Abstract
The objective of this study is to identify and characterize the genetic variants related to the glomerular filtration rate (GFR) linkage on 2q37. Of the positional candidate genes, we selected IRS1 and resequenced its 2-kb promoter region and exons for sequence variants in 32 subjects. A total of 11 single nucleotide polymorphisms (SNPs) were identified. To comprehensively cover the 59-kb-long intron-1, eight additional tagging SNPs were selected from the HapMap. All the 19 SNPs were genotyped by TaqMan Assay in the entire data set (N = 670; 39 families). Association analyses between the SNPs and GFR and type 2 diabetes–related traits were performed using the measured genotype approach. Of the SNPs examined for association, only the Gly(972)Arg variant of IRS1 exhibited a significant association with GFR (P = 0.0006) and serum triglycerides levels (P = 0.003), after accounting for trait-specific covariate effects. Carriers of Arg972 had significantly decreased GFR values. Gly(972)Arg contributed to 26% of the linkage signal on 2q. Expression of IRS1 mutant Arg972 in human mesangial cells significantly reduced the insulin-stimulated phosphorylation of IRS1 and Akt kinase. Taken together, the data provide the first evidence that genetic variation in IRS1 may influence variation in GFR probably through impaired insulin receptor signaling.
Diabetic nephropathy (DN) is a serious microvascular complication of both type 1 and type 2 diabetes (T2DM). Elevated urinary albumin excretion and decreased glomerular filtration rate (GFR) are risk factors for cardiovascular morbidity and mortality and progression to end-stage renal failure in individuals with diabetes. End-stage renal disease secondary to DN accounts for ∼50% of cases in most developed countries. The pathogenesis of DN is multifactorial. Familial clustering of DN and related traits such as albuminuria and GFR indicate that genes play a major role in its susceptibility (1). Despite the recent efforts to identify and characterize DN susceptibility genes, the genes involved in susceptibility to the development and or progression of DN and in particular GFR have yet to be identified.
Equations that estimate GFR facilitate the diagnosis, evaluation, and management of chronic kidney disease. GFR varies between individuals and is influenced by genetic and environmental factors and their interactions. Both genome-wide linkage and association analytical approaches have been used to localize the susceptibility genes/variants that influence variation in GFR (2–11). However, the functional significance of the finding from such studies is yet to be explored. To localize genes contributing to variation in GFR, we recently performed a genome-wide linkage screen using GFR data (N = 453) from the Mexican American participants of the San Antonio Family Diabetes/Gallbladder Study (SAFDGS) and identified the strongest linkage of GFR to occur on chromosome 2q35–37 near the markers D2S1363-D2S427 (LOD = 3.8; LODc = 3.3) after incorporating genotype by environment (T2DM) interaction effects (5). Identification of linkage of GFR on 2q is detailed in the Supplementary Data. We hypothesize that there is a gene(s) located on chromosome 2q35–37 that may be related to the linkage signal identified.
Of the positional candidate genes in the critical interval, IRS1 gene is located 200 kb telomeric to the marker D2S1363. When phosphorylated, IRS1 acts as a docking protein for multiple Src homology-2 (SH2)–containing proteins such as phosphatidylinositol-3-kinase (PI3K), growth factor receptor–binding protein-2, and tyrosine phosphatase SHP2. These proteins in turn further activate downstream effectors that mediate the biological effects of insulin in several tissues including kidney (12,13). Mice deficient in IRS1 by targeted disruption of its gene display insulin resistance and impaired glucose tolerance (14), and mice heterozygous for defects in genes for both IRS1 and the insulin receptor (15) or IRS1 and glucokinase (16) develop overt diabetes. Genetic variation in IRS1, particularly Gly(972)Arg (rs1801278) variant, plays a contributory role in insulin resistance and diabetes susceptibility (17) probably through impaired insulin receptor signaling (18–20). Genetic variants (rs2943641; rs2943650) located 500 kb upstream of IRS1 were recently reported to be associated with insulin resistance, hyperinsulinemia, dyslipidemia, and T2DM (21,22).
Given the functional significance of IRS1 together with our finding that it is a positional candidate gene in the GFR linkage region on chromosome 2q, we determined whether genetic variation in the IRS1 gene contributes to variation in GFR in the Mexican American participants of SAFDGS.
RESEARCH DESIGN AND METHODS
The SAFDGS family member recruitment and data collection procedures were reported previously (23). In brief, probands were Mexican Americans with T2DM with low income, and all first-, second-, and third-degree relatives of probands were invited to participate in the study. A variety of metabolic, hemodynamic, anthropometric, and demographic variables were collected from about 700 individuals drawn from 39 large Mexican American families. Diabetes status was defined by the 1999 criteria of the World Health Organization (i.e., fasting glucose levels ≥126 mg/dL and/or 2-h glucose levels ≥200 mg/dL).
Of the individuals examined at SAFDGS, GFR data were available for 453 individuals distributed across 29 families. GFR values were estimated as described previously (5) by the two recently recalculated GFR prediction equations (24): the recalculated simplified four-variable Modification of Diet in Renal Disease (GFR-MDRD) study equation (25) and the Cockcroft-Gault (GFR-CG) equation (26) adjusted for body surface area and corrected for the bias in the MDRD data. The quantitative trait values of triglycerides (TGL) and albumin-to-creatinine ratio were log transformed and used in the association analyses since their raw data were nonnormally distributed. The Institutional Review Board of the University of Texas Health Science Center at San Antonio approved all procedures, and all subjects gave informed consent.
Molecular variants identification and genotyping.
Exons and 2-kb putative promoter region of IRS1 gene were screened for DNA sequence variants in 32 individuals, who contributed positively to the linkage, by direct sequencing of PCR products amplified from genomic DNA isolated from peripheral blood leukocytes. Oligonucleotide primers used for PCR and sequencing are available upon request. Sequencing was performed using ABI Prism Big Dye Terminator Cycle Sequencing kit (Applied Biosystems [ABI]) and a capillary sequencer (Model 3730xl; ABI). Genotyping of all the SNPs was performed using the TaqMan assay (ABI), which was carried out on a GeneAmp PCR system 9700 (ABI), and fluorescent signals were detected on an ABI PRISM 7700 sequence detector (ABI). To assure accuracy of the genotyping, coded blind replicate samples from 50 subjects were included in each assay.
Statistical genetic analysis.
