Abstract
Autophagy is one of the intracellular systems responsible of protein trafficking (degradation/recycling) in eukaryotic cells. Whereas this ubiquitous process contributes to the cytosolic homeostasis, its deregulation is often associated to various pathologies (neurodegenerative diseases, cancer, pathologies with altered inflammatory response, etc.). The present paper gives an overview of autophagy, especially in inflammation, including mechanisms, regulation, functions and future therapies.
Keywords: Animals; Apoptosis; Autophagy; drug effects; physiology; Bacterial Infections; metabolism; pathology; Homeostasis; Humans; Inflammation; metabolism; pathology; Lysosomes; drug effects; metabolism; Proteasome Endopeptidase Complex; metabolism; Protein Kinase Inhibitors; pharmacology; Proteins; metabolism; Sirolimus; analogs & derivatives; pharmacology; Stress, Physiological; metabolism; pathology; Virus Diseases; metabolism; pathology
Keywords: autophagy, inflammation, infectious diseases, cancer, cell death, drugs
1. Introduction
Biological mechanisms are controlled by a tight balance of positive and negative signals responsible of anabolism and catabolism, also called homeostasis. At the cell level, the equilibrium between macromolecule anabolism and catabolism guarantees the cell or the tissue integrity. In this context, the unsettling of this functional equilibrium can lead to various events including cell death or survival (adequate or inadequate), growth of cancer cells, inflammation, or the difficulty of the cells to adapt their metabolism to their nutritional environment (1, 2). This review is dedicated to the mechanisms developed by the cells to control their homeostasis in physiological and stress conditions. After a brief description of the main cellular degradation systems, this paper focuses on the recent knowledge of autophagy processes (molecular and regulatory mechanisms) and their roles during inflammation.
2. Ubiquitine-proteasome and lysosomal systems represent the main cellular mechanisms which control the protein trafficking
To regulate their homeostasis and therefore their survival death, eukaryotic cells exhibit two main degradation pathways including ubiquitine-proteasome and lysosomal systems (Figure 1). The ubiquitine-proteasome system is associated with the degradation of most of the short-lived proteins (3••) and clearly appears as a selective catabolic process. The most common source of proteasome substrates is mainly cytosolic proteins and missfolded proteins from the endoplasmic reticulum, retrotranslocated into the cytosol. Less frequently, the proteins can be transferred to the proteasome via endocytosis (4–6) which requires a GTPase dynamin-1 activity and includes caveolar and clathrin-mediated endocytosis.
Figure 1.
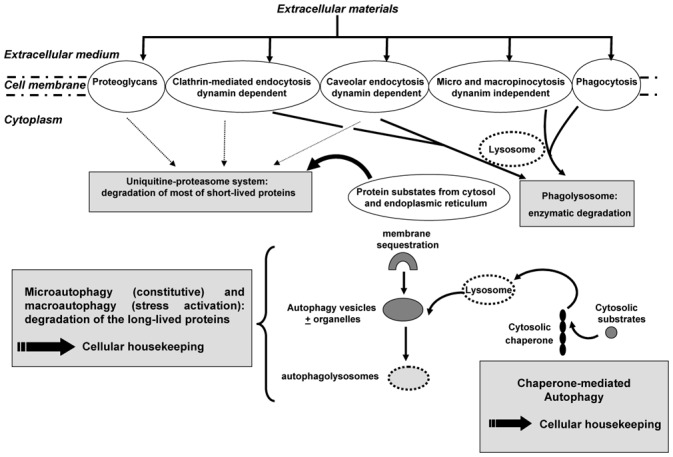
Schematic diagram of the main cellular mechanisms controlling the protein degradation pathways in eukaryotic cells
The second system of protein degradation is composed by the lysosomal system in which proteins or lipids from both extra- and intra-cellular compartments are delivered to the lytic organelles of the cells (lysosomes) and includes in pinocytosis (macro and micro) and phagocytosis (see below) (7). The pinocytosis is associated with membrane ruffling and is not blocked by specific mutations in dynamin-2 in contrast to caveolar endocytosis (8). The role of proteoglycans into the protein degradation has been also widely reported. For instance, syndecan-4 has been shown to act as a co-receptor for several growth factors including FGF-2 (9). Indeed, Tkachenko et al (9) revealed that syndecan-4 uptake, induced by treatment