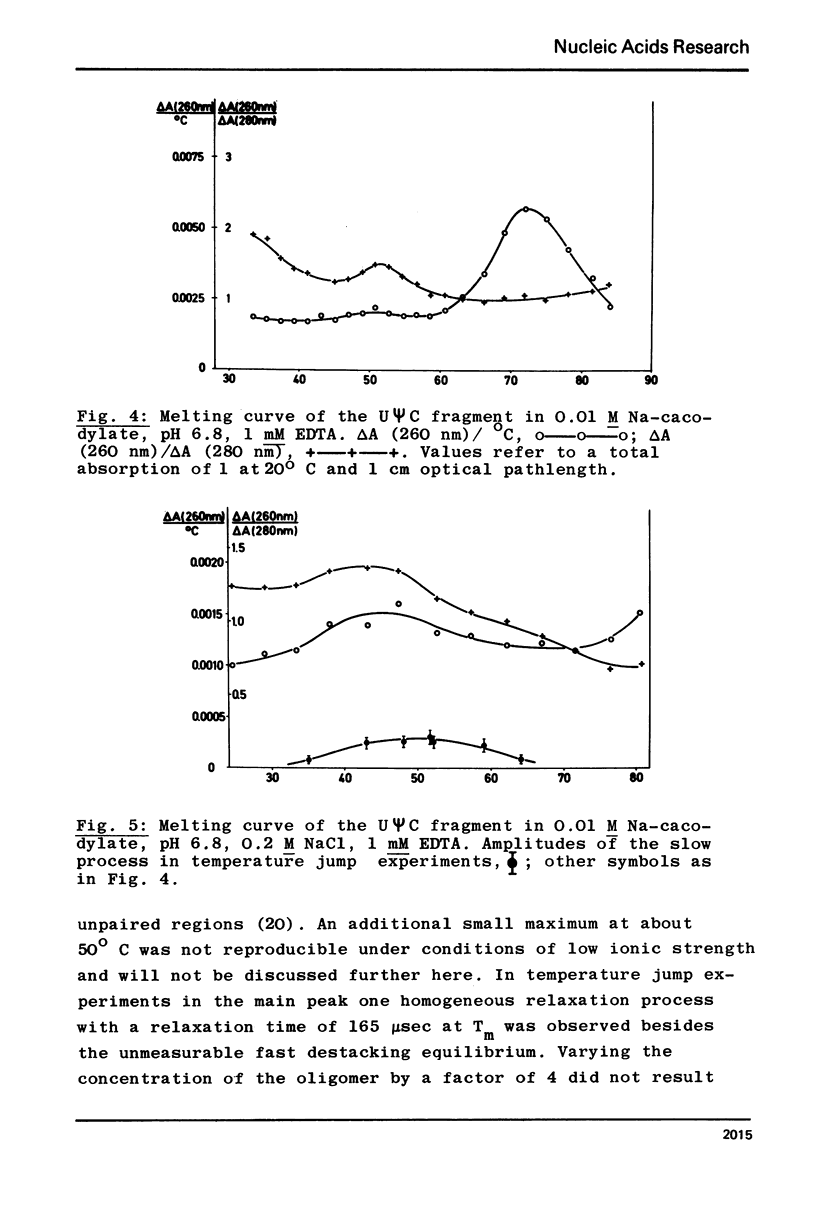
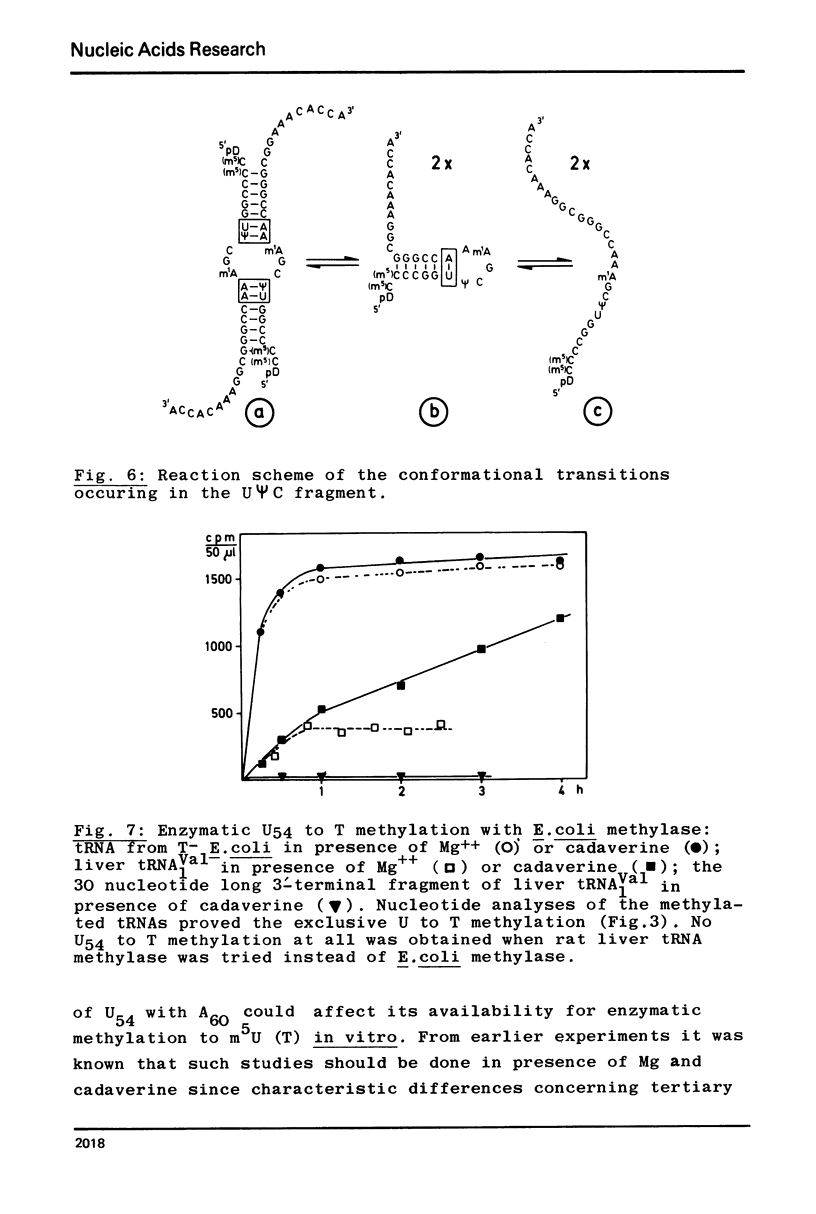
In contrast to all other known tRNAs, mammalian tRNAVal1 contains two adenosines A59 and A60, opposite to U54 and ψ55 in the UψCG sequence of the TψC loop, which could form unusual A:U (or A:ψ) pairs in addition to the five “normal” G:C pairs. In order to measure the number of G:C and A:U (A:ψ) pairs in the TψC stem, we prepared the 30 nucleotide long 3′-terminal fragment of this tRNA by “m7G-cleavage”. From differentiated melting curves and temperature jump experiments it was concluded that the TψC stem in this fragment is in fact extended by an additional A60:U54 pair. A dimer of this fragment with 14 base pairs was characterized by gel electrophoresis and by the same physical methods. An additional A:U pair in the tRNAVal1 fragment does not necessarily mean that this is also true for intact tRNA. However, we showed that U54 is far less available for enzymatic methylation in mammalian tRNAVal1 compared to tRNA from T−E. coli. This clear difference in U54 reactivity, together with the identification of an extra A60:U54 pair in the UψCG containing fragment suggests the presence of a 6 base pair TψC stem and a 5 nucleotide TψC loop in this tRNA.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coutts S. M., Gangloff J., Dirheimer G. Conformational transitions in tRNA Asp (brewer's yeast). Thermodynamic, kinetic, and enzymatic measurements on oligonucleotide fragments and the intact molecule. Biochemistry. 1974 Sep 10;13(19):3938–3948. doi: 10.1021/bi00716a019. [DOI] [PubMed] [Google Scholar]
- Coutts S. M., Riesner D., Römer R., Rabl C. R., Maass G. Kinetics of conformational changes in tRNA Phe (yeast) as studied by the fluorescence of the Y-base and of formycin substituted for the 3'-terminal adenine. Biophys Chem. 1975 Oct;3(4):275–289. doi: 10.1016/0301-4622(75)80020-2. [DOI] [PubMed] [Google Scholar]
- Gillum A. M., Urquhart N., Smith M., RajBhandary U. L. Nucleotide sequence of salmon testes and salmon liver cytoplasmic initiator tRNA. Cell. 1975 Nov;6(3):395–405. doi: 10.1016/0092-8674(75)90189-0. [DOI] [PubMed] [Google Scholar]
