Abstract
The Major Facilitator Superfamily (MFS) is the largest known superfamily of secondary carriers found in the biosphere. It is ubiquitously distributed throughout virtually all currently recognized organismal phyla. This superfamily currently (2012) consists of 74 families, each of which is usually concerned with transport of a certain type of substrate. Many of these families, defined phylogenetically, do not include even a single member that is functionally characterized. In this article we probe the evolutionary origins of these transporters, providing evidence that they arose from a single 2 TMS hairpin structure that triplicated to give a 6 TMS unit that duplicated to a 12 TMS protein, the most frequent topological type of these permeases. We globally examine MFS protein topologies, focusing on exceptional proteins that deviate from the norm. Nine distantly related families appear to have members with 14 TMSs where the extra two are usually centrally localized between the two 6 TMS repeat units. They probably have arisen by intragenic duplication of an adjacent hairpin. This alternative topology probably arose multiple times during MFS evolution. Convincing evidence for MFS permeases with fewer than 12 TMSs was not forthcoming, leading to the suggestion that all 12 TMSs are required for optimal function. Some homologues appear to have 13, 14, 15 or 16 TMSs, and the probable locations of the extra TMSs were identified. A few MFS permeases are fused to other functional domains or are fully duplicated to give 24 TMS proteins with dual functions. Finally, the MFS families with no known function were subjected to genomic context analyses leading to functional predictions.
Keywords: Transport proteins, secondary active transport, MFS, multi-drug resistance, uptake/efflux, evolution, superfamily
Introduction
Before 1993, several families of secondary carriers of similar topology were recognized, but there was no evidence that these were related by common descent. Among these were a family of sugar porters including the mammalian glucose facilitators, two families of drug and multi-drug efflux pumps, a family of metabolic uptake porters, a family of organo-phosphate-ester:inorganic phosphate antiporters and a family of oligosaccharide porters that included the well studied lactose permease of E. coli. In that year, our bioinformatic efforts provided the first evidence that these families were related, and we coined the term “Major Facilitator Superfamily” (MFS) [1]. It included uniporters, symporters and antiporters.
Five years later, the MFS was expanded to include 12 more families [2] and following another year of intense research, it was expanded to a total of 34 families [3]. Today, we recognize 74 MFS families. Each well characterized family within this superfamily transports a different set of related compounds (e.g., simple monosaccharides, oligosaccharides, amino acids, peptides, vitamins, enzyme cofactors, drugs, chromophores, nucleobases, nucleosides, nucleotides, iron chelates, organic and inorganic anions and cations, etc. [4–7]. However, for 17 families, no functionally characterized member is known. These are called Unknown Major Facilitators (UMFs). No evidence as to the substrates transported or the directions of transport have been postulated.
The MFS and its many recognized families are tabulated and described within the IUBMB-approved Transporter Classification Database (TCDB; www.tcdb.org) [8, 9]. For details about the mechanisms of MFS transport and high resolution structures of MFS porters, see [7, 10–15].
Results
2 TMS Repeats in MFS transporters
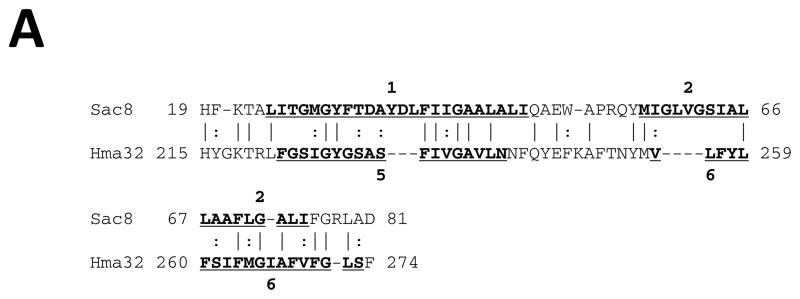
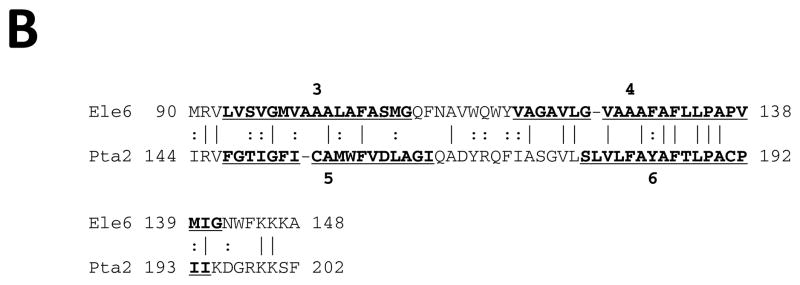
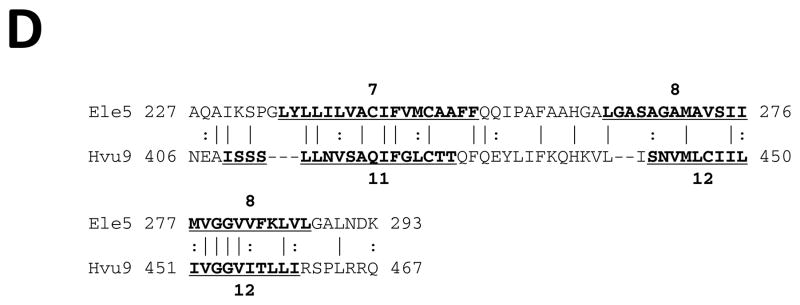
Early structural and sequence similarity studies revealed that MFS proteins exhibit two 6 TMS repeat units (see [1] and references cited therein). It has been suggested that a 3 TMS precursor gave rise to the 6 TMS repeat element [29, 30]. However, the evidence supporting this possibility was weak, and the symmetry of the hydrophobic regions observed in WHAT [23] and AveHAS [31] plots, which is often a good indication of repeat units, tentatively suggested a 2 TMS repeat unit in both of the two 6 TMS halves of these homologues. Novel programs, such as AncientRep (see Methods), and recent advances in the tools used to look for internal repeats, have significantly improved our ability to identify ancient intragenic duplications. Here we provide strong statistical evidence that a 2 TMS precursor gave rise to MFS proteins.
Horizontal and vertical comparisons of TMSs 1–2 with 3–4, 3–4 with 5–6, 7–8 with 11–12 and 9–10 with 11–12 yielded comparison scores of 8 to 10 S.D. (Figs. 1A–D). When looking for 3 TMS repeats, and TMSs 1, 2 and 3 were compared with TMSs 4, 5 and 6, 1,2 paired most frequently with 5,6 (data not shown). These results confirmed the 2 TMS prediction based on the comparison scores and TMS alignments presented in Figures 1A–D. These data suggest strongly that a 2 TMS triplication and not a 3 TMS duplication gave rise to all MFS superfamily members.
Figure 1.