The genotypic data were checked for Mendelian consistency using the program SimWalk2 (27). All polymorphisms were tested for Hardy-Weinberg Equilibrium. Allele frequencies were estimated using maximum likelihood techniques, which account for pedigree structure. Linkage disequilibrium (LD) between SNPs was estimated using the r2 values. We performed association analysis in our complex pedigree-based data using the measured genotype approach (MGA) within the variance components (VC) analytical framework (28,29). The VC-based approach accounts for the nonindependence among family members. In this analytical technique, VCs are modeled as random effects (e.g., additive genetic effects and random environmental effects), whereas the effects of measured covariates such as age and sex are modeled as fixed effects on the trait mean. The marker genotypes were incorporated in the mean effects model as a measured covariate, assuming additivity of allelic effects (28–30). The effect of this measured genotype (i.e., association parameter) together with other covariate effects and VCs were estimated by maximum likelihood techniques. For association analysis between genotypes and GFR, age and sex terms in addition to other covariates were used in consideration of both biological and/or statistical significance. For GFR-CG analysis, age, age2, sex, diabetes, duration of diabetes, systolic blood pressure (SBP), and antihypertensive medications were used as covariates. For GFR-MDRD, age, sex, BMI, diabetes, duration of diabetes, SBP, and antihypertensive medications were used as covariates.
The hypothesis of no association is tested by comparing the likelihood of a model in which the effect of the measured genotype is estimated with a model where the effect of the measured genotype was fixed at zero. Twice the difference in the log-likelihoods of these models yields a test statistic that is asymptotically distributed, approximating a χ2 distribution with one degree of freedom. Before MGA was performed, the quantitative transmission disequilibrium test (31) was used to examine hidden population stratification. Given the LD patterns among the examined 13 SNPs, we obtained a P value after adjusting for multiple testing using an effective number of SNPs calculated following the method of Li and Ji (32). For a statistical threshold of ≤0.05, the required experiment-wide significance threshold, using the effective number of SNPs (which was 7.66) to adjust for multiple testing was 0.00667. Because the issue of correction for multiple testing is not straight forward with regard to multiple correlated traits, there were no additional attempts to correct for multiple testing because the corresponding models are not independent. All statistical techniques described above were implemented in the computer program SOLAR.
The Bayesian quantitative trait nucleotide (BQTN) analytical technique was used to analyze the IRS1 SNP and GFR data further, with the use of the program SOLAR. This technique has been detailed elsewhere (33). Given complete SNP data for a gene, this statistical technique can be used to identify the sequence variants that are either potentially functional or that exhibit the highest disequilibria with such potential functional sites. The BQTN model is a simple extension of the classical variance component model, which aims to disentangle the genetic architecture of a quantitative trait. Because a candidate gene may contain a number of SNPs that could generate several possible competing models of quantitative trait nucleotide (QTN) action, we used a Bayesian model selection/model averaging approach to analyze the SNP data simultaneously to estimate the probability that each SNP has a direct effect on the phenotype (33,34).
To determine whether the associated SNP(s) found by the QTN analysis can account for our reported linkage of GFR at chromosome 2q35–37, we combined the QTN analysis with our identity-by-descent–based variance component linkage analysis. If a variant, or set of variants, in the IRS1 gene contributes to the observed linkage signal, linkage analysis conditional on a fixed effect, MGA of the polymorphism will yield a LOD score near zero. Alternatively, if the associated polymorphism is in less than complete LD with the true functional site, linkage analysis will yield a reduced, but nonzero LOD score. This method has been described in detail elsewhere (29).
Western blot analysis.
The plasmids carrying IRS1 gene with glycine at codon 972 (wild type) and arginine at codon 972 (mutant) were provided by Dr. Sesti (35). Plasmids were transfected into human kidney mesangial cells using the FuGene HD transfection reagent as recommended (Roche Diagnostics). Cells (90–95% confluence) were washed twice with PBS, and media were replaced with 0.45 mL of OPTI-MEM I (Invitrogen). Precomplex of the plasmid DNA (0.5 μg/well) with FuGene (0.5 μL/well) in Opti-MEM was mixed (50 μL/well) and incubated at room temperature for 5 min. DNA and FuGene complexes were added to each well and incubated at 37°C with 5% CO2. Cells were then incubated for 6 h followed by addition of 0.5 mL of fresh media with 34% serum to a final concentration of 17%. After 48 h of transfection, cells were made quiescent by serum deprivation for 24 h, followed by treatment with insulin (100 μM) for 0, 5, and 10 min. Cells were harvested and lysed with radioimmunoprecipitation assay buffer. Equal amounts of total cellular proteins were fractioned by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with the following antibodies: anti-IRS1 (Upstate), antiphospho-IRS1 (Tyr 941) (Santa Cruz), anti-Akt, antiphospho-Akt (Thr 308) (Cell Signaling), and antiactin (Sigma). Bound antibodies were visualized using SuperSignal West Pico kit (Thermo Scientific). Densitometric analysis was performed using the National Institutes of Health ImageJ program.