- Gross H. J., Simsek M., Raba M., Limburg K., Heckman J., Raj Bhandary U. L. 2'-O-methyl ribothymidine: a component of rabbit liver lysine transfer RNA. Nucleic Acids Res. 1974 Jan;1(1):35–43. doi: 10.1093/nar/1.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F., Sawyer R. C., Dahlberg J. E. A primer ribonucleic acid for initiation of in vitro Rous sarcarcoma virus deoxyribonucleic acid synthesis. J Biol Chem. 1975 May 10;250(9):3487–3497. [PubMed] [Google Scholar]
- Jank P., Shindo-Okada N., Nishimura S., Gross H. J. Rabbit liver tRNA1Val:I. Primary structure and unusual codon recognition. Nucleic Acids Res. 1977 Jun;4(6):1999–2008. doi: 10.1093/nar/4.6.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith G., Picaud F., Weissenbach J., Ebel J. P., Petrissant G., Dirheimer G. The primary structure of rabbit liver tRNA Phe and its comparison with known tRNA Phe sequences. FEBS Lett. 1973 May 1;31(3):345–347. doi: 10.1016/0014-5793(73)80138-3. [DOI] [PubMed] [Google Scholar]
- MANS R. J., NOVELLI G. D. A convenient, rapid and sensitive method for measuring the incorporation of radioactive amino acids into protein. Biochem Biophys Res Commun. 1960 Nov;3:540–543. doi: 10.1016/0006-291x(60)90171-6. [DOI] [PubMed] [Google Scholar]
- Marcu K., Mignery R., Reszelbach R., Roe B., Sirover M., Dudock B. The absence of ribothymidine in specific eukaryotic transfer RNAs. I. Glycine and threonine tRNAs of wheat embryo. Biochem Biophys Res Commun. 1973 Nov 16;55(2):477–483. doi: 10.1016/0006-291x(73)91111-x. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Zachau G. Partial degradation of transfer RNAs and transfer RNA fragments by spleen phosphodiesterase as studied by disc electrophoretic methods. Biochim Biophys Acta. 1972 Sep 14;277(3):523–538. doi: 10.1016/0005-2787(72)90095-0. [DOI] [PubMed] [Google Scholar]
- Piper P. W., Clark B. F. The nucleotide sequences of cytoplasmic methionine and valine tRNAs from mouse myeloma cells. FEBS Lett. 1974 Oct 1;47(1):56–59. doi: 10.1016/0014-5793(74)80425-4. [DOI] [PubMed] [Google Scholar]