Global Topological Analyses of MFS Porters
The Major Facilitator Superfamily (TC# 2.A.1)
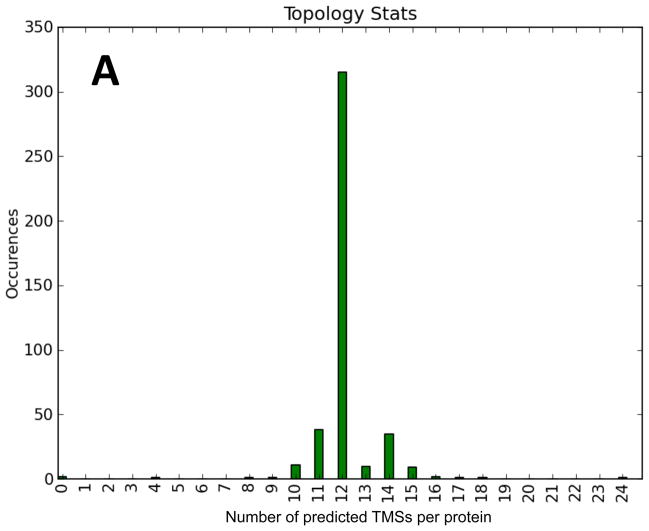
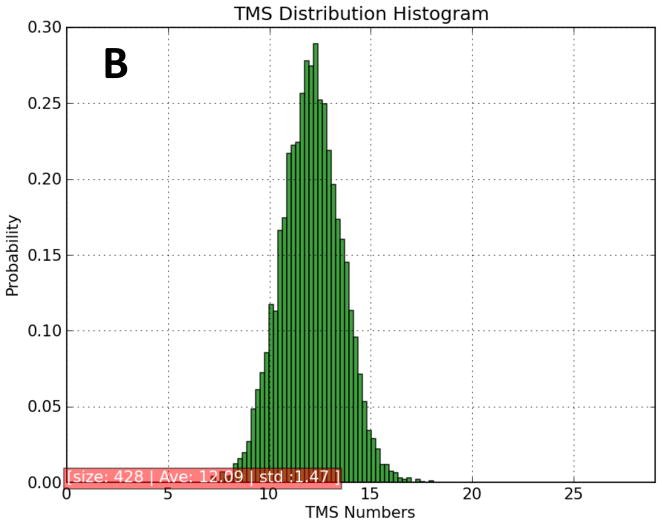
The predicted topologies of MFS porters in TCDB (TC# 2.A.1) were examined using the novel TMStats program (V.S. Reddy and M.H. Saier, accompanying manuscript). None of the 428 porters examined appeared to have fewer than 12 TMSs, and 75% were predicted to have 12. Many MFS porters have been shown experimentally to have 12 TMSs per polypeptide chain, but some (e.g., members of the drug:H+ antiporter-2 (DHA2) family (TC# 2.A.1.3) have been shown to have 14 TMSs with the extra two TMSs sandwiched in between the two 6 TMS repeat units [32–34]. It has been presumed that all members of the DHA2 family have a 14 TMS topology, while all members of the related DHA1 family (TC# 2.A.1.2) have 12 TMSs, but this has not been demonstrated.
We have conducted topological predictions for the proteins listed under TC# 2.A.1 (MFS) using the TMStats program. The distribution of predicted topological types is shown in Figure 2A, and the histogram is shown in Figure 2B. Proteins predicted to have 1 or 4 TMSs proved to be Membrane Fusion Protein (MFP; TC# 8.A.1) family members that serve as auxiliary constituents of some MFS efflux pumps [34, 35]. They were excluded from this study using the “No auxiliary proteins” option of TMStats.
Figure 2.
Eight proteins were predicted to have 10 TMSs, but on closer inspection, all could be interpreted as 11 or 12 TMS proteins except for one that proved to be N-terminally truncated due to incomplete sequencing and erroneous initiation codon selection. Thus, it seems likely that none of the MFS permeases examined has just 10 TMSs.
Thirty-five proteins were predicted to have 11 TMSs. One was from the sugar porter (SP) family (Family 1) and six were from the DHA1 family (Family 2). In each of these cases, the program (based on HMMTOP [25]), missed one small peak of hydrophobicity, explaining the prediction of 11 TMSs. Five Anion:Cation Symporter (ACS, TC# 2.A.1.14) family members and ten out of 20 organic cation transporters (OCT; TC# 2.A.1.19) were predicted by the program to have 11 TMSs. In all cases, the hydropathy plots could be interpreted in terms of 12 TMSs with one small peak of hydrophobicity having been missed by HMMTOP. Interestingly, in members of both of these families, putative TMSs 1 and 2 are separated by large hydrophilic loops, and hydrophobic peak 1 is most frequently missed, causing the program to predict 11 TMSs. Thus, we could not gain convincing evidence for an MFS porter of 10 or 11 TMSs.
Three proteins in the DHA2 family were predicted to have 12 TMSs (3.7, 3.21 and 3.28). However in 3.7 and 3.28, two additional C-terminal peaks of hydrophobicity, not counted by HMMTOP, were identified. 3.21 exhibits 12 putative TMSs with no additional C-terminal peaks, but alignment with its closest homologues revealed that TMSs 1 and 2 are lacking because the wrong initiation codon had been selected. Thus, all members of MFS family DHA2 appear to have 14 TMSs. In view of these findings, we suggest that all 12 TMSs of all examined MFS permeases are required for function.
Nine permeases were predicted to have 13 TMSs. Of these, three were from the DHA2 family 3, and two were from the ACS family 14 (14.2 and 14.12). In the two ACS family proteins, the topologies could be interpreted in terms of 12 TMSs, but an extra N-terminal TMS may exist in both proteins, accounting for the 13 TMS prediction. Of the three DHA2 family members predicted to have 13 TMSs, all may have 14 TMSs.
Thirty-one porters were predicted to have 14 TMSs. Twenty-five of these are from the DHA2 (2.A.1.3) family, showing that a majority of the members of this family are predicted to exhibit 14 TMSs. In addition, NrtP (TC# 2.A.1.8.3) of the nitrate/nitrite porter (NNP) family exhibits 14 putative TMSs. This protein aligns with the 12 TMS nitrate porter of the diatom, Cylindrotheca fusiformis (TC# 2.A.1.8.4) throughout its 12 TMS length, but NtrP has a central insertion with two additional TMSs not present in the latter protein. Moreover, all top hits obtained with NrtP as the query sequence in NCBI PSI-BLAST searches exhibited 14 putative TMSs, with the extra 2 centrally located. The same proved to be true for the sialic acid transporter, NanT of E. coli (TC# 2.A.1.12.1) with the extra 2 TMSs between the two 6 TMS repeat units. Although the lactate/pyruvate porter, Jen1 of S. cereviscae (TC# 2.A.1.12.2), has 12 TMSs, all top hits obtained with NCBI-BLAST and NanT as the query sequence had 14 TMSs. AmpG of E. coli (TC# 2.A.1.25.2), which transports peptidoglycan degradation products (muropeptides) and penicillin, exhibits 14 clear peaks of hydrophobicity. Its closest hit with TC-BLAST was TC# 2.A.1.25.3 which displays 12 TMSs. These two proteins align well, but AmpG has two extra TMSs in the C-terminal position. All top hits to AmpG obtained in an NCBI-BLAST search exhibited 14 peaks of hydrophobicity.
The lysophospholipid importer of E. coli (TC# 2.A.1.42.2) was predicted to have 14 TMSs, but 12 of these are in the transporter domain while the additional two putative TMSs are in the fused acyl ACP synthetase domain [36]. This MFS permease therefore has the usual 12 TMS topology. All four members of the vacuolar basic amino acid transporter (V-BAAT; TC# 2.A.1.48) family in TCDB exhibit an apparent 14 TMS topology, and their closest TC hits outside the V-BAAT family were the 14 TMS DHA2 family members. In fact, an NCBI search with any one of these proteins as the query yielded additional proteins predicted to have 14 TMSs. Family 48 includes members only with 14 TMSs, with extra TMSs 7 and 8 being sandwiched in between the two 6 TMS repeat units (Table 1).
Table 1.