RESULTS
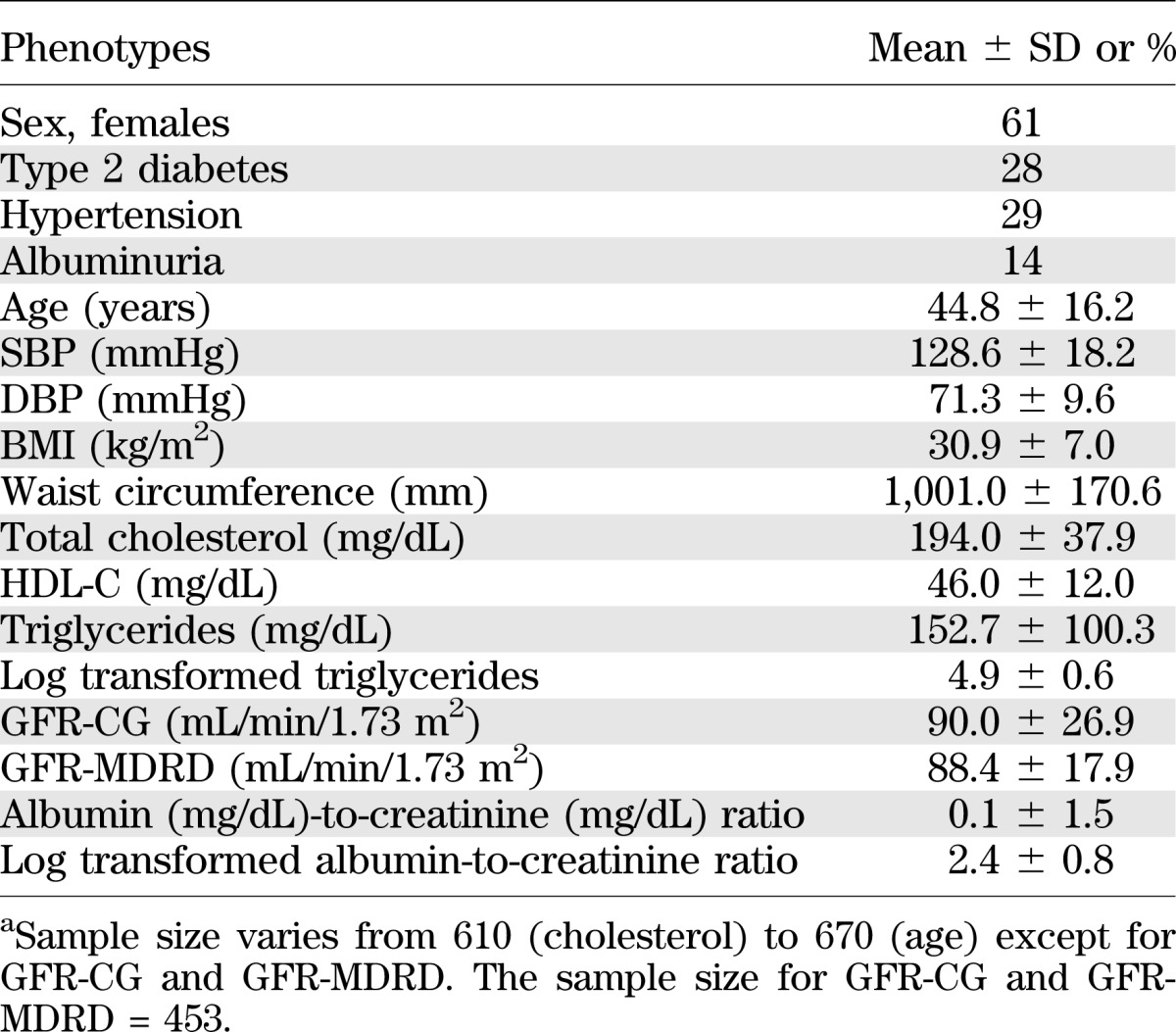
The clinical characteristics of the study participants are shown in Table 1. Of the genotyped individuals, the available phenotypic data for each phenotype varied from 610 subjects for total cholesterol to 670 subjects for age. In the total sample, 29, 28, and 14% of the subjects had hypertension, T2DM, and albuminuria, respectively (Table 1). However, the GFR data were available for only 453 subjects. Of the individuals with GFR data, 29% had T2DM.
TABLE 1.
Clinical characteristics of the genotyped SAFDGS participants used for the current studya
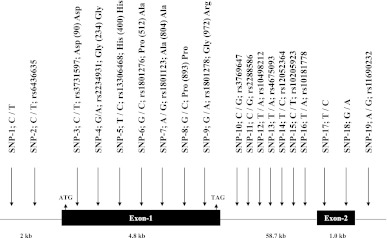
IRS1 (NM_005544) is composed of two exons, and the full-length protein is encoded by exon-1 (Fig. 1). To identify DNA polymorphisms in IRS1, we sequenced both exons and 2-kb putative promoter region of IRS1 in 32 subjects who contributed positively to the linkage of GFR. In total, we identified 11 SNPs including seven in the coding region, and two each in the 3′ UTR and putative promoter regions (Fig. 1). Of the seven SNPs identified in the coding region, five are synonymous (Asp90Asp, Gly234Gly, His400His, Ala804Ala, Pro893Pro) and two are nonsynonymous SNPs (Pro512Ala, Gly(972)Arg). In addition, we chose eight tagging SNPs (SNPs-10–16, 19) from HapMap database for the analysis to comprehensively cover ∼59-kb-long region of intron 1 of IRS1 (Fig. 1). All the 19 SNPs (11 identified by resequencing and 8 from the public database) shown in Fig. 1 (SNP-1 to SNP-19) were genotyped in the entire data set (N = 670; 39 families) by TaqMan assay.
FIG. 1.
Schematic diagram of human IRS1 (NM_005544) gene structure on 2q36 and the location of the SNPs identified and genotyped in SAFDGS. The exons of the IRS1 are represented by solid box and the intron by a thin line. SNPs with the base change and their reference sequence (rs) numbers are indicated by the vertical arrow.
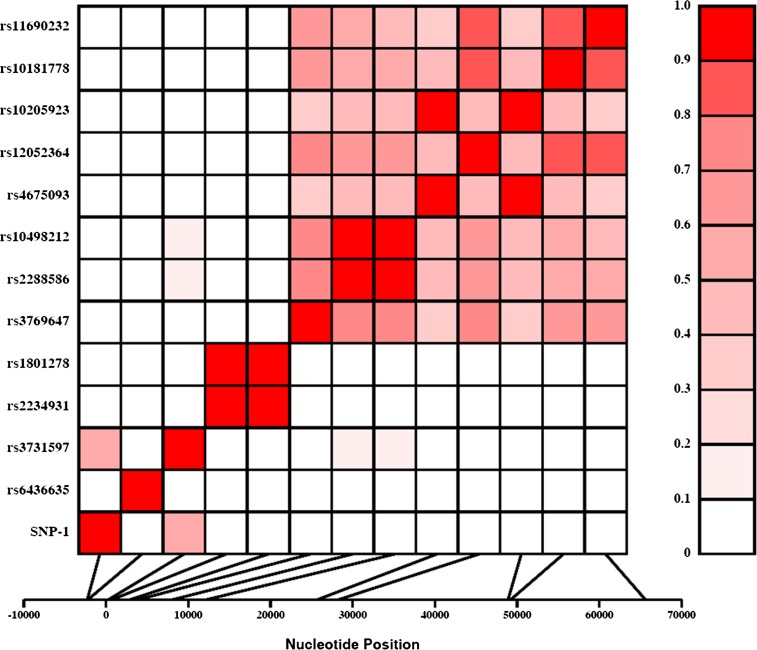
Based on the genotypic data, six were excluded from further analysis because one (SNP-7) failed to be in Hardy-Weinberg Equilibrium and the minor allele frequencies of five SNPs (SNPs-5, 6, 8, 17, and 18) were less than 0.5% (Fig. 1). We estimated the pairwise LD (r2) between all the 13 SNPs and found that the pairwise LD ranged from 0 to 1.0 (Fig. 2). As can be seen from Fig. 2, the relatively high pairwise LDs (r2 > 0.8) were found for the following SNPs pairs: rs2234931-rs1801278 (r2 = 1.0), rs2288586-rs10498212 (r2 = 0.97), rs4675093-rs10205923 (r2 = 0.92), rs12052364-rs10181778 (r2 = 0.88), and rs12052364-rs11690232 (r2 = 0.84).
