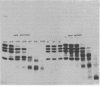
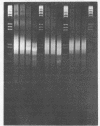
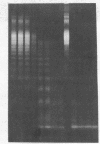
Abstract
Chromatin 'core particles' have been digested with trypsin to varying extents. The resulting particles are homogeneous by the criterion of ultracentrifuge boundary analysis. Sedimentation coefficients are lowered as cleavages are introduced into the histones, showing that an unfolding of the core particle occurs. This unfolding is further characterised by a lower melting temperature together with a premelting phase, higher molar ellipticity in the circular dichroism spectra at 280 nm and increased kinetics of digestion by both micrococcal nuclease and DNase I. Differences are also observed in the products of nuclease digestion. The most consistent interpretation of the data involves an unfolding process whereby free rods of DNA are released to extend from a nucleoprotein core.
Full text
PDF




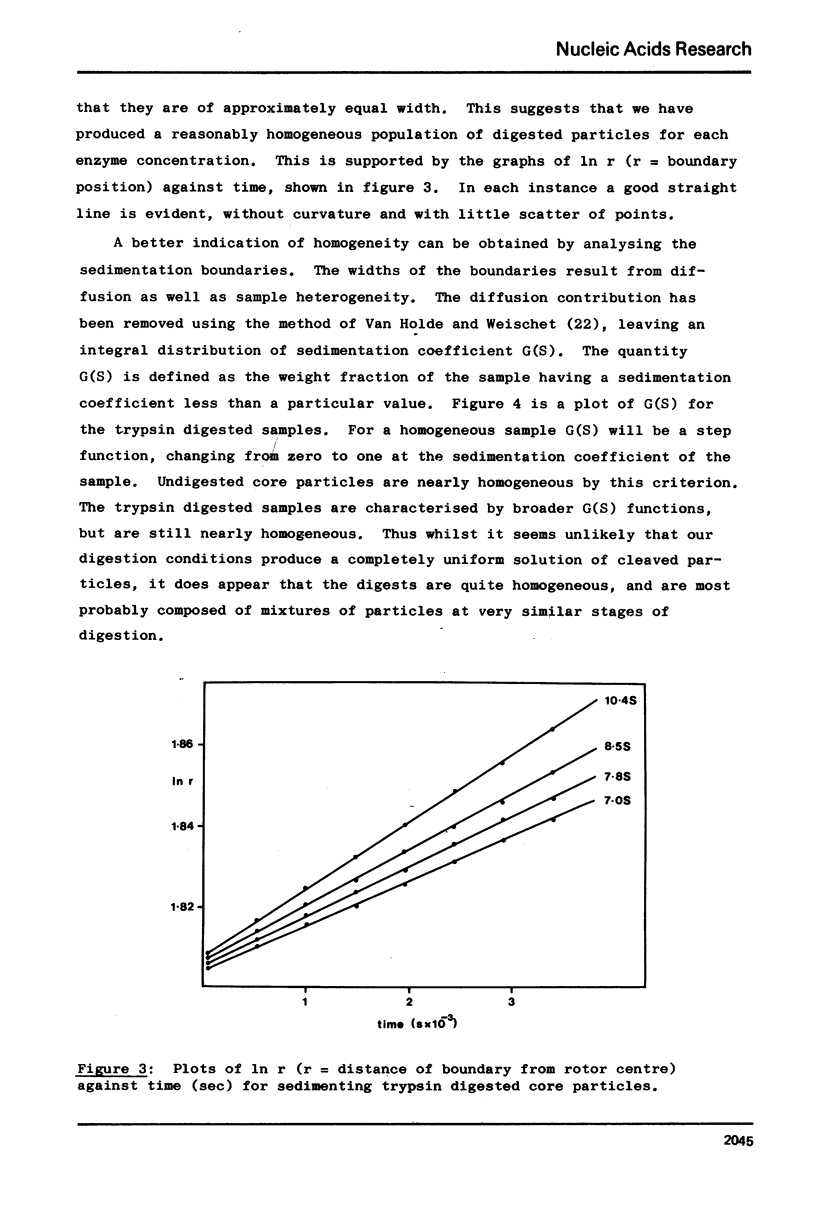
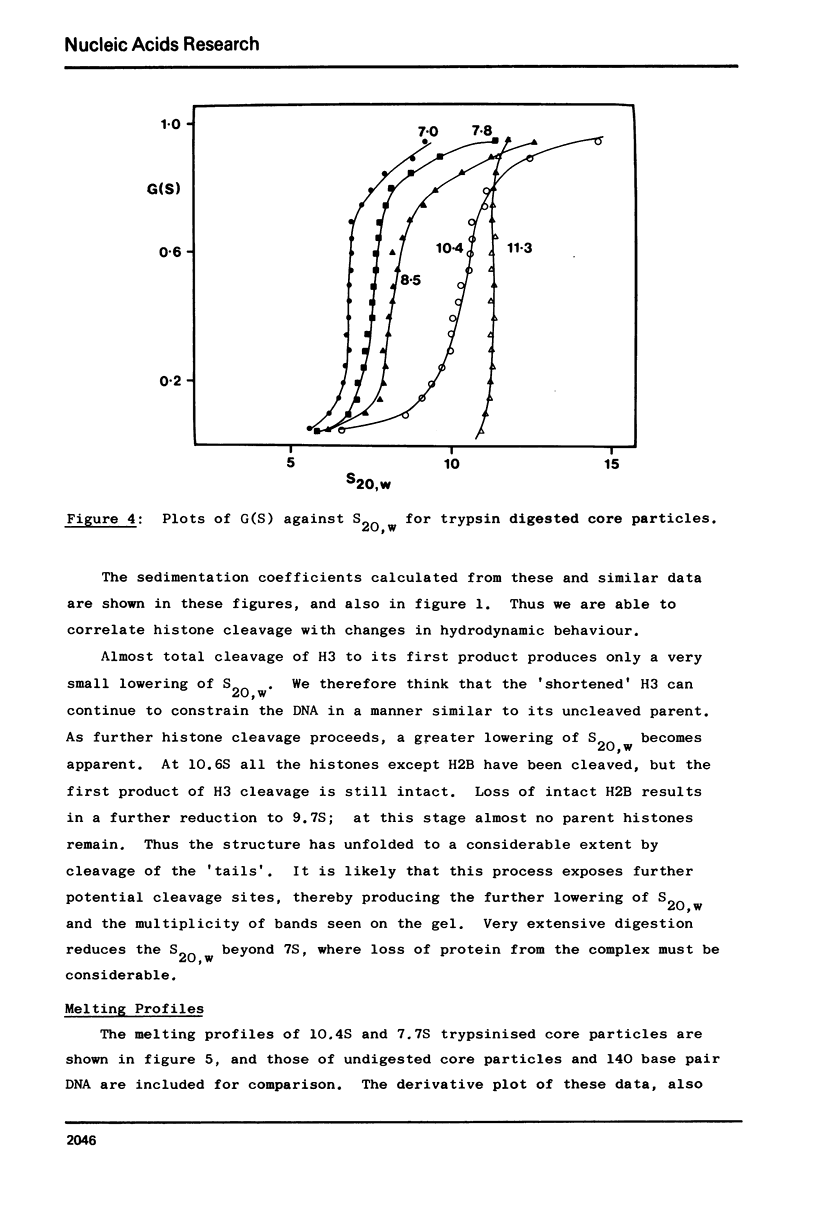
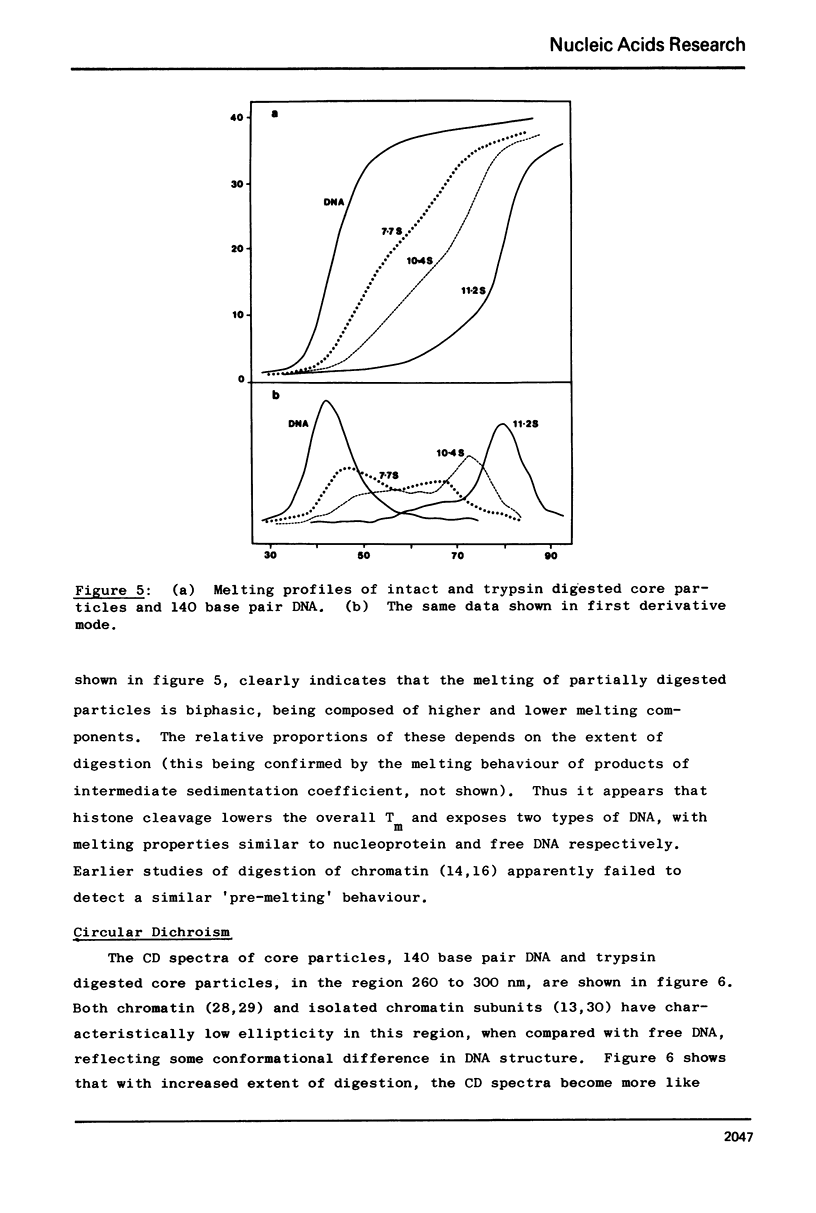
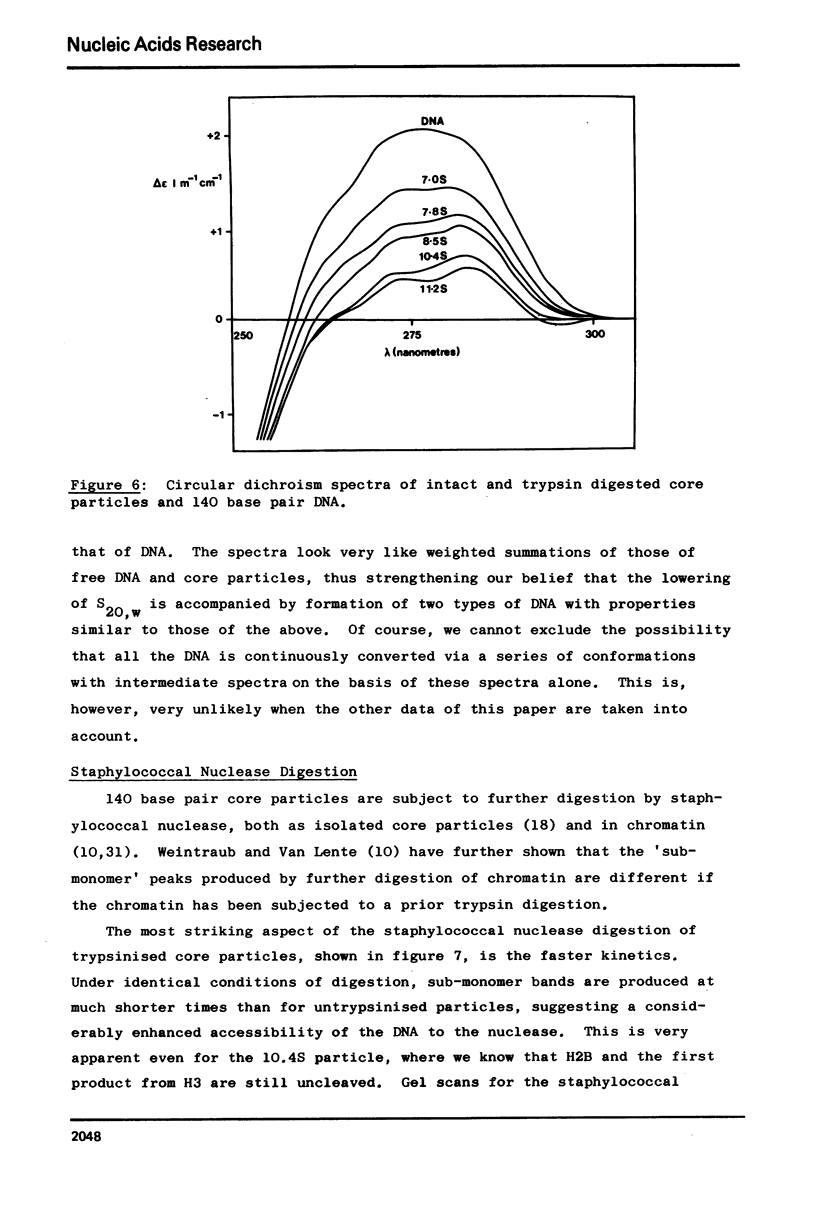