MFS protein topologies with more than 12 putative TMSs.
| Family TC# | Family Abb’n | # TMSs | Topology |
|---|---|---|---|
| 2.A.1.3 | DHA2 | 14 or 16? | 6+2+6 or 6+2+6+2 |
| 2.A.1.8 | NNP | 12, 14 or 24 | 6+6 or 6+2+6; 24 (12+12) |
| 2.A.1.12 | SHS | 12, 14 or 15 | 6+6 or 6+2+6 or 6+2+6+1 |
| 2.A.1.14 | ACS | 12 or 13 | 6+6 or 1+6+6 |
| 2.A.1.16 | SIT | 14 or 15 | 6+2+6 or 6+2+6+1 |
| 2.A.1.25 | PAT | 12 or 14 | 6+6 or 6+6+2 |
| 2.A.1.48 | V-BAAT | 14 | 6+2+6 |
| 2.A.2 | GPH | 12 or 14 | 6+6 or 6+2+6 |
| 2.A.17 | POT | 12 or 14 | 6+6 or 6+2+6 |
| 2.A.71 | FBT | 12 or 14 | 6+6 or 6+2+6 |
| Fusion Proteins | |||
| 2.A.1.42.1 | LplT | 14 | 12 TMS porter fusion to a 2 TMS acyltransferase/acyl-ACP synthetase (12+2) |
| 2.A.1.43.1 | PMP | 18 | 12 TMS porter fusion to a 6 TMS YedZ domain (12+6) |
| 2.A.1.8.1 | NNP | 24 | Two 12 TMS transporters fused together (12+12) |
Arn3 (TC# 2.A.1.16.1), Enb1 (TC# 2.A.1.16.2), Taf1 (TC# 2.A.1.16.3) and Arn1 (TC# 2.A.1.16.4), all from S. cerevisiae within the siderophore-iron transporter (SIT) family, are predicted to have 15 TMSs according to the WHAT and HMMTOP programs. The extra 3 TMSs may be in the centers, between the two repeat units (TMSs 7 and 8) as well as at the C-termini (TMS 15). Finally, VBA2 (TC# 2.A.1.48.2) could have 14, or less likely, 15 TMSs. 16 TMSs were predicted for LfrA of Mycobacterium smegmatis (TC# 2.A.1.3.3) and QepA of E. coli (TC# 2.A.1.3.33). It is possible that these proteins have TMSs in a 6+2+6+2 arrangement, but two adjacent putative TMSs near the C-terminus of each protein are fairly hydrophilic and could be inter-TMS hydrophilic loops.
TC# 2.A.1.34.1 is a sensor kinase/MFS fusion protein from Anaeromyxobacter. It is predicted to have 17 TMSs. However, the N-terminal sensor kinase domain exhibits 5–6 putative TMSs while the transporter domain has 12. TC# 2.A.1.43.2 has 18 putative TMSs, but this protein is a fusion between an N-terminal 12 TMS MFS permease and a C-terminal 6 TMS YedZ domain [37]. Thus, both of these proteins exhibit the usual 12 TMS MFS topology. Finally, 2.A.1.8.11 is a 24 TMS fusion protein (NarK1-NarK2) with two full length MFS carriers, each of 12 TMSs [38]. NarK1 is a NO3−:H+ symporter while NarK2 is a NO3−/NO2− antiporter. These three fusion proteins therefore do not represent exceptions to the 12 TMS rule. The two transporter domains in this double sized porter exhibit negative cooperativity with respect to NO3− affinity [38].
Other MFS Families
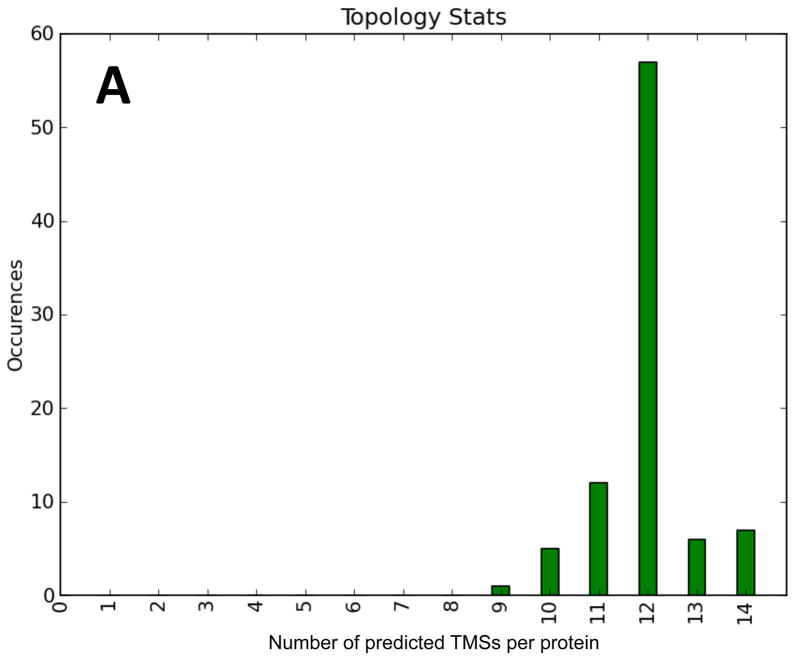
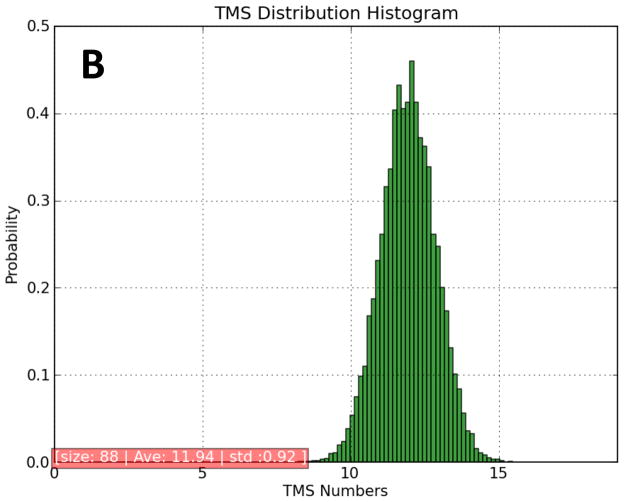
In addition to the MFS families listed under TC# 2.A.1, there are six families that have been shown to be distant members of the MFS. These include families with TC#s 2.A.2 (GPH), 2.A.12 (AAA), 2.A.17 (POT), 2.A.48 (RFC), 2.A.60 (OAT) and 2.A.71 (FBT). The 88 proteins included (those within these families represented in TCDB on 11/4/11) yielded the distribution shown in Figure 3A as well as the histogram shown in Figure 3B. Only one protein, the folate carrier of C. elegans (folt1; TC# 2.A.48.2.2), was predicted to have 9 TMSs in a 5 + 4 arrangement. Examination of the hydropathy plot revealed that peaks 1 and 7 were small. While peak 1 was not predicted to be a TMS, peak 7 was. Alignment of the protein sequence with all of its close orthologues showed that folt1 is shorter than the rest, containing an internal deletion corresponding to TMSs 9 and 10 in the 12 TMS homologues. Thus, assuming that a sequencing error was responsible for this unique deletion, the native folt1 protein probably has 12 TMSs. The possibility that this protein is the translated product of a pseudogene cannot be excluded.
Figure 3.
Most of the proteins predicted to possess 10 or 11 TMSs could be interpreted in terms of the expected 12 TMS topology. Thus, peaks of hydrophobicity, revealed by the WHAT program, were not predicted to be TMSs by the HMMTOP program. Because these proteins proved to be homologous throughout their lengths with close homologues that appear to possess 12 TMSs, we conclude that these proteins also have 12 TMSs.