FIG. 2.
LD between SNP pairs within the IRS1 gene. SNPs are labeled on the y-axis, and the locations (bp) within the gene are shown on the x-axis. Pairwise LD is estimated using r2 values and depicted in the figure by the color intensity of the shaded box, as shown in the legend. The diagonal represents a comparison of each SNP against itself (i.e., r2 = 1.0). (A high-quality color representation of this figure is available in the online issue.)
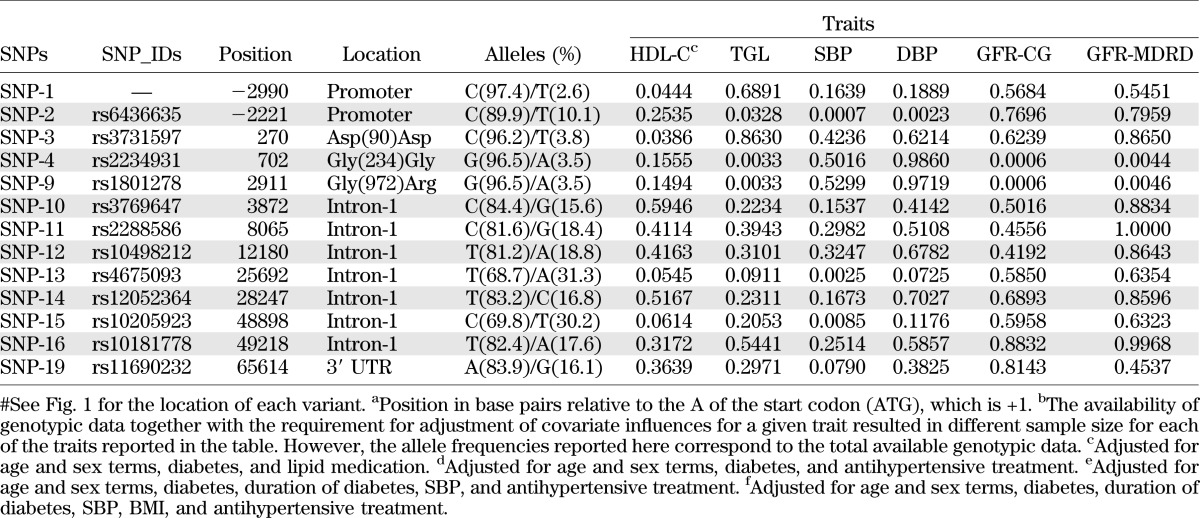
Association analysis between genotypic and GFR data were performed using MGA. The β-coefficients and P values of the covariates used are given in the Supplementary Data. The location, allele frequencies, and association analyses of 13 SNPs are summarized in Table 2. Of the 13 SNPs examined for association, the G to A substitution at codon 972 changing the amino acids from Glycine to Arginine (Gly(972)Arg) with minor allele frequencies of about 4% was found to be significantly associated with GFR-CG (P = 0.0006). Given the sample size of 431 for this analysis (Table 2 and Table 3), genetic effects that account for 2.9% of the total phenotypic variation in GFR-CG could be detected with 80% power with an experiment-wide P value < 0.05 (i.e., effective number of SNPs = 7.66). In this study, the Gly(972)Arg polymorphism explains 3.3% of the phenotypic variation in GFR-CG. The Gly(234)Gly (rs2234931), which was in complete LD (r2 = 1; Fig. 2) with Gly(972)Arg variant (rs1801278) also exhibited similar association with GFR. The Gly(972)Arg and Gly(234)Gly variants were also found to be significantly associated with GFR-MDRD (P = 0.0046) (Table 2). These associations continue to be significant after adjusting for multiple testing. It should be noted that no individuals were homozygous for the rare allele in either of these SNPs.
TABLE 2.
Association analysis between the IRS1 polymorphisms and GFR and T2DM-related traits
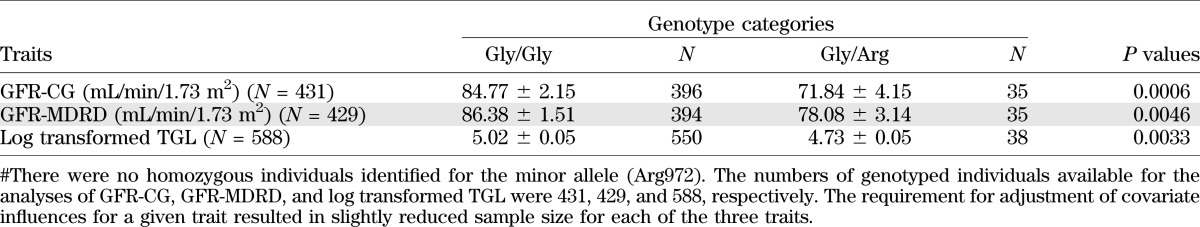
TABLE 3.
Mean values of GFR by Gly(972)Arg genotype categories
Subsequently, we conducted BQTN analysis with eight SNPs, after excluding five SNPs (SNP-4-rs2234931; SNP-12-rs10498212, SNP-15-rs10205923, SNP-16-rs10181778, SNP-19-rs11690232) that exhibited redundancy owing to relatively high pairwise LD (r2 > 0.80) (Fig. 1 and Fig. 2). Of the eight SNPs, SNPs 1–3, 9–11, 13, and 14 resulted in the examination of 256 plausible additive gene action models; the Gly(972)Arg variant provided strongest evidence for the observed association with GFR-CG, with estimated posterior probability of >0.99 (data not shown). Such a high posterior probability of effect is suggestive of a direct functional effect for this variant. As revealed by the MGA, the mean GFR values by genotypic categories differed significantly (Table 3). The carriers of Arg972 variant had significantly decreased GFR-CG and GFR-MDRD values.