with FGF2, requires the integrity of plasma membrane lipid rafts for its initiation and occurs in a clathrin and dynamin independent manner. Thus, two internalization pathways can be envisaged depending on the basal expression of syndecans and clathrin (10). De facto, the protein degradation is controlled by a tight balance between surface proteoglycans and clathrin coated pits (11). The lysosomal system can be also divided into two main pathways depending on the origin of the degraded materials. Thus, it is possible to distinguish the phagocytosis mechanism mediating the degradation of extracellular materials (Figure 2a) and the autophagy processes which lead to the sequestration and the degradation of cytoplasmic components including organelles via the lysosomes (12•). These different pathways are associated with the non-selective degradation of long-lived proteins. Two different types of autophagy can be distinguished. The first one, the macroautophagy, usually called as autophagy, begins by the formation of cell membrane cisterna of nonlysosomal origin which encloses a portion of cytoplasm (with or without organelles) and constitutes an autophagosome. Therefore, the membrane of autophagosome fuses with the lysosomal membrane and then constitutes the autophagolysosome (acronyme: autolysosome) where hydrolytic enzymes degrade the cytoplasmic content (inner membrane, organelles, etc.) (Figure 2b, c) (13, 14). The envelope of autophagosome is then characterized by a double membrane. Thus, the macroautophagy garantees the turnover of the most long-lived proteins which can be recycled by the cells. A similar sequestration and lysosomal degradation process occurs in the second type of autophagy named microautophagy. This process is closely related to macroautophagy excepted for two points: (i) transfer of cytosolic components into the lysosome is mediated by direct invaginations of the lysosomal membrane; (ii) it represents a constitutive mechanism in contrast to macroautophagy which is activated in stress conditions (15). The third mechanism of autophagy is a selective degradation system in which cytoplasmic subtrates containing a defined motif (a pentapeptide sequence related to KFERQ) are specifically recognized and transported by a cytosolic chaperone to the lysosomal compartment (16). The molecular chaperone is a complex formed by the constitutive form of the heat shock protein of cognate 70 Kda (hsc70), the heat shock protein of 40 kDa (hsp40), the hsp90, the hsp70 cochaperone protein (hip), the hsp70-hsp90 organizing protein (hop) and the Bcl2-associated athanogene 1 protein (bag-1) (17). Therefore, this process is called chaperone-mediated autophagy and controlled the degradation of a very large list of cytoplasmic subtstrates (18). This list includes metabolic proteins (pyruvate kinase, aspartate aminotranferase, etc), proteins involved in cell proliferation and differentiation (phospholipid-binding proteins such as annexin II and IV, or paired box transcription factors) and the lipocalin superfamily of proteins such as α2 microglobulin (19). Thus, the balance between the whole of these mechanisms is responsible of a very tightly regulated control of the cellular housekeeping.
Figure 2. Morphological feature of phagocytose and autophagy in eukaryotic cells.
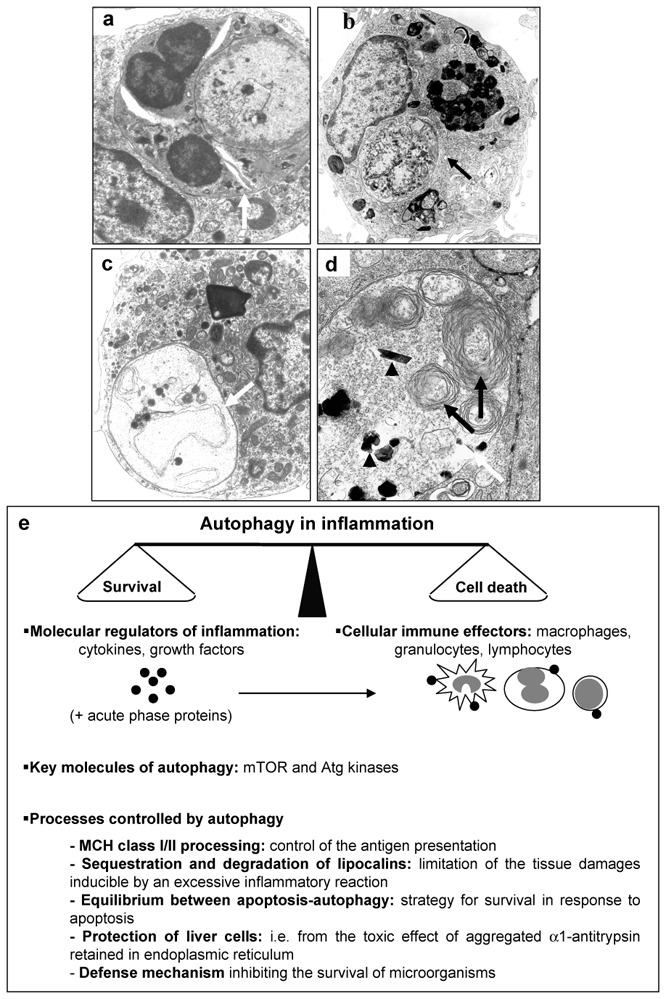
(a) Phagocytose of a polynuclear (white arrow) by a macrophage; (b, c) typical feature of autophagy (white and black arrows) into macrophages in culture; (d) macrophage in culture degrading calcium phosphate ceramic, autophagic vesicle (white arrow) (containing cell endogenous materials (black arrow) and exogenous materials (black arrow head). Original magnification: (a, c) X 7,500; (b) X 5,000; (d) X 11,000; (e) cellular/molecular protagonists involved in autophagy associated with inflammation and the main processes controlled by autophagy during inflammatory process.