Fifty seven of the 88 homologues were predicted to have 12 TMSs. Interestingly, every member of the AAA family (2.A.12) was predicted to have 12 TMSs. Six and seven homologues of these six families were predicted to have 13 and 14 TMSs, respectively. After examining these proteins in greater detail, we concluded that some of these 13 proteins have 12 TMSs, but most have 14. In every case where 14 TMSs were predicted, the extra two were present in the middle of these proteins, between the two 6 TMS repeat units. Among the proteins with 14 clear peaks of hydrophobicity (14 probable TMSs) were 2.A.2.6.2, 2.A.17.1.1-4 and 2.A.71.1.1-4. We conclude that in the GPH, POT and FBT families, both the 12 and 14 TMS homologues exist, and in general, these segregate according to TC subfamily. It should be noted that several MFS families tabulated under TC# 2.A.1 also have 14 TMSs, sometimes exclusively, but frequently among 12 TMS homologues. These results confirm the conclusion that the 14 TMS topology evolved from the 12 TMS topology many times in different families.
Variations in 14 TMS Topologies
Several MFS (2.A.1) homologues have a modified 12 TMS topology with 1–4 extra TMSs predicted. 2.A.1.14.2 has an extra predicted N-terminal TMS in an arrangement: 1+6+6. The same is true for 2.A.1.40.1-3. These three proteins probably have 13 TMSs where TMS 1 is distant from TMS 2 (the first TMS in the first 6 TMS half).
Few proteins were predicted to have 15 or 16 TMSs. All members of family 16 (SIT) were predicted to have 15 TMSs. However, the penultimate putative TMS was only weakly hydrophobic while being more strongly amphipathic. Direct experimental evidence will be required to determine the topologies of these C-terminal domains.
In summary, MFS permeases are remarkably constant in topology, having 12 TMSs with high frequency, 14 TMSs with lower frequencies, and 13, 15 or 16 TMSs with very low frequencies (Table 1). When 13 or 15 TMSs are predicted, the extra one is likely to be N-terminal or more frequently, C-terminal to the standard 12 or 14 TMS topological arrangements, respectively. Very few homologues may have 16 TMSs in a 6+2+6+2 arrangement (see Table 1).
Functional Predictions for MFS Families of Unknown Specificities
Small gene size, high gene density, intronless coding regions and simple operon organization in bacterial genomes render functional predictions feasible [39, 40]. Our previous molecular genetics studies based on operon context have proven to be successful in identifying the substrates of transporters of unknown function [36, 37, 41, 42]. Further, D.A. Rodionov and his coworkers have demonstrated the success of these predictions using the same databases and procedures (see [43–45] as well as earlier publications from this research group). Operon context analyses and identification of transcription factor-controlled regulons were facilitated by the use of the SEED database [26] along with RegPrecise and RegPredict [27, 28]. SEED identifies close homologues using the PSI-BLAST algorithm [46, 47].
Sixteen families listed under TC# 2.A.1 include no member with known substrate(s). These families are called Unknown Major Facilitator (UMF) families (UMF1-16). The former UMF1 family (2.A.1.24) now includes a member of known function and therefore was excluded from these studies. Below we use the SEED and RegPredict databases as well as relationships with other families in TCDB to predict functions.
UMF2 (2.A.1.26)
The UMF2 protein of E. coli, ycaD with 12 TMSs, is found adjacent to the ycaM gene, coding for a protein with 13 putative TMSs, a member of the APC superfamily. All functionally characterized members of the APC family (2.A.3) within the APC superfamily (Wong et al., manuscript in preparation) are known to take up amino acids, polyamines and organocations. Adjacent to and divergently transcribed in many organisms, we find an operon encoding pyruvate-formate lyase and its activator, as well as a probable formate efflux system. In other co-regulated operons, the enzyme, acetyl-CoA carboxylase, which catalyzes the formation of the precursor of fatty acid biosynthesis, malonyl-CoA, can be found. These results indicate that the transporter, YcaD, functions in a capacity related to the synthesis, utilization or degradation of acetyl-CoA. It could be a pyruvate uptake system, an exporter for a byproduct of acetyl-CoA synthesis or an exporter for an end product of metabolism of the substrate of the APC uptake porter.
A palindromic DNA sequence, CCATGAAATTTTTCGACTGAA, was identified in front of the ycaDM operon in various enterobacterial genomes. This sequence was identified in front of six such operons within the RegPredict training set. This fact reveals that the protein binding to this site is restricted in scope and probably regulates a specific branch of the metabolome. It was by this method that we were able to identify the acetyl-CoA carboxylase gene which apparently is co-regulated with the ycaDM operon.
UMF3 (2.A.1.29)
Only two UMF3 family members proved to be sufficiently similar in sequence to warrant being included within the same SEED subsystem. In the gene neighborhood of the Aeropyrum pernix homologue, a gene cluster encodes a complete ABC peptide uptake system (TC# 3.A.1.5) with all five expected constituents. The presence of a peptide uptake system as well as a glutamate decarboxylase within the same operon with each of the two archaeal UMF3 family members suggests that the MFS transporter could be an exporter for an amino acid or amino acid derivative such as the product of the α-decarboxylation reaction, γ-aminobutyric acid.
UMF4 (2.A.1.33)
YqgE of B. subtilis is encoded in an operon also encoding the essential cell division protein, FtsI, a peptidoglycan synthetase (EC# 2.4.1.129) which exhibits a penicillin binding protein (PBP3) transpeptidase domain. This enzyme is a member of the DD peptidase family which includes transpeptidases, carboxypeptidases and endopeptidases. The byproduct of the FtsI transpeptidase reaction (crosslinking extracytoplasmic peptidoglycan cell wall strands) is D-alanine. Possibly YqgE is a D-alanine uptake porter. In some organisms, YqgE homologues are encoded in operons with a Pst phosphate transporter of the ABC superfamily (TC# 3.A.1.7.1); other times, these genes are not co-expressed but are in the same gene neighborhood.
UMF5 (2.A.1.46)
Examining the gene neighborhoods of the two UMF5 representatives in TCDB from Bordetella pertussis and Tropheryma whipplei did not reveal extensive gene conservation and thus, no functional predictions were made.
UMF6 (2.A.1.47)
Streptococcus pneumoniae and S. thermophilus each encodes a UMF6 family member that is present in an operon with two lantibiotic peptide biosynthetic enzymes. It seems likely that these transporters export the bacteriocidal peptides produced by these biosynthetic enzymes. All such UMF6 homologues appear to derive from Firmicutes and Molecutes which are known to produce bacteriocins. However, several UMF6 homologues may export other substrates, such as other peptides with differing modes of action.
UMF7 (2.A.1.51)
UMF7 homologues are found in many bacteria. In the α-proteobacterial genuses, Brucella, Rhizobium and Agrobacterium, they colocalize with (1) a LysR transcriptional regulator, most of which function in the regulation of amino acid/nitrogen metabolism, (2) a BacA peptide/bleomycin uptake transporter of Rhizobium meliloti, (3) a putative polysaccharide deacetylase and (4) an EngA-type GTPase. The BacA protein (TC# 9.A.18.1.2) is homologous to the microcin peptide uptake permease, SbmA (TC# 9.A.18.1.1). On this basis we suggest that UMF7 may function in some aspect of amino acid metabolism, possibly catalyzing the efflux of peptide hydrolysis products: amino acids or amino acid derivatives.
