Activity-based protein profiling (ABPP) is a chemical proteomic platform for characterizing enzyme activities in native biological systems.[1–3] Original small-molecule probes for ABPP were designed to target large numbers of enzymes that share mechanistic and/or structural features. These efforts have yieled activity-based probes for many enzyme classes, including serine[4–5] and cysteine[6] hydrolases,oxidoreductases,[7] metalloproteases,[8] histone deacetylases,[9] kinases,[10] and glycosidases.[11–12] ABPP has been applied to discover enzyme activities that are deregulated in biological processes such as cancer[13] and infectious disease[14]. Configuring ABPP to operate in a competitive mode has further enabled the development of selective inhibitors to probe the function of disease-relevant enzymes in cell and animal models.[5–6][15] As biological studies using ABPP have evolved, the need for target-selective activity-based probes has also become apparent. The specificity of such probes opens up new biological applications, including direct spatial and temporal visualization of active enzymes in cells and tissues.[16–17] While attractive in principle, the development of protein-selective, activity-based imaging probes poses substantial technical challenges. Such a probe should ideally possess several features, including high selectivity for a single enzyme target, a reporter tag for imaging the probe-labeled enzyme, and suitable cell permeability and pharmacokinetic properties for in vivo studies. These objectives have, so far, been realized for only a handful of probes that label proteolytic enzymes[18] and whether they can be achieved for probes that target additional types of enzymes remains unknown.
We recently used competitive ABPP to develop potent and selective covalent carbamate inhibitors for the integral membrane serine hydrolase KIAA1363[19] (also known as AADACL1 or NCEH1), which is highly expressed in aggressive human cancer cell lines and primary tumors.[19–22] In cancer cells, KIAA1363 regulates a set of pro-tumorigenic ether lipids and disruption of this metabolic pathway with the KIAA1363 inhibitor JW480 impairs cancer cell migration and tumor growth in vivo.[19] Here we have asked whether carbamate inhibitors could be converted into selective chemical probes for spatial and temporal imaging of KIAA1363 activity in cancer cells. There is a particular need for activity-based imaging probes for KIAA1363, since this enzyme is subject to extensive and variable post-translational modification (primarily glycosylation),[21] which has, so far, impeded efforts to develop antibodies for immunofluorescence imaging. Our established structure-activity relationship[19] suggested that the naphthyl-containing carbamoylating arm of JW480 could potentially be replaced with a hydrophobic fluorophore group without substantial losses in potency or selectivity for KIAA1363 (Figure 1a).
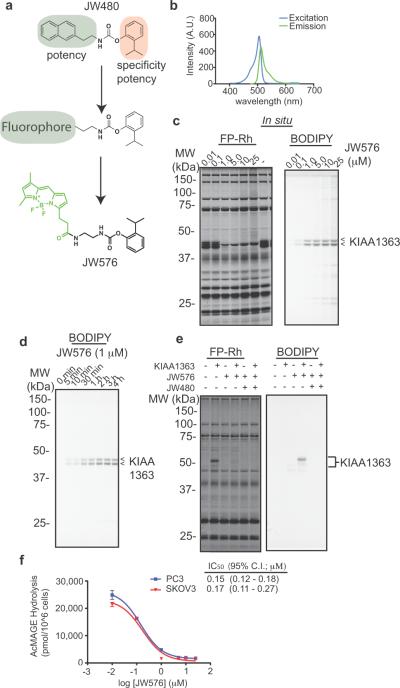
Figure 1.
Development of a fluorescent activity-based probe that selectively targets the serine hydrolase KIAA1363. (a) Design of JW576, an activity-based probe that selectively targets KIAA1363. (b) Excitation and emission spectra of JW576. (c) Competitive ABPP profiling of PC3 cells in situ. (d) Time-course of KIAA1363 labeling by JW576 (1 μM) in PC3 cells. (e) Labeling of recombinant KIAA1363 by JW576. Mock- or KIAA1363-transfected COS7 cells were treated with JW576 (0.1 μM) with or without JW480 competitor (1 μM) for 2 hr. (f) Inhibition of 2-AcMAGE hydrolysis activity of KIAA1363 in PC3 and SKOV3 cells treated in situ with JW576.
Of the many types of fluorophores that could be incorporated into a KIAA1363-selective probe, we chose the BODIPY class owing to its similar overall size and hydrophobicity compared to the naphthyl moiety in JW480 (Figure 1a). In the resulting molecule, JW576, the BODIPY fluorophore was appended to the parent carbamate structure through a propylamide linker (Figure 1a, Supp. Scheme. 1). Excitation and emission spectra of JW576 revealed maxima at 505 nm and 512 nm, respectively (Figure 1b). These results indicated that the embedded fluorophore retains the spectral properties of the parent BODIPY compound and should be suitable for fluorescence detection of KIAA1363 and any other potential JW576-reactive proteins.