3. Molecular aspects of macroautophagy: a strongly conserved lysosomal pathway
In the present section, the term autophagy will be used to refer to macroautophagy. As autophagy is a strongly conserved lysosomal pathway, its molecular mechanisms have been explored using genetic studies on yeast (18, 19), drosophila (20) and Caenorhabditis elegans (21) during the past decade. ATP is the first necessary molecule for the cells to perform autophagy. Indeed, autophagy sequestration and other post-sequestering events are dependent and sensitive to the levels of intracellular ATP (22). Several genes specifically dedicated to autophagy have been identified and are called APG for AutoPhaGy (18) and AUT for AUTophagocytosis genes (23). To clarify the data available in the literature, the nomenclature of the genes involved during autophagy have been recently unified by Klionsky et al (19) who proposed the term of ATG for AuTophaGy-related genes. Most of these genes encode proteins which exert their activities at the step of autophagosome formation (24•), explaining the closed association of the Atg proteins with the membranes implicated in autophagy. During the step of autophagosome formation, the different proteins (i.e. : Atg12 and Atg5) are covalently conjugated by a ubiquitine-like reaction and the very large complex (around 800 kDa) binds thereafter to the isolation membrane until the formation of autophagosome. Some of these proteins exhibit a protease activity such as Atg4B (autophagin-1), involved in the cleavage/recycling of Atg8 and then participates to the cell degradation by autophagy (25, 26). In this context, autophagy implies a specific recruitment of Atg proteins from the cytosol to the membranes which initiates the sequestering membranes. The involvement of the ATG genes in the autophagosome formation has been clearly demonstrated using deficient embryonic stem (ES) cells. Thus, Mizushima et al (27••) generated ATG5−/− mouse ES cells and demonstrated that the Atg12-Atg5 complex plays a key role in the formation of the isolation membrane while these deficient cells showed defects of autophagy pathway. These data confirmed those obtained in Saccharomyces cerevisiae (28). Moreover, Mizushima et al (27••) restored the autophagosome formation after transfection of the deficient ES cells with GFP-Atg5 which allowed, for the first time, the visualization of the autophagosome formation in real time. If the Atg12-Atg5 complex is essential in the first steps of autophagosome formation, several proteins are needed for the late formation of autophagic vacuoles. The small GTP binding protein Rab7 belongs to these proteins (29), as revealed by similar experiments as Atg5. Indeed, the role of Rab7 has been addressed by inhibiting Rab7 function using RNA interference methods and the overexpression of Rab7 dominant negative. The GTP membrane-bound forms of Rab protein recruit specific cytosolic proteins targeting vesicles to appropriate sites on the acceptor membranes. Thus, autophagy is regulated by GTPase.
These data demonstrate that macroautophagy regulating mechanisms are controlled by a very dense and specific protein network. Therefore, the discovery of these different autophagy-related genes and proteins provides new markers for autophagosomes and leads to the development of biochemical methods to monitor autophagy activity (30) completing the standard method (electron microscopy).