UMF8 (2.A.1.52)
Eight UMF8 homologues were examined in enteric bacteria. In E. coli and its close relatives, UMF8 was in the same operon with a transcriptional regulator of the GntR family as well as a probable L-xylulose-5-phosphate-3-epimerase. In other bacteria such as Proteus mirabilis and Yersinia pestis the transporter again appeared to be in operons that also encoded either a hexulose-6-phosphate isomerase or an L-xylulose-5-phosphate-3-epimerase. An E. coli operon convergently transcribed from the one bearing the UMF8 homologue encodes two putative glucuronide transporters (TC# 2.A.2.3) and an α-glucosidase as well as a putative outer membrane sugar porin (TC# 1.B.21.2.1). All of these results indicate that UMF8 is a transporter for one or more sugars that feed into the pentose phosphate pathway. Candidates include gluconate, glucuronate, galactonate, galacturonate, and certain pentoses.
UMF9 (2.A.1.54)
The archaeal UMF9 protein in TCDB, present in Methanococcus maripaludis, is encoded in a gene cluster that also encodes several enzymes that probably function in methanogenesis. Immediately adjacent to UMF9 is a possible uroporphyrinogen decarboxylase, an enzyme involved in heme biosynthesis, but this enzyme is homologous to methyl-cobalamin:coenzyme M methyltransferase, an enzyme involved in methane production from methanol, dimethylamine and trimethylamine. Convergently transcribed from this two gene operon is a four gene operon that encodes a dimethylamine methyltransferase corrinoid protein, two proteins similar in sequence to methylcobalamin:coenzyme M methyltransferase and a large (593 aa) protein with a ferredoxin domain, probably involved in electron transfer. Thus, UMF9 could be an uptake system for substrates of methanogenesis such as di- and tri-methylamine as well as methanol.
UMF10 (2.A.1.59)
UMF10 proteins are found in both bacteria and archaea. The archaeal homologs occur in poorly conserved operons with few clues as to the potential transport substrates. The bacterial proteins often occur in different types of gene clusters with potential implications as to function. For example, in Nostoc punctiforme, Chlorobium tepidum, and Chlorobaculum parvum, the UMF10 homologues occur together with 8 TMS rhomboid intramembrane proteases that probably catalyze protein hydrolysis. They may also function in bacterial transmembrane protein translocation [48]. Sensor histidine kinase/response regulator pairs are also encoded. The sensor kinases are in the same family with QseC, a bacterial adrenergic receptor that activates virulence gene expression in response to interkingdom cross-signaling [49]. This sensor kinase/response regulator pair also regulates the master switch of the bacterial flagellum, flhDC [50].
UMF11 (2.A.1.62)
UMF11 proteins, sometimes annotated as macrolide efflux systems in the NCBI protein databank, are found in many Firmicutes of the genera Bacillus, Staphylococcus and Lactobacillus. They occur in gene clusters, often within operons, that include an azoreductase, responsible for the oxidation of diamines to azo compounds (R-N=N-R′), and homologs of the periplasmic YdcF S-adenosyl methionine binding protein that in some organisms plays a role in vancomycin resistance [51]. Since vancomycin contains multiple amino groups, it is possible that the UMF11 homolog and the azoreductase play a role in vancomycin resistance. In several Firmicutes, these genes may be under the control of a LysR transcriptional regulator, and in some of them, putative glycine/betaine ABC uptake systems, arginine/ornithine antiporters and aspartate ammonia-lyases can also be found. Thus, since these gene clusters are concerned with nitrogen metabolism, bacterial UMF11 homologs are likely to play a role in the transport of amino acids or their derivatives.
UMF12 (2.A.1.63)
In several archaea and bacteria (e.g., Methanosarcina mazei and Thermoanaerobacter pseudethanolicus) the UMF12 homologues are present within operons that also encode the five constituents of a complete ABC peptide uptake porter of the PepT family (TC# 3.A.1.5). TC-BLAST searches revealed that these proteins are probably peptide rather than oligosaccharide transporters, both of which can be found in the PepT family. Possibly these UMF12 transporters export amino acids and/or their derivatives.
In other archaea, including several Pyrococcus species, the UMF12 homologs are encoded within gene clusters that are involved in amino acid metabolism. Enzymes encoded include indolepyruvate oxidoreductase, concerned with aromatic amino acid interconversions, asparaginase (isoaspartyl aminopeptidase), which cleaves asparagine to aspartate and ammonia, and another MFS permease most similar to the efflux pumps of the Dha1 family (TC# 2.A.1.2). These observations suggest that like UMF11 proteins, UMF12 homologs are involved in the transport of amino acids. Finally, in Deinococcus species, UMF12 homologs are sandwiched in between endonucleases (Family III) and putative zinc-ribbon nucleic acid binding proteins. Possibly these Deinococcal transporters are involved in nucleotide or oligonucleotide transport.
UMF13 (2.A.1.64)
Members of the UMF13 family are found only in Streptococci. They are encoded within operons that also encodes radical SAM domain proteins, which create free radicals in reactions dependent on 4Fe-4S centers and S-adenosyl methionine. Arginase/agmatinase, which removes the guanadinium group from both of these compounds, is also encoded. Upstream of this operon and transcribed in the same direction are a MutR transcription factor and a lysine N-methylase. However, a space between the lysine N-methylase gene and the radical SAM domain gene is large enough (~200 bp) to indicate independent transcription. Similarly, downstream of the UMF13-encoding operon is a two cistron operon that encodes acetolactate synthase and alpha-acetolactate decarboxylase, involved in acetoin and butanediol production. The coexpression of UMF13 homologs with arginase and agmatinase suggests that UMF13 could be an uptake porter for arginine and/or agmatine.
UMF14 (2.A.1.65)
UMF14 proteins are found only in animals and were not investigated further.
UMF15 (2.A.1.66)
Archaeal UMF15 homologs appear to function in the uptake of α- and β-galactosides. In some organisms (Thermosipho, Pyrococcus and Thermococcus), the operons encoding UMF15 homologues also encode a putative α-galactosidase and galactokinase. The β-galactosidase gene is convergently transcribed, and divergently transcribed from this monocistronic operon, is a gene encoding galactose-1-phosphate uridylyl transferase. In other archaea (Staphylothermus and Halothermathrix), the transporter gene and the β-galactosidase gene are either cotranscribed or convergently transcribed. In one case (Thermofilum) these two genes are cotranscribed while the galactose metabolic genes occur in different operons in the same gene cluster. There is little doubt that these UMF15 proteins transport galactosides. In many organisms including E. coli, the galactose regulon includes several operons that are not co-localized on the bacterial chromosome.
Also within the UMF15 family (TC# 2.A.1.66.2) is a gene from Leptospira interrogans encoding a putative 4-hydroxybenzoate uptake porter, MFS_1. Its gene occurs in an operon that also encodes 2,3-diketo-5-methylthiopentyl-1-phosphate enolase-phosphatase of the methionine salvage pathway. This enzyme uses S-adenosyl methionine (SAM) as a substrate. It is possible that this transporter catalyzes the uptake of SAM. In Comamonas sp. strain E6, a homologue, Pmd, may be a protocatechuate (PCA) uptake porter [52]. The UMF15 family is also found in eukaryotes, e.g., in the Stromenophile, Thalassiosira pseudonana.
UMF16 (2.A.1.67)
UMF16 is found in a variety of actinobacteria including species of Streptomyces and Mycobacterium. In several of these organisms, the gene encoding the UMF16 homolog is present in what appears to be an operon where the downstream gene is an FAD-binding-4 superfamily oxidase. These enzymes include sugar-1,4-lactone oxidases, one of which is D-arabanose-1,4-lactone oxidase, the enzyme that catalyzes the final step in the biosynthesis of D-erythroascorbate, an antioxidant made by fungi and actinobacteria. Homologous to D-arabinose-1,4-lactone oxidase is L-gulonolactone oxidase which converts L-gulonolactone to L-xylo-hex-2-ulono-1,4-lactone, the precursor of ascorbate [53]. Among the actinobacteria that have this gene arrangement are Catenulispora acidiphila, Streptomyces griseus, S. coelicolor, Thermobispora bispora, Streptosporangium roseum and Thermomonospora curvata. These proteins probably function in a single pathway. Since the oxidase catalyzes the final step in L-ascorbate or D-erythroascorbate biosynthesis, we propose that UMF16 is an L-ascorbate and/or D-erythroascorbate exporter, although it may have an uptake function for one of the several potential precursors of erythroascorbate and/or ascorbate biosynthesis including D-arabinose, D-ribose, L-galactose, L-gulose or myo-inositol [54].