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt W. F., Böhm L., Von Holt C. Proteolytic degradation of histones and site of cleavage in histone F2al and F3. FEBS Lett. 1975 Mar 1;51(1):88–93. doi: 10.1016/0014-5793(75)80860-x. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Cotter R. I. The molecular weight of nucleosome protein by laser light scattering. FEBS Lett. 1976 Nov;70(1):209–211. doi: 10.1016/0014-5793(76)80759-4. [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Walker I. O. The modification of deoxyribonucleohistone by trypsin and chymotrypsin. Eur J Biochem. 1973 May 2;34(3):519–526. doi: 10.1111/j.1432-1033.1973.tb02789.x. [DOI] [PubMed] [Google Scholar]
- Foe V. E., Wilkinson L. E., Laird C. D. Comparative organization of active transcription units in Oncopeltus fasciatus. Cell. 1976 Sep;9(1):131–146. doi: 10.1016/0092-8674(76)90059-3. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Murphy R. F., Bonner J. Structure of transcriptionally active chromatin. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4404–4408. doi: 10.1073/pnas.72.11.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Howarth O. W., Clark V. M., Pardon J. F., Richards B. M. The existence of random coil N-terminal peptides - 'tails' - in native histone complexes. FEBS Lett. 1976 Feb 1;62(1):7–10. doi: 10.1016/0014-5793(76)80004-x. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Howarth O. W., Clark V. W., Pardon J. F., Richards B. M. An investigation of the conformational and self-aggregational processes of histones using 1H and 13C nuclear magnetic resonance. Biochemistry. 1975 Oct 21;14(21):4590–4600. doi: 10.1021/bi00692a006. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel R., Fasman G. D. Chromatin and nucleosome structure. Nucleic Acids Res. 1976 Aug;3(8):1839–1855. doi: 10.1093/nar/3.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Tatchell K., Van Holde K. E., Richards B. M. Low-angle neutron scattering from chromatin subunit particles. Nucleic Acids Res. 1975 Nov;2(11):2163–2176. doi: 10.1093/nar/2.11.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards B., Pardon J., Lilley D., Cotter R., Wooley J., Worchester D. The sub-structure of nucleosomes. Cell Biol Int Rep. 1977 Jan;1(1):107–116. doi: 10.1016/0309-1651(77)90017-0. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Shaw B. R., Corden J. L., Sahasrabuddhe C. G., Van Holde K. E. Chromatographic separation of chromatin subunits. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1193–1198. doi: 10.1016/s0006-291x(74)80410-9. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih T. Y., Fasman G. D. Conformation of deoxyribonucleic acid in chromatin: a circular dichroism study. J Mol Biol. 1970 Aug 28;52(1):125–129. doi: 10.1016/0022-2836(70)90182-8. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Histones H3 and H4 interact with the ends of nucleosome DNA. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4400–4404. doi: 10.1073/pnas.73.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Modification of chromatin by trypsin. The role of proteins in maintainance of deoxyribonucleic acid conformation. Biochemistry. 1972 May 23;11(11):2003–2008. doi: 10.1021/bi00761a002. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Sober H. A. Circular dichroism of calf liver nucleohistone. Biochemistry. 1970 Aug 4;9(16):3103–3109. doi: 10.1021/bi00818a001. [DOI] [PubMed] [Google Scholar]
- Sugano N., Okada S. Physicochemical properties of chromatin digested with trypsin. Biochim Biophys Acta. 1975 Feb 24;383(1):78–85. doi: 10.1016/0005-2787(75)90247-6. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. Cleavable cross-links in the analysis of histone-histone associations. FEBS Lett. 1975 Oct 15;58(1):353–358. doi: 10.1016/0014-5793(75)80296-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Van Lente F. Dissection of chromosome structure with trypsin and nucleases. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4249–4253. doi: 10.1073/pnas.71.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]