We first evaluated whether JW576 specifically targeted KIAA1363 by performing competitive ABPP with the serine hydrolase-directed probe fluorophosphonate-rhodamine (FP-Rh).[23] PC3 cancer cells, which have high endogenous KIAA1363 levels,[19] were treated with JW576 in situ (0.01–25 μM for 4 hr), after which cells were homogenized, treated with FP-Rh (1 μM), and analyzed by gel-based ABPP. JW576 was found to selectively inhibit both ~40–45 kDa glycoforms of KIAA1363 with an IC50 value of 0.34 ± 0.15 μM (Figure 1c). Re-scanning of the gel on the BODIPY fluorescence channel confirmed that JW576 inhibition of FP-Rh labeling of KIAA1363 was paralleled by the appearance of a JW576-KIAA1363 covalent adduct (Figure 1c). Importantly, scanning for BODIPY fluorescence also permitted detection of any other JW576-protein adducts, which were only observed at trace levels and at high concentrations of JW576 (> 5 μM) above the IC50 value for KIAA1363 labeling (Figure 1c). Similar profiles were observed for JW576 in PC3 cell lysates and in other cancer cell lines (Supp Fig 1). KIAA1363 inhibition and labeling by JW576 (1 μM) were also found to be time-dependent, with near-complete target modification and negligible off-target interactions observed within an hour in living PC3 cells (Figure 1d). We also confirmed specific labeling in COS7 cells transfected with either a mock- or KIAA1363-expressing plasmid, where JW576-labeled KIAA1363 signals were abrogated by pre-treatment with JW480 (Figure 1e). Finally, we found that JW576 blocked KIAA1363 activity in cancer cells using a C16:0 2-acetyl MAGE substrate hydrolysis assay (Figure 1f and Supp Fig 2), which provided similar in situ IC50 values (0.15–0.17 μM) as competitive ABPP assays. Collectively, these data confirmed that JW576 serves as a potent and selective fluorescent inhibitor for KIAA1363 in cancer cells.
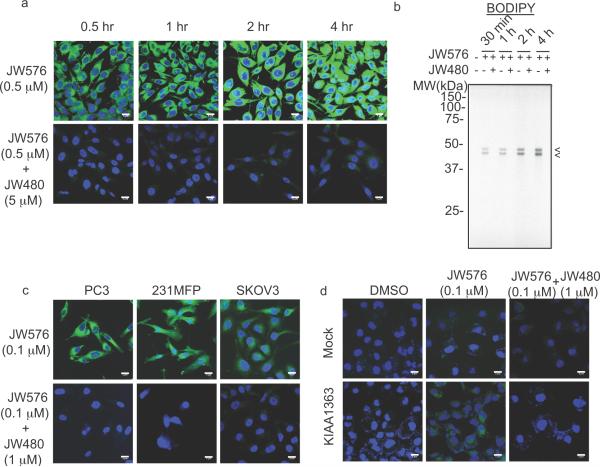
We next assessed whether JW576 could be used to image KIAA1363 activity in cancer cells by confocal fluorescence microscopy. PC3 cells were treated with JW576 (0.5 μM) for 0.5–4 hr and imaged on BODIPY and DAPI channels to detect retained JW576 signals and nuclear DNA, respectively. Significant, time-dependent increases in BODIPY fluorescence were observed throughout the cancer cell body (Figure 2a, top), and these signals were blocked by co-treatment with 10X JW480 inhibitor (Figure 2a, bottom). These results indicated that the observed BODIPY fluorescence signals reflect specific labeling of KIAA1363 by JW576, as opposed to non-specific retention of the compound. Also supporting this conclusion, gel-based ABPP showed specific, time-dependent and JW480-sensitive labeling of KIAA1363 by JW576 (Figure 2b). With the parameters for fluorescence imaging established, we performed confocal microscopy experiments in three aggressive human cancer cell lines from distinct tumors of origin - prostate (PC3), breast (231MFP) and ovarian (SKOV3) - all of which are known to express high levels of KIAA1363.[13,19] In each case, strong, JW480-sensitive intracellular staining was observed for JW576-treated cells (Figure 2c). Similarly strong intracellular staining was detected in COS7 cells transfected with a KIAA1363 cDNA, but not in mock-transfected COS7 cells (Figure 2d). These results establish the utility of JW576 as a fluorescent imaging probe for direct visualization of active KIAA1363 in cells and indicate that this enzyme is localized predominantly to intracellular membrane compartments in cancer cells.