Homology Between MFS and APC Superfamily Members
Surprisingly, all three proteins under 2.A.1.40 showed excellent scores (as low as e−10) with members of the NCS2 (2.A.40) family. NCS2 proteins, like 2.A.1.40 proteins, exhibit 13 putative TMSs as the most frequent topology. 12 and 14 TMSs are predicted for some homologues. However, 2.A.40 family members brought up 2.A.53 (SulP) family member with scores as low as 10−10. A representative is provided in Figure 4. In both proteins (2.A.1.40.1; AzgA of Aspergillus nidulans versus 2.A.53.3.4, a SulP homologue of Chloroflexus auranticus), a comparison score of 14 S.D. was obtained using the GSAT program. Many other high scoring pairs were also observed. Both the NCS2 and SulP families are in the APC superfamily, strongly implying that the MFS and APC superfamilies are related. SulP family members, like NCS2 and some MFS permeases exhibited up to 14 peaks of hydrophobicity in an apparent 6 + 2 + 6 arrangement (see Table 1).
Figure 4.
Discussion
In this article, we describe the current status of the Major Facilitator Superfamily (MFS) and present new bioinformatic research results that demonstrate that the probable origin of the 6 TMS repeat unit in these proteins involved triplication of a 2 TMS hairpin structure. The standard and most prevalent topology for MFS porters is 12 TMSs, but we found that several families include members with 14 TMSs, an established topology for QacA of Staphylococcus aureus, a member of the DHA2 family (TC# 2.A.1.3) [35]. In most such cases, and probably in all DHA2 family members, the extra 2 TMSs separate the two 6 TMS repeat units. These 2 extra TMSs appear to be homologous to the primordial 2 TMS element that we believe was the precursor of all MFS permeases, and it may have arisen by triplication of the adjacent hairpin structure. We tentatively suggest that TMSs 5 and 6 duplicated to give 7 and 8, based on sequence similarities.
Because some MFS families have both 12 and 14 TMS members, and because the evolutionary distances between the different families are substantial, we propose that the 14 TMS topology has arisen from the 12 TMS precursors several times independently (see Table 1). In addition, genetically engineered loss of the central two TMSs in a 14 TMS drug efflux pump changed its specificity, but was still functional for cation (but not drug) transport [32, 55]. Since an equivalent internal partial duplication is rare in other transport protein families, specific genetic conditions or selective pressures must be responsible for this recurrent event. It will be interesting to know what these conditions and pressures were.
Topological analyses revealed the surprising topological inflexibility of the MFS. We could not confirm the presence of a single member with less than 12 TMSs, although the results do not exclude such a possibility. Nor do we have examples of “split” proteins, where two or more polypeptide chains comprise a single permease. This is surprising in view of the fact that one can experimentally split MFS permeases without loss of activity [56, 57]. Perhaps the intact permeases, included within a single polypeptide chain, facilitates both substrate recognition and the conformational changes that comprise the transport catalytic cycle [58, 59]. Split porters may have diminished activity and therefore would have been selected against during evolutionary time.
When topological variation within the MFS did occur, the extra TMSs, in addition to the two central TMSs in most 14 TMS proteins, were either N-terminal or C-terminal (Table 1). The latter option occurred more frequently, and the presence of a single N-terminal putative TMS was exceptionally rare. Never did we find more than one extra N-terminal TMS. However, extra C-terminal TMSs, although also rare, occurred with higher frequency, and either one or two putative TMSs were identified. A few cases of gene fusion were noted. In one case, a 6 TMS YedZ cytochrome b domain was fused C-terminal to an MFS carrier. This protein is present in a magnetotactic bacterium, where the transporter might be a magnetosome iron uptake porter. Then the YedZ domain could reduce part of the Fe3+ to Fe2+ in preparation for the production of magnetic magnetite crystals [37, 60, 61]. Another fusion involved a lysophospholipid uptake permease (12 TMSs) fused to an acyltransferase/acyl-ACP synthetase (2 TMSs) [36]. A well studied two permease fusion has been characterized where one porter catalyzes nitrate uptake while the other catalyzes nitrate/nitrite exchange. One can imagine that the former is most useful in initiating nitrate uptake while the latter is functional after intracellular nitrite builds up in the cytoplasm. The two permeases are known to exhibit negative cooperativity [38, 62]. This may favor one over the other, depending on substrate availability.
The functional predictions presented in this report provide the very first evidence for the substrate specificities of members of the “Unknown Major Facilitators” (UMFs) for which no member of the family has been characterized. These predictions could be made with various levels of confidence. For example, galactoside uptake permeases (UMF15) were predicted with high confidence. Others were predicted to take up acetyl-CoA (UMF2), D-alanine (UMF4), one or more sugar substrates of the pentose-phosphate pathway (PPP; i.e., sugar acids or pentoses) [63] (UMF8) and archaeal substrates for methanogenesis (di/trimethylamine and methanol; UMF9). Members of several UMF families may export amino acids or their metabolic products (UMF3, UMF7, UMF10, UMF11, UMF12 and UMF13) or bacteriocidal peptides (UMF6; see Table 2). One of these (UMF11) may also export drugs such as vancomycin, while another (UMF12) may also include members that transport nucleotides. Like the UMF8 family, members of the UMF16 family may take up a precursor of ascorbate or one of its related antioxidants or export end products of ascorbate/erythroascorbate biosynthesis (Table 2).
Table 2.
Summary of functional predictions for Uncharacterized Major Facilitator (UMF) families with unknown specificities.