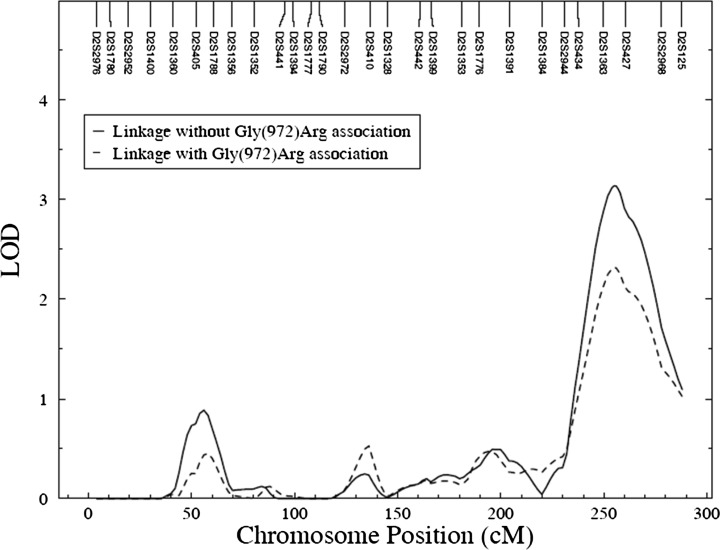
To determine whether Gly(972)Arg could account for the initial linkage signal, we performed linkage analysis on chromosome 2 conditional on the measured genotype effect. Because the sample size available (N = 431) for the Gly(972)Arg genotypic data was slightly smaller than the sample size (N = 453) used for the original linkage study (5), we repeated the linkage analysis of GFR-CG with genotype-by-diabetes interaction influences. As was done in our original linkage report (5), the LOD score obtained from the linkage analysis was adjusted with 2df to 1df, denoted as LODc score. More details on the adjustment of 2df to 1df are provided in the Supplementary Data. The LODc obtained in the current study was 3.1 (N = 431) compared with the original report of 3.3 (N = 453). By inclusion of the Gly(972)Arg polymorphism in the model, the LODc score went down from 3.1 to 2.3; thus, about 26% of the evidence for linkage of GFR at 2q was explained by this variant (Fig. 3). Taken together, these data suggest that the Gly(972)Arg contribute to the decrease in GFR and that Gly(972)Arg may be one of the several potential functional sites regulating GFR or it is in LD with the true functional variant(s) in IRS1 or genes flanking IRS1.
FIG. 3.
Linkage analysis conditional on the Gly(972)Arg polymorphism on chromosome 2q.
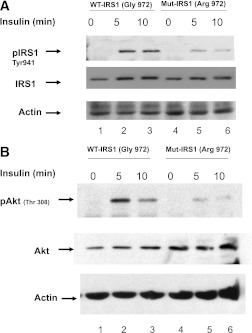
To further determine the functional mechanism by which Arg972 of variant of IRS1 may contribute to variation in GFR, we transfected the human renal mesangial cells with the expression vector pcDNA3 carrying wild-type (Gly972) and mutant (Arg972) IRS1 (35) and studied their effect on the insulin-induced tyrosine (941) phosphorylation of IRS1 and threonine (308) phosphorylation of Akt kinase, a critical downstream effector of PI 3-kinase. Transfected cells were treated with 100 nmol/L insulin for 0, 5, and 10 min and lysed. Equal amount of total proteins were subjected to SDS-PAGE and probed with the following antibodies: anti-IRS1, antiphospho-IRS1, anti-Akt, antiphospho-Akt, and actin. In cells expressing Arg972 variant of IRS1, insulin-stimulated phosphorylation of IRS1 was significantly decreased by 57.3% at 5 min and by 65.8% at 10 min of insulin treatment compared with cells expressing Gly972 variant of IRS1 (Fig. 4A). Compared with cells expressing Gly972 variant of IRS, insulin-induced Thr308-Akt phosphorylation was also significantly reduced in cells expressing Arg972 variant of IRS1 by 75.9% at 5 min and by 88% at 10 min of insulin treatment (Fig. 4B).
FIG. 4.
Effect of Arg972 on the insulin-induced phosphorylation of IRS1 and Akt in human mesangial cells. A: Tyrosine (941) phosphorylation of IRS1. Human mesangial cells were transfected with plasmids carrying wild type (WT; Gly972) and mutant (Mut; Arg972) IRS1 and treated with 100 nM insulin for 0, 5, and 10 min. Equal amount of total cellular proteins were separated on SDS-PAGE gel and probed with anti-IRS1 and antiphospho-IRS1. B: Threonine (308) phosphorylation of Akt kinase. Human mesangial cells were transfected with plasmids carrying wild-type (Gly972) and mutant (Arg972) IRS1 and treated with 100 nM insulin for 0, 5, and 10 min. Equal amount of total cellular proteins were immunoprecipitated with anti-Akt kinase and separated on SDS-PAGE gel and probed with anti-Akt and antiphospho-Akt kinase. Representative images of three independent experiments are shown.
Association analysis was also extended to available phenotypic data including SBP, diastolic blood pressure (DBP), TGL, HDL-cholesterol (HDL-C), albumin-to-creatinine ratio, and T2DM. It is noteworthy that the two variants [Gly(972)Arg and Gly(234)Gly] that exhibited significant associations with the GFR measures were also found to be significantly associated with TGL (P = 0.0033) (Table 2). In addition, a promoter variant of IRS1 (SNP-2; rs6436635) with minor allele frequency of 10.1% was significantly associated with TGL (P = 0.0328), SBP (P = 0.0007), and DBP (P = 0.0023) (Table 2). In addition, a promoter variant (SNP-1) with a minor allele frequency of 2.6% and a synonymous variant (SNP-3; Asp90Asp) with minor allele frequency of 3.8% also showed statistically significant association with HDL-C (P = 0.0444 and P = 0.0386, respectively). A pair of intronic region SNP (rs4675093-rs10205923), which were in strong LD (r2 = 0.92), also exhibited significant association with SBP (P = 0.0025 and 0.0085, respectively). These SNPs also showed suggestive association with HDL-C (Table 2).
DISCUSSION
There is a large interindividual variation in GFR in patients with diabetes and in the general population. It is known that variation in GFR is influenced by environmental and genetic factors as well as their interactive influences (2–11). In an effort to identify and characterize GFR susceptibility gene(s), we localized a genetic region on chromosome 2q35–37 that exhibited linkage to GFR (5). As we report in this study, this locus, which does not confer susceptibility to T2DM (5), exerts a differential influence on GFR in diabetic and nondiabetic individuals. Of the positional candidate genes, IRS1 is a strong susceptibility candidate gene to influence GFR since the IRS1 protein plays a pivotal role in modulating tissue response to insulin including in the kidney. A structural defect in this protein as a result of genetic variation may alter kidney function.