- Piper P. W. The primary structure of the major cytoplasmic valine tRNA of mouse myeloma cells. Eur J Biochem. 1975 Feb 3;51(1):295–304. doi: 10.1111/j.1432-1033.1975.tb03929.x. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Staphylococcal transfer ribonucleic acids. II. Sequence analysis of isoaccepting glycine transfer ribonucleic acids IA and IB from Staphylococcus epidermidis Texas 26. J Biol Chem. 1974 Aug 10;249(15):4787–4796. [PubMed] [Google Scholar]
- Römer R., Riesner D., Coutts S. M., Maass G. The coupling of conformational transitions in alanine specific transfer ribonucleic acid from yeast studied by a modified differential absorption technique. Eur J Biochem. 1970 Jul;15(1):77–84. doi: 10.1111/j.1432-1033.1970.tb00978.x. [DOI] [PubMed] [Google Scholar]
- Sigler P. B. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng. 1975;4(00):477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- Simsek M., Petrissant G., Rajbhandary U. L. Replacement of the sequence G-T-phi-C-G(A)- by G-A-U-C-G- in initiator transfer RNA of rabbit-liver cytoplasm. Proc Natl Acad Sci U S A. 1973 Sep;70(9):2600–2604. doi: 10.1073/pnas.70.9.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Oshima T., Saneyoshi M., Nishimura S. Replacement of ribothymidine by 5-methyl-2-thiouridine in sequence GT psi C in tRNA of an extreme thermophile. FEBS Lett. 1974 Jul 1;43(1):59–63. doi: 10.1016/0014-5793(74)81105-1. [DOI] [PubMed] [Google Scholar]
- Wildenauer D., Gross H. J., Riesner D. Enzymatic methylations: III. Cadaverine-induced conformational changes of E. coli tRNA fMet as evidenced by the availability of a specific adenosine and a specific cytidine residue for methylation. Nucleic Acids Res. 1974 Sep;1(9):1165–1182. doi: 10.1093/nar/1.9.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W., Zachau H. G. A specific chemical chain scission of tRNA at 7-methylguanosine. FEBS Lett. 1970 Dec;11(3):160–164. doi: 10.1016/0014-5793(70)80518-x. [DOI] [PubMed] [Google Scholar]