| Family | TC# | Prediction | Organismal Type(s) |
|---|---|---|---|
| UMF-1 | 2.A.1.24 | Drug Resistance | B, E |
| UMF-2 | 2.A.1.26 | Precursor or metabolite of acetyl-CoA (pyruvate uptake?). | B |
| UMF-3 | 2.A.1.29 | Exporter of γ-aminobutyric acid (GABA) or another amino acid derivative. | B, A |
| UMF-4 | 2.A.1.33 | D-alanine uptake | B |
| UMF-5 | 2.A.1.46 | None. Possible drug resistance efflux pump (based on sequence similarity). |
B, A, E |
| UMF-6 | 2.A.1.47 | Lantibiotic export. | B |
| UMF-7 | 2.A.1.51 | Amino acid or amino acid derivative export. | B |
| UMF-8 | 2.A.1.52 | Uptake of sugars or sugar acids including galactosides. | B |
| UMF-9 | 2.A.1.54 | Uptake of methanol or methylamines in archaea. Most similar to amino acid uptake porters of the Pht family (2.A.1.53) in bacteria. |
B, A |
| UMF-10 | 2.A.1.59 | May transport amino acids and their derivatives. | B, A |
| UMF-11 | 2.A.1.62 | May transport amino acids and their derivatives. | B |
| UMF-12 | 2.A.1.63 | Most similar to members of the UMF-11, UMF-13 and DHA3 families. May export amino acids and their derivatives and/or drugs; a few may transport nucleotides. |
B, A, E |
| UMF-13 | 2.A.1.64 | May transport amino acids and their derivatives. | B (Streptococci) |
| UMF-14 | 2.A.1.65 | None. | B, E |
| UMF-15 | 2.A.1.66 | Galactoside uptake. | A |
| UMF-16 | 2.A.1.67 | L-ascorbate and/or D-erythroascorbate exporters or uptake porters for precursors of these compounds. | B |
Methods
Identification of Internal Repeats
The AncientRep (AR) program (V.S. Reddy and M.H. Saier, accompanying manuscript) was used to identify internal repeats. AR offers two techniques for finding repeat units: ‘horizontal and ‘vertical’. Horizontal searches look for internal repeats within a single protein, whereas vertical searches locate TMS repeat units across a list of homologues. All comparisons are performed using GSAT/GAP (V.S. Reddy and M.H. Saier, accompanying manuscript), and scores are given in standard deviations (S.D.). The best comparison scores were manually confirmed and analyzed further using the GSAT/GAP program [16]. The GSAT program was derived from the GAP algorithm (V.S. Reddy and M.H. Saier, accompanying manuscript). The GSAT/GAP program was set at default settings with a gap creation penalty of 8 and a gap extension penalty of 2 with 500 random shuffles. A length of 60 amino acyl residues, the average size of a typical protein domain, and 10 S.D., corresponding to a probability of 10−24 that the level of similarity arose by chance, is considered sufficient to establish homology, resulting from divergent evolution between two proteins or internal repeat units [9, 17–19]. Optimization of the GSAT/GAP alignment was performed on sequences by maximizing the number of identities, minimizing gaps, and removing non-aligned sequences at the ends. Optimization usually yields a higher comparison score that better represents the level of similarity between two shorter internal sequences. Horizontal comparisons are more convincing when several good scores are obtained (5 SDs and up, 60 or more residues in length and under 15% gaps.). Vertical searching is useful when horizontal scores are insufficient. Good vertical results will reveal repeat units that are poorly conserved within a single protein, but are more easily recognized in one or more of its homologues. The vertical approach is valid only if homology is established between the pair in question by virtue of the superfamily principle [20]. The reliability of these quantitative methods have been discussed and compared with other approaches by Matias et al., 2010 [21] and Wang et al., 2009 [22]. The basis for concluding that the observed degrees of sequence similarities necessary to give 10 SD for a stretch of >60 aas are due to divergent rather than convergent evolution has been considered [9, 19].
Topological Analyses
For topological analyses of single protein sequences, the WHAT and HMMTOP programs were used [23–25]. Statistical analyses, based on predictions made by the WHAT and HMMTOP programs, were performed using the TMStats program (V.S. Reddy and M.H. Saier, accompanying manuscript). The TMStats histogram represents the probability of finding a protein with a particular number of TMSs within the selected TC hierarchy. These probabilities are extrapolated by generating a microcosmic representation of the user’s actual selection, and consist of 100,000 computer-generated proteins. TMS predictions are performed using HMMTOP [24, 25]. Next to the histogram is a bar graph that illustrates the actual distribution of TMSs as found in the selected proteins derived from TCDB. A TMS average is displayed alongside these graphs, accompanied by a score in S.D. to represent the TMS distribution in the selected TC systems. The final items in the report are tabulated data containing a list of TCIDs sorted by their corresponding TMS count. Clicking on the corresponding member brings the user to the entry in TCDB for further analysis.
Functional Analyses
To propose possible related functions, genome context analyses were performed using The SEED-Viewer, which allows the exploration of over 1,500 curated genomes in order to find homologous genes, their operon context, and consequently their known or putative roles in other organisms [26]. This was done alongside RegPrecise and RegPredict, which allow for the identification of transcription factor-controlled regulons [27, 28].
Acknowledgments
The work reported in this paper was supporter by NIH Grant 2 RO1 GM077402-05A1. We thank Carl Welliver for his assistance in the preparation of this manuscript.
References
- 1.Marger MD, Saier MH., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 2.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 4.Chen DE, Podell S, Sauer JD, Swanson MS, Saier MH., Jr The phagosomal nutrient transporter (Pht) family. Microbiology. 2008;154:42–53. doi: 10.1099/mic.0.2007/010611-0. [DOI] [PubMed] [Google Scholar]
- 5.Lorca GL, Barabote RD, Zlotopolski V, Tran C, Winnen B, Hvorup RN, Stonestrom AJ, Nguyen E, Huang LW, Kim DS, et al. Transport capabilities of eleven gram-positive bacteria: comparative genomic analyses. Biochim Biophys Acta. 2007;1768:1342–1366. doi: 10.1016/j.bbamem.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yen MR, Chen JS, Marquez JL, Sun EI, Saier MH. Multidrug resistance: phylogenetic characterization of superfamilies of secondary carriers that include drug exporters. Methods Mol Biol. 2010;637:47–64. doi: 10.1007/978-1-60761-700-6_3. [DOI] [PubMed] [Google Scholar]
- 7.Saier MH, Jr, Paulsen IT. Phylogeny of multidrug transporters. Semin Cell Dev Biol. 2001;12:205–213. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 8.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemieux MJ. Eukaryotic major facilitator superfamily transporter modeling based on the prokaryotic GlpT crystal structure. Mol Membr Biol. 2007;24:333–341. doi: 10.1080/09687680701496507. [DOI] [PubMed] [Google Scholar]
- 11.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holyoake J, Caulfeild V, Baldwin SA, Sansom MS. Modeling, docking, and simulation of the major facilitator superfamily. Biophys J. 2006;91:L84–86. doi: 10.1529/biophysj.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radestock S, Forrest LR. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J Mol Biol. 2011;407:698–715. doi: 10.1016/j.jmb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Guan L, Freites JA, Kaback HR. Opening and closing of the periplasmic gate in lactose permease. Proc Natl Acad Sci U S A. 2008;105:3774–3778. doi: 10.1073/pnas.0800825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Nie Y, Kaback HR. Residues gating the periplasmic pathway of LacY. J Mol Biol. 2009;394:219–225. doi: 10.1016/j.jmb.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yen MR, Choi J, Saier MH., Jr Bioinformatic analyses of transmembrane transport: novel software for deducing protein phylogeny, topology, and evolution. J Mol Microbiol Biotechnol. 2009;17:163–176. doi: 10.1159/000239667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dayhoff MO, Barker WC, Hunt LT. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 19.Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolittle RF. Of urfs and orfs: a primer on how to analyze derived amino acid sequences. University Science Books; Mill Valley, CA: 1986. [Google Scholar]