To date, several DNA sequence variants have been identified in human IRS1. Of those, the main focus has been on Gly(972)Arg variant, since Almind et al. (36) first identified this polymorphism to contribute to T2DM in a Danish cohort. Subsequent case control studies in different ethnic groups have examined the association between the Gly(972)Arg variant of IRS1 and T2DM and related traits. Although the association results have been controversial, a meta-analysis combining 27 studies, which included 3,408 cases and 5,419 control subjects, suggested that the carriers of the Gly(972)Arg variant have a 25% increased risk for developing T2DM (17). In addition, a marked ethnic difference in the distribution of Gly(972)Arg variant was also observed. The Arg972 variant of IRS1 was found to be more common in Caucasians (20%) (37) compared with African Americans (11%) (38), Asians (4%) (39), and Mexican Americans (4%) (40). In this study, we observed a frequency of about 4% for Arg972, which is similar to the one reported in another Mexican American population (40). Similarly, Gly(972)Arg failed to show an association with T2DM as reported (40).
Of the 13 SNPs examined for association with GFR, only the Gly(972)Arg variant of IRS1 exhibited significant association with GFR estimated by two formulae. The BQTN analysis using information from eight nonredundant SNPs also demonstrated that the Gly(972)Arg variant could influence phenotypic variation in the levels of GFR with estimated posterior probability of >0.99. This high posterior probability effect reflects the potential functional impact of this variant on GFR. It is interesting that the carriers of Arg972 had significantly decreased GFR values. The subsequent conditional linkage/QTN analysis revealed that the Gly(972)Arg variant explained ∼26% of the linkage signal for GFR on 2q. Taken together, our data demonstrate that the Gly(972)Arg variant significantly contributes to decrease in GFR.
The functional mechanism by which Arg972 variant contributes to decline in GFR remains speculative. Several studies have demonstrated that expression of the 972Arg variant of IRS1 in cultured adipocytes, muscle, and pancreatic b cells caused a defect not only in Tyr-941 phosphorylation of IRS1 but also in binding of the p85 regulatory subunit of PI3K to the IRS1, resulting in a decrease in IRS1-associated PI3K activity and subsequent activation of the Ser/Thr kinase Akt (19,35,41–44). Hribal et al. (20) generated transgenic mice overexpressing Arg972 and confirmed a significant attenuation in insulin signaling pathways downstream of IRS1 in the liver, skeletal muscle, and adipose tissue.
In the current study, we studied the effect of Arg972 on the insulin-stimulated tyrosine phosphorylation of IRS1 and threonine phosphorylation of Akt in human renal mesangial cells. We show that expression of Arg972 in mesangial cells significantly reduces insulin-induced phosphorylation of IRS1 and Akt kinase compared with wild-type controls. Impaired IR/IRS1 signaling in mesangial cells provides a potential mechanism by which Arg972 variant of IRS1 contributes to decreased GFR. Alterations in the contractile property of mesangial cells as a result of impaired IR/IRS1 signaling by Arg972 variant may lead to alterations in glomerular hemodynamics, thereby influencing GFR. Because insulin has been shown to be a survival factor for mesangial cells (45), impaired insulin signaling by the Arg972 variant may lead to apoptosis of these cells, thereby contributing to structural changes in the glomerulus resulting in a decrease in surface area available for filtration. Functional studies are necessary to prove the hypothesis that the Arg972 variant of IRS1 not only impairs IR signaling to protect mesangial cells against apoptosis but also impairs contractile function of mesangial cells. Impaired nitric oxide (NO) production in vascular endothelial cells by the Arg972 variant may also contribute to the decreased GFR. Human umbilical vein endothelial cells isolated from carriers of the Arg972 variant of IRS1 have impaired eNOS expression in response to insulin (46). Because the impaired IR/IRS1 signaling is a potential mechanism for the observed association, we also performed association analysis for the 13 SNPs using fasting glucose data (N = 541) after excluding T2DM individuals that are on antidiabetic medications. Although the Gly(972)Arg variant failed to show an association, the promoter variant (rs6436635) was found to be nominally associated (P = 0.035) with glucose levels.
The present data also show an association of Gly(972)Arg variant with TGL in our cohort, and the carriers of Arg972 had decreased TGL values. Our finding of lower TGL concentrations in carriers of Arg972 contradicts previous observations in other populations at high risk to develop atherosclerotic cardiovascular disease (47,48). However, our findings are in agreement with others who reported that patients with T2DM (49) and patients with insulin resistance (50) have low concentrations of TGL. The functional significance of promoter variants, Asp90Asp, and a pair of intronic variants (rs4675093 and rs10205923) that exhibit an association with cardiovascular risk factors such as HDL-C, TGL, SBP, and DBP in this study remains to be determined.
Despite several strengths, our study has potential limitations. To our knowledge, the two estimates of GFR (GFR-CG and GFR-MDRD) have not been validated in the Mexican American population. Whereas the time-consuming and expensive direct measurement of GFR is difficult to perform (e.g., inulin, iothalamate clearance) in large family studies, the estimated GFR using the abbreviated MDRD and GC formulae has been proposed as the best validated means for transforming serum creatinine measurements into GFR (25,51) and used in several linkage studies (2–11). Another limitation is that because our resequencing effort was based on only 32 individuals, it is possible that we might have missed rare variants specific to this population. However, given the recent emphasis on the role of rare variants in complex disease causation compared with the common variants examined using the genome-wide association studies, it is very interesting to note that the Gly(972)Arg associated with decline in GFR values has a frequency of 4.0%. We tried to replicate the association between Gly(972)Arg variant of IRS1 and GFR in two independent family studies: the San Antonio Family Heart Study (Mexican Americans, N = 844) and the Joslin Study on the Genetics of Type 2 Diabetes (Caucasians, N = 825). Unfortunately, we failed to replicate the findings.
In conclusion, we provide evidence for the first time that Arg972 variant of IRS1 is associated with variation in GFR; carriers of Arg972 had significantly decreased GFR values and Arg972 variant accounts for 26% of the linkage of GFR on 2q37. It is noteworthy that we show that expression of Arg972 in human mesangial cells reduces the insulin-stimulated phosphorylation of IRS1 and Akt kinase. Together, it is possible to speculate that the associated variant is one of multiple potential functional variants influencing variation in GFR in the IRS1 gene or is in LD with a potential functional variant(s) in the genes flanking IRS1. There is evidence for long-range LD in the human genome (52). However, in the absence of knowledge on such long-range LD of the SNP of interest in this study, it is important to note that genetic variants located 500 kb upstream of IRS1 were recently shown to be strongly associated with impaired metabolic profiles including insulin resistance and T2DM. Furthermore, these variants also impaired the IR/IRS1 signaling (21,22). Therefore, a thorough screening of the critical region on 2q37 may identify additional variants that could potentially relate to the GFR linkage finding in this study. Identification of susceptibility genes that contribute to the decline in GFR may facilitate prediction, prevention, and development of improved treatments of this devastating complication of diabetes.