Figure 2.
Imaging of KIAA1363 activity in cancer cells treated with JW576. (a) Confocal imaging of JW576-treated PC3 cells. (b) Gel analysis of PC3 cells treated as in (a) with JW576 in the presence or absence of 10× JW480. (c) Confocal imaging of prostate (PC3), breast (231MFP) and ovarian (SKOV3) cancer cells treated with JW576 ± 10× JW480 competitor for 2 hr. (d) JW576 treatment of mock- or KIAA1363-transfected COS7 cells in the presence or absence of 10× JW480 for 2 hr. Arrows in (b) indicate labeled-KIAA1363. Scale bars = 5 μm.
We sought to determine the subcellular membrane structure(s) that contain KIAA1363 activity by treating cancer cells with JW576 (0.1 μM, 2 hr) and then counterstaining for markers of various subcellular organelles, including the nucleus (DAPI), plasma membrane (wheat germ agglutinin (WGA)), and the endoplasmic reticulum (ER-tracker). Confocal imaging of JW576-treated cells on wavelengths specific to each marker revealed substantial co-localization of KIAA1363 activity with the ER, and very little signal overlap in the nucleus or at the cell surface (Figure 3a, b). Beyond these three measurable locations, puncate patterns of JW576 staining were also observed on intracellular membraneous structures non-overlapping with the ER (Figure 3b, arrows), indicative of some localization of KIAA1363 to other organelles (possibly endosomal compartments). In an effort to more completely describe the sub-cellular localization of KIAA1363 activity, we performed quantitative co-localization analysis. After gating each fluorescence channel to exclude autofluorescent background, we were able identify the locations in the cell where significant overlap between two fluorescence channels occurred (Figure 3c). Quantifying the percentage of each fluorescent signal that overlapped with another marker confirmed substantial (~95%) localization of the KIAA1363 signal to the ER. In contrast, KIAA1363 activity showed minimal overlap with WGA or DAPI signals (Figure 3d, upper panel), which also did not, as expected, overlap with each other (Figure 3d, lower panel). These experiments have thus identified the ER as the principle sub-cellular compartment where KIAA1363 resides in cancer cells. It would be interesting to determine, in future studies, if KIAA1363's distribution in cancer cells is dynamically regulated by signaling pathways or other cellular events, and JW576 should offer a suitable imaging probe for these types of studies.
Figure 3.
JW576 enables temporal tracking of active KIAA1363 in cancer cells. (a & b) Confluent SKOV3 cells were treated with JW576 (0.1 μM, 2 hr). Sub-cellular localization of active KIAA1363 populations was analyzed by staining either the plasma membrane with wheat-germ agglutinin (a) or the endoplasmic reticulum (ER) with ER-tracker Red (b) followed by confocal imaging. Images show each fluorescence channel separately and the overlay of all three. White arrows in the zoomed inlay of (b) highlight punctate JW576-labeled regions not stained by ER-Tracker. (c) Quantitative colocalization analysis of JW576 with the ER (top) and plasma membrane (bottom). Areas of significant signal overlap are false-colored white. (d) Quantification of JW576 (top) and DAPI (bottom) colocalization with the indicated sub-cellular markers. Scale bars = 20 μm.
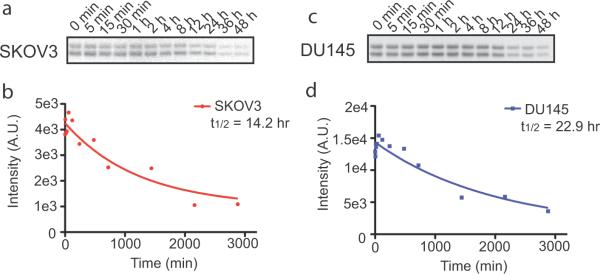
In addition to spatial mapping of active KIAA1363 in cancer cells, we wondered whether JW576 could be used for temporal tracking of the cellular turnover of KIAA1363. For this study, we followed the general protocols for traditional pulse-chase experiments, but used JW576 treatment in place of metabolic incorporation of radiolabeled amino acids. Two different human cancer cell lines (SKOV3 and DU145) were pulse-treated with JW576 (5 μM) for 10 minutes, washed with fresh media, and then incubated with media containing an excess of JW480 to quench any unreacted KIAA1363. Cell proteomes were then harvested at the indicated times over 48 hours and KIAA1363 activity levels measured by gel-based ABPP (Figure 4a, c). Time-dependent reductions in KIAA1363 signals were observed in both cell lines and fitting the quantified band intensities to an exponential decay model provided half-life estimates of 14.2 and 22.9 hours for KIAA1363 in SKOV3 and DU145 cells, respectively (Figure 4b, d).