- 21.Matias MG, Gomolplitinant KM, Tamang DG, Saier MH., Jr Animal Ca2+ release-activated Ca2+ (CRAC) channels appear to be homologous to and derived from the ubiquitous cation diffusion facilitators. BMC Res Notes. 2010;3:158. doi: 10.1186/1756–0500–3–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, Dukarevich M, Sun EI, Yen MR, Saier MH., Jr Membrane porters of ATP-binding cassette transport systems are polyphyletic. J Membr Biol. 2009;231:1–10. doi: 10.1007/s00232-009-9200-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol. 2001;3:501–502. [PubMed] [Google Scholar]
- 24.Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 25.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 26.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novichkov PS, Laikova ON, Novichkova ES, Gelfand MS, Arkin AP, Dubchak I, Rodionov DA. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 2010;38:D111–118. doi: 10.1093/nar/gkp894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, Arkin AP, Mironov AA, Dubchak I. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 2010;38:W299–307. doi: 10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirai T, Heymann JA, Maloney PC, Subramaniam S. Structural model for 12-helix transporters belonging to the major facilitator superfamily. J Bacteriol. 2003;185:1712–1718. doi: 10.1128/JB.185.5.1712-1718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hvorup RN, Saier MH., Jr Sequence similarity between the channel-forming domains of voltage-gated ion channel proteins and the C-terminal domains of secondary carriers of the major facilitator superfamily. Microbiology. 2002;148:3760–3762. doi: 10.1099/00221287-148-12-3760. [DOI] [PubMed] [Google Scholar]
- 31.Zhai Y, Saier MH., Jr A web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J Mol Microbiol Biotechnol. 2001;3:285–286. [PubMed] [Google Scholar]
- 32.Jin J, Guffanti AA, Beck C, Krulwich TA. Twelve-transmembrane-segment (TMS) version (DeltaTMS VII-VIII) of the 14-TMS Tet(L) antibiotic resistance protein retains monovalent cation transport modes but lacks tetracycline efflux capacity. J Bacteriol. 2001;183:2667–2671. doi: 10.1128/JB.183.8.2667-2671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin J, Guffanti AA, Bechhofer DH, Krulwich TA. Tet(L) and tet(K) tetracycline-divalent metal/H+ antiporters: characterization of multiple catalytic modes and a mutagenesis approach to differences in their efflux substrate and coupling ion preferences. J Bacteriol. 2002;184:4722–4732. doi: 10.1128/JB.184.17.4722-4732.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen IT, Brown MH, Littlejohn TG, Mitchell BA, Skurray RA. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci U S A. 1996;93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvat EM, Zhang YM, Tran CV, Zhang Z, Frank MW, Rock CO, Saier MH., Jr Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J Biol Chem. 2005;280:12028–12034. doi: 10.1074/jbc.M414368200. [DOI] [PubMed] [Google Scholar]
- 37.von Rozycki T, Schultzel MA, Saier MH., Jr Sequence analyses of cyanobacterial bicarbonate transporters and their homologues. J Mol Microbiol Biotechnol. 2004;7:102–108. doi: 10.1159/000078653. [DOI] [PubMed] [Google Scholar]
- 38.Goddard AD, Moir JW, Richardson DJ, Ferguson SJ. Interdependence of two NarK domains in a fused nitrate/nitrite transporter. Mol Microbiol. 2008;70:667–681. doi: 10.1111/j.1365-2958.2008.06436.x. [DOI] [PubMed] [Google Scholar]
- 39.Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 40.Shlykov MA, Zheng WH, Chen JS, Saier MH., Jr Bioinformatic characterization of the 4-Toluene Sulfonate Uptake Permease (TSUP) family of transmembrane proteins. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamem.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Feige JN, Chang AB, Anderson IJ, Brodianski VM, Vitreschak AG, Gelfand MS, Saier MH., Jr A transporter of Escherichia coli specific for L- and D-methionine is the prototype for a new family within the ABC superfamily. Arch Microbiol. 2003;180:88–100. doi: 10.1007/s00203-003-0561-4. [DOI] [PubMed] [Google Scholar]
- 42.Castillo R, Saier MH. Functional Promiscuity of Homologues of the Bacterial ArsA ATPases. Int J Microbiol. 2010;2010:187373. doi: 10.1155/2010/187373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eitinger T, Rodionov DA, Grote M, Schneider E. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol Rev. 2011;35:3–67. doi: 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 44.Jeanguenin L, Lara-Nunez A, Rodionov DA, Osterman AL, Komarova NY, Rentsch D, Gregory JF, 3rd, Hanson AD. Comparative genomics and functional analysis of the NiaP family uncover nicotinate transporters from bacteria, plants, and mammals. Funct Integr Genomics. 2011 doi: 10.1007/s10142-011-0255-y. [DOI] [PubMed] [Google Scholar]
- 45.Suvorova IA, Tutukina MN, Ravcheev DA, Rodionov DA, Ozoline ON, Gelfand MS. Comparative genomic analysis of the hexuronate metabolism genes and their regulation in gammaproteobacteria. J Bacteriol. 2011;193:3956–3963. doi: 10.1128/JB.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freeman M. Rhomboid proteases and their biological functions. Annu Rev Genet. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- 49.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clarke MB, Sperandio V. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2005;57:1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x. [DOI] [PubMed] [Google Scholar]
- 51.Chao KL, Lim K, Lehmann C, Doseeva V, Howard AJ, Schwarz FP, Herzberg O. The Escherichia coli YdcF binds S-adenosyl-L-methionine and adopts an alpha/beta-fold characteristic of nucleotide-utilizing enzymes. Proteins. 2008;72:506–509. doi: 10.1002/prot.22046. [DOI] [PubMed] [Google Scholar]
- 52.Kamimura N, Aoyama T, Yoshida R, Takahashi K, Kasai D, Abe T, Mase K, Katayama Y, Fukuda M, Masai E. Characterization of the protocatechuate 4,5-cleavage pathway operon in Comamonas sp. strain E6 and discovery of a novel pathway gene. Appl Environ Microbiol. 2010;76:8093–8101. doi: 10.1128/AEM.01863-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velisek J, Cejpek K. Biosynthesis of food constituents: Vitamins. 2. Water-soluble vitamins: Part 2 - a Review. Czech Journal of Food Science. 2007;25:101–118. [Google Scholar]
- 54.Wolucka BA. Biosynthesis of D-arabinose in mycobacteria - a novel bacterial pathway with implications for antimycobacterial therapy. Febs J. 2008;275:2691–2711. doi: 10.1111/j.1742-4658.2008.06395.x. [DOI] [PubMed] [Google Scholar]
- 55.Wang W, Guffanti AA, Wei Y, Ito M, Krulwich TA. Two types of Bacillus subtilis tetA(L) deletion strains reveal the physiological importance of TetA(L) in K(+) acquisition as well as in Na(+), alkali, and tetracycline resistance. J Bacteriol. 2000;182:2088–2095. doi: 10.1128/jb.182.8.2088-2095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Kaback HR. Proximity relationships between helices I and XI or XII in the lactose permease of Escherichia coli determined by site-directed thiol cross-linking. J Mol Biol. 1999;291:683–692. doi: 10.1006/jmbi.1999.2948. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W, Guan L, Kaback HR. Helices VII and X in the lactose permease of Escherichia coli: proximity and ligand-induced distance changes. J Mol Biol. 2002;315:53–62. doi: 10.1006/jmbi.2001.5206. [DOI] [PubMed] [Google Scholar]
- 58.Fluman N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim Biophys Acta. 2009;1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 59.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brokx SJ, Rothery RA, Zhang G, Ng DP, Weiner JH. Characterization of an Escherichia coli sulfite oxidase homologue reveals the role of a conserved active site cysteine in assembly and function. Biochemistry. 2005;44:10339–10348. doi: 10.1021/bi050621a. [DOI] [PubMed] [Google Scholar]
- 61.Jogler C, Schuler D. Genomics, genetics, and cell biology of magnetosome formation. Annu Rev Microbiol. 2009;63:501–521. doi: 10.1146/annurev.micro.62.081307.162908. [DOI] [PubMed] [Google Scholar]
- 62.Wood NJ, Alizadeh T, Richardson DJ, Ferguson SJ, Moir JW. Two domains of a dual-function NarK protein are required for nitrate uptake, the first step of denitrification in Paracoccus pantotrophus. Mol Microbiol. 2002;44:157–170. doi: 10.1046/j.1365-2958.2002.02859.x. [DOI] [PubMed] [Google Scholar]
- 63.Saier MH., Jr . Enzymes in metabolic pathways: a comparative study of mechanism, structure, evolution, and control. Harper & Row; New York: 1987. [Google Scholar]