ACKNOWLEDGMENTS
This study was supported by a grant-in-aid award (0855175F) (F.T.) from the American Heart Association, the Carl W. Gottschalk Research Scholar Award from the American Society of Nephrology (F.T.), the Norman S. Coplon Award from Satellite Healthcare (F.T.), and the Diabetes Action Research and Education Foundation (F.T.). This work was also supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (DK-42273, DK-47482, and DK-53889 [to R.D.]) and VA-Merit Review and the National Center for Research Resources Contracts UL1 RR025767 and KL2 RR025766 for the Institute for Integration of Medicine and Science.
No potential conflicts of interest relevant to this article were reported.
F.T. researched data, contributed to the discussion, wrote the manuscript, reviewed and edited the manuscript, and performed the studies. S.P., J.B., R.D., and H.E.A. researched data, contributed to the discussion, and reviewed and edited the manuscript. J.S. and B.B. researched data and contributed to the discussion. R.A., N.H.A., and S.F. researched data and reviewed and edited the manuscript. T.L.V. researched data. V.S.F. and L.A. contributed to the discussion and reviewed and edited the manuscript. F.T. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank the members of SAFDGS for their participation and cooperation; Dr. Andrzej Krolewski, Senior Investigator at the Joslin Diabetes Center and an Associate Professor of Medicine at Harvard Medical School, for sharing the data to replicate candidate variants in his Joslin Study cohort on the Genetics of Type 2 Diabetes; the General Clinical Research Center; and the South Texas Healthcare System.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1078/-/DC1.
REFERENCES
- 1.Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic factors in diabetic nephropathy. Clin J Am Soc Nephrol 2007;2:1306–1316 [DOI] [PubMed] [Google Scholar]
- 2.Fox CS, Yang Q, Cupples LA, et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study. J Am Soc Nephrol 2004;15:2457–2461 [DOI] [PubMed] [Google Scholar]
- 3.Hunt SC, Coon H, Hasstedt SJ, et al. Linkage of serum creatinine and glomerular filtration rate to chromosome 2 in Utah pedigrees. Am J Hypertens 2004;17:511–515 [DOI] [PubMed] [Google Scholar]
- 4.Placha G, Poznik GD, Dunn J, et al. A genome-wide linkage scan for genes controlling variation in renal function estimated by serum cystatin C levels in extended families with type 2 diabetes. Diabetes 2006;55:3358–3365 [DOI] [PubMed] [Google Scholar]
- 5.Puppala S, Arya R, Thameem F, et al. Genotype by diabetes interaction effects on the detection of linkage of glomerular filtration rate to a region on chromosome 2q in Mexican Americans. Diabetes 2007;56:2818–2828 [DOI] [PubMed] [Google Scholar]
- 6.Schelling JR, Abboud HE, Nicholas SB, et al. Family Investigation of Nephropathy and Diabetes Research Group Genome-wide scan for estimated glomerular filtration rate in multi-ethnic diabetic populations: the Family Investigation of Nephropathy and Diabetes (FIND). Diabetes 2008;57:235–243 [DOI] [PubMed] [Google Scholar]
- 7.Freedman BI, Bowden DW, Rich SS, et al. Genome-wide linkage scans for renal function and albuminuria in Type 2 diabetes mellitus: the Diabetes Heart Study. Diabet Med 2008;25:268–276 [DOI] [PubMed] [Google Scholar]
- 8.Köttgen A. Genome-wide association studies in nephrology research. Am J Kidney Dis 2010;56:743–758 [DOI] [PubMed] [Google Scholar]
- 9.Rao M, Mottl AK, Cole SA, et al. Meta-analysis of genome-wide linkage scans for renal function traits. Nephrol Dial Transplant 2012;27:647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köttgen A, Glazer NL, Dehghan A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 2009;41:712–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köttgen A, Pattaro C, Böger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet 2010;42:376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahn CR, Folli F. Molecular determinants of insulin action. Horm Res 1993;39(Suppl. 3):93–101 [DOI] [PubMed] [Google Scholar]
- 13.Virkamäki A, Ueki K, Kahn CR. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J Clin Invest 1999;103:931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araki E, Lipes MA, Patti ME, et al. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature 1994;372:186–190 [DOI] [PubMed] [Google Scholar]
- 15.Brüning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell 1997;88:561–572 [DOI] [PubMed] [Google Scholar]
- 16.Terauchi Y, Iwamoto K, Tamemoto H, et al. Development of non-insulin-dependent diabetes mellitus in the double knockout mice with disruption of insulin receptor substrate-1 and beta cell glucokinase genes. Genetic reconstitution of diabetes as a polygenic disease. J Clin Invest 1997;99:861–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jellema A, Zeegers MP, Feskens EJ, Dagnelie PC, Mensink RP. Gly972Arg variant in the insulin receptor substrate-1 gene and association with Type 2 diabetes: a meta-analysis of 27 studies. Diabetologia 2003;46:990–995 [DOI] [PubMed] [Google Scholar]
- 18.McGettrick AJ, Feener EP, Kahn CR. Human insulin receptor substrate-1 (IRS-1) polymorphism G972R causes IRS-1 to associate with the insulin receptor and inhibit receptor autophosphorylation. J Biol Chem 2005;280:6441–6446 [DOI] [PubMed] [Google Scholar]
- 19.Sentinelli F, Filippi E, Cavallo MG, Romeo S, Fanelli M, Baroni MG. The G972R variant of the insulin receptor substrate-1 gene impairs insulin signaling and cell differentiation in 3T3L1 adipocytes; treatment with a PPARgamma agonist restores normal cell signaling and differentiation. J Endocrinol 2006;188:271–285 [DOI] [PubMed] [Google Scholar]
- 20.Hribal ML, Tornei F, Pujol A, et al. Transgenic mice overexpressing human G972R IRS-1 show impaired insulin action and insulin secretion. J Cell Mol Med 2008;12:2096–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet 2009;41:1110–1115 [DOI] [PubMed] [Google Scholar]