Figure 4.
Temporal tracking of KIAA1363 turnover with JW576. (a & b) KIAA1363 protein half-life determination in SKOV3 cancer cells was determined by pulsed treatment with JW576 (5 μM, 10 min) Quantification of gel-resolved, labeled KIAA1363 (b) was performed with ImageJ software and fluorescence intensity values were fit to single-phase exponential decay models (a). (c & d) KIAA1363 protein half-life determination in DU145 cells as above in (a & b). Exponential decay curves were generated in Prism 5 software. Data shown are representative of thee separate experiments.
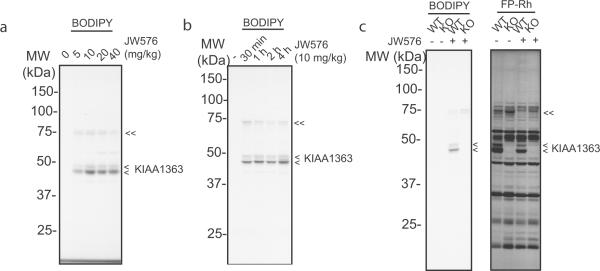
An ideal target-selective, activity-based probe would not only allow for visualization of enzyme activity in cells, but also in animal models. We tested whether JW576 could label KIAA1363 in C57BI/6J mice by treating animals with escalating doses of the compound (5–40 mg/kg, i.p.) for 4 hr. Mice were then sacrificed and proteomes from heart, a tissue that expresses high levels of KIAA1363,[24] were analyzed by gel-based ABPP. Scanning on the BODIPY channel revealed significant labeling of KIAA1363 at all tested doses (Figure 5a). The rate of in vivo labeling of KIAA1363 by JW576 was fast, with near-maximal signals being observed as early as 30 min post-dosing (Figure 5b). JW576 maintained good selectivity for KIAA1363 in vivo, with only a single, faint additional probe target (70 kDa protein) being detected in the heart proteome (Figures 4a, b). We confirmed that the 40–45 kDa JW576-labeled protein doublet corresponded to KIAA1363 by repeating in vivo treatments in wild-type (WT) and mKIAA1363 knockout (KO) mice (Figure 5c). We also analyzed these KIAA1363-WT and KO heart proteomes by competitive ABPP with FP-Rh, which revealed substantial reductions in KIAA1363 activity in WT mice treated with JW576 and complete absence of KIAA1363 signals in KO mice (Figure 5c). These competitive ABPP gels also provided evidence that the 70 kDa protein labeled by JW576 was itself a serine hydrolase (Figure 5c), likely corresponding to the blood carboxylesterase ES1, which is a known off-target of JW480,[19] the parent carbamate inhibitor from which JW576 was derived. Overall, these studies confirm that JW576 can be used to rapidly label KIAA1363 activity with good selectivity in vivo.
Figure 5.
JW576 labels KIAA1363 in vivo. (a) C57BI/6J mice were treated with the indicated doses of JW576 via intraperitoneal injection for 4 hrs, after which animals were sacrificed and heart tissue removed and analyzed for JW576-labeled proteins by in gel-fluorescence scanning. (b) Time-course analysis of heart tissues from C57BI/6J mice treated with JW576 (10 mg/kg) and processed as in (a). (c) Competitive ABPP of heart proteomes from KIAA1363-wild type (WT) and knockout (KO) mice treated with JW576 (10 mg/kg, 1 hr). Scanning on BODIPY (left) and FP-Rh (right) channels confirmed selective labeling of KIAA1363 by JW576 in vivo. Single and double arrows in (a–c) mark JW576-labeled KIAA1363 and an off-target serine hydrolase (likely ES1), respectively.