- 22.Kilpeläinen TO, Zillikens MC, Stančákova A, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet 2011;43:753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puppala S, Dodd GD, Fowler S, et al. A genomewide search finds major susceptibility loci for gallbladder disease on chromosome 1 in Mexican Americans. Am J Hum Genet 2006;78:377–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levey AS, Coresh J, Greene T, et al. Chronic Kidney Disease Epidemiology Collaboration Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann Intern Med 2006;145:247–254 [DOI] [PubMed] [Google Scholar]
- 25.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D, Modification of Diet in Renal Disease Study Group A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Ann Intern Med 1999;130:461–470 [DOI] [PubMed] [Google Scholar]
- 26.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron 1976;16:31–41 [DOI] [PubMed] [Google Scholar]
- 27.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 1996;58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- 28.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet 1986;50:181–194 [DOI] [PubMed] [Google Scholar]
- 29.Almasy L, Blangero J. Exploring positional candidate genes: linkage conditional on measured genotype. Behav Genet 2004;34:173–177 [DOI] [PubMed] [Google Scholar]
- 30.Richardson DK, Schneider J, Fourcaudot MJ, et al. Association between variants in the genes for adiponectin and its receptors with insulin resistance syndrome (IRS)-related phenotypes in Mexican Americans. Diabetologia 2006;49:2317–2328 [DOI] [PubMed] [Google Scholar]
- 31.Abecasis GR, Cookson WOC, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet 2000;8:545–551 [DOI] [PubMed] [Google Scholar]
- 32.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005;95:221–227 [DOI] [PubMed] [Google Scholar]
- 33.Blangero J, Goring HH, Kent JW, Jr, et al. Quantitative trait nucleotide analysis using Bayesian model selection. Hum Biol 2005;77:541–559 [DOI] [PubMed] [Google Scholar]
- 34.Curran JE, Jowett JB, Elliott KS, et al. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet 2005;37:1234–1241 [DOI] [PubMed] [Google Scholar]
- 35.Federici M, Hribal ML, Ranalli M, et al. The common Arg972 polymorphism in insulin receptor substrate-1 causes apoptosis of human pancreatic islets. FASEB J 2001;15:22–24 [DOI] [PubMed] [Google Scholar]
- 36.Almind K, Bjørbaek C, Vestergaard H, Hansen T, Echwald S, Pedersen O. Aminoacid polymorphisms of insulin receptor substrate-1 in non-insulin-dependent diabetes mellitus. Lancet 1993;342:828–832 [DOI] [PubMed] [Google Scholar]
- 37.Baroni MG, Arca M, Sentinelli F, et al. The G972R variant of the insulin receptor substrate-1 (IRS-1) gene, body fat distribution and insulin-resistance. Diabetologia 2001;44:367–372 [DOI] [PubMed] [Google Scholar]
- 38.Lei HH, Coresh J, Shuldiner AR, Boerwinkle E, Brancati FL. Variants of the insulin receptor substrate-1 and fatty acid binding protein 2 genes and the risk of type 2 diabetes, obesity, and hyperinsulinemia in African-Americans: the Atherosclerosis Risk in Communities Study. Diabetes 1999;48:1868–1872 [DOI] [PubMed] [Google Scholar]
- 39.Ito K, Katsuki A, Furuta M, et al. Insulin sensitivity is not affected by mutation of codon 972 of the human IRS-1 gene. Horm Res 1999;52:230–234 [DOI] [PubMed] [Google Scholar]
- 40.Celi FS, Negri C, Tanner K, et al. Molecular scanning for mutations in the insulin receptor substrate-1 (IRS-1) gene in Mexican Americans with Type 2 diabetes mellitus. Diabetes Metab Res Rev 2000;16:370–377 [DOI] [PubMed] [Google Scholar]
- 41.Hribal ML, Federici M, Porzio O, et al. The Gly→Arg972 amino acid polymorphism in insulin receptor substrate-1 affects glucose metabolism in skeletal muscle cells. J Clin Endocrinol Metab 2000;85:2004–2013 [DOI] [PubMed] [Google Scholar]
- 42.Porzio O, Federici M, Hribal ML, et al. The Gly972→Arg amino acid polymorphism in IRS-1 impairs insulin secretion in pancreatic beta cells. J Clin Invest 1999;104:357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almind K, Inoue G, Pedersen O, Kahn CR. A common amino acid polymorphism in insulin receptor substrate-1 causes impaired insulin signaling. Evidence from transfection studies. J Clin Invest 1996;97:2569–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura R, Araki E, Ura S, et al. Impact of natural IRS-1 mutations on insulin signals: mutations of IRS-1 in the PTB domain and near SH2 protein binding sites result in impaired function at different steps of IRS-1 signaling. Diabetes 1997;46:929–936 [DOI] [PubMed] [Google Scholar]
- 45.Hiromura K, Monkawa T, Petermann AT, Durvasula RV, Shankland SJ. Insulin is a potent survival factor in mesangial cells: role of the PI3-kinase/Akt pathway. Kidney Int 2002;61:1312–1321 [DOI] [PubMed] [Google Scholar]
- 46.Federici M, Pandolfi A, De Filippis EA, et al. G972R IRS-1 variant impairs insulin regulation of endothelial nitric oxide synthase in cultured human endothelial cells. Circulation 2004;109:399–405 [DOI] [PubMed] [Google Scholar]
- 47.Baroni MG, D’Andrea MP, Montali A, et al. A common mutation of the insulin receptor substrate-1 gene is a risk factor for coronary artery disease. Arterioscler Thromb Vasc Biol 1999;19:2975–2980 [DOI] [PubMed] [Google Scholar]
- 48.Marini MA, Frontoni S, Mineo D, et al. The Arg972 variant in insulin receptor substrate-1 is associated with an atherogenic profile in offspring of type 2 diabetic patients. J Clin Endocrinol Metab 2003;88:3368–3371 [DOI] [PubMed] [Google Scholar]
- 49.Ossei-Gerning N, Mansfield MW, Stickland MH, Grant PJ. Insulin receptor substrate-1 gene polymorphism and cardiovascular risk in non-insulin dependent diabetes mellitus and patients undergoing coronary angiography. Clin Lab Haematol 1997;19:123–128 [DOI] [PubMed] [Google Scholar]
- 50.Jellema A, Mensink RP, Kromhout D, Saris WH, Feskens EJ. Metabolic risk markers in an overweight and normal weight population with oversampling of carriers of the IRS-1 972Arg-variant. Atherosclerosis 2003;171:75–81 [DOI] [PubMed] [Google Scholar]
- 51.Lamb EJ, Tomson CR, Roderick PJ, Clinical Sciences Reviews Committee of the Association for Clinical Biochemistry Estimating kidney function in adults using formulae. Ann Clin Biochem 2005;42:321–345 [DOI] [PubMed] [Google Scholar]
- 52.Saunders MA, Slatkin M, Garner C, Hammer MF, Nachman MW. The extent of linkage disequilibrium caused by selection on G6PD in humans. Genetics 2005;171:1219–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]