In summary, we report herein a versatile fluorescent probe for visualizing KIAA1363 activity in cancer cells and in vivo. This agent JW576 is potent (sub-μM IC50 values in cells) and selective for KIAA1363 and was used: 1) in imaging experiments to determine that KIAA1363 is largely, but not exclusively localized to the ER in cancer cells, and 2) in pulse-chase experiments to estimate a half-life of 14–23 hrs for KIAA1363 in cancer cells. That JW576 is also capable of labeling KIAA1363 activity in vivo sets the stage for future studies where this enzyme's cellular and subcellular localization can be imaged in normal tissues and tumors (e.g., in mouse xenograft models). For such studies, the specificity of KIAA1363 signals can be confirmed by pre-treatment with JW480 alongside other carbamate inhibitors that do not target KIAA1363 (but still inhibit ES1, a common off-target for carbamates[24]). Considering that KIAA1363 activity is highly elevated in aggressive human cancer cells and primary tumors,[19–22] we anticipate that JW576 should serve as a valuable functional probe to answer questions about KIAA1363 expression and localization during tumorigenesis, tumor progression, and metastasis. More generally, our studies have extended the use of target-selective fluorescent activity-based probes beyond cysteine proteases[16–17] to include an enzyme from the serine hydrolase class. Considering the recent emergence of selective, covalent inhibitors for many other serine hydrolases,[15][24] some of which are also carbamates,[24] we suspect that the strategy described herein should prove useful for developing activity-based imaging probes for several enzymes from this class.
Supplementary Material
Footnotes
We would like to thank A. Adibekian and the Cravatt lab for insightful discussions and W. Kiosses and the TSRI Microscopy Core for technical assistance. This research was supported by the California Institute for Regenerative Medicine (predoctoral fellowship to J.W.C.), the American Association for Cancer Research (Centennial predoctoral fellowship to R.E.M.), the Damon Runyon Cancer Research Foundation (HHMI postdoctoral fellowship to R.E.M.), the US National Institutes of Health (CA087660) and the Skaggs Institute for Chemical Biology.
References
- [1].Jessani N, Cravatt BF. Curr Opin Chem Biol. 2004;8:54–59. doi: 10.1016/j.cbpa.2003.11.004. [DOI] [PubMed] [Google Scholar]
- [2].Cravatt BF, Wright AT, Kozarich JW. Annu Rev Biochem. 2008;77:383–413. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- [3].Berger AB, Vitorino PM, Bogyo M. Am J Pharmacogenomics. 2004;4:371–381. doi: 10.2165/00129785-200404060-00004. [DOI] [PubMed] [Google Scholar]
- [4].Liu, Patricelli MP, Cravatt BF. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kidd D, Liu Y, Cravatt BF. Biochemistry. 2001;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- [6].Greenbaum D, Baruch DA, Hayrapetian L, Darula Z, Burlingame A, Medzihradszky KF, Bogyo M. Mol Cell Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]
- [7].Adam GC, Sorensen EJ, Cravatt BF. Nat. Biotechnol. 2002;20:805–809. doi: 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]
- [8].Sieber SA, Niessen S, Hoover HS, Cravatt BF. Nat. Chem. Biol. 2006;2:274–281. doi: 10.1038/nchembio781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Salisbury CM, Cravatt BF. Proc Natl Acad Sci U S A. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Biochemisty. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- [11].Hekmat O, Kim YW, Williams SJ, He S, Withers SG. J. Biol. Chem. 2005;280:35126–35135. doi: 10.1074/jbc.M508434200. [DOI] [PubMed] [Google Scholar]
- [12].Vocadlo DJ, Bertozzi CR. Angew Chem Int Ed Engl. 2004;43:5338–5342. doi: 10.1002/anie.200454235. [DOI] [PubMed] [Google Scholar]
- [13].Nomura DK, Long JZ, Niessen S, Hoover HS, Ng S, Cravatt BF. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Greenbaum DC, Baruch A, Grainger M, Bozdech Z, Medzihradszky KF, Engel J, DeRisi J, Holder AA, Bogyo M. Science. 2002;298:2002–2006. doi: 10.1126/science.1077426. [DOI] [PubMed] [Google Scholar]
- [15].Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Nat Chem Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Nat Med. 2009;15:967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lee J, Bogyo M. ACS Chem Biol. 2010;5:233–243. doi: 10.1021/cb900232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fonovic M, Bogyo M. Exper Rev Proteomics. 2008;5:721–730. doi: 10.1586/14789450.5.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chang JW, Nomura DK, Cravatt BF. Chem Biol. 2011;18:476–484. doi: 10.1016/j.chembiol.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chiang KP, Niessen S, Saghatelian A, Cravatt BF. Chem Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- [21].Jessani N, Liu Y, Humphrey M, Cravatt BF. Proc Natl Acad Sci U S A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, Cravatt BF. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- [23].Patrcelli MP, Giang DK, Stamp LM, Burbaum JJ. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- [24].Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Proc Natl Acad Sci U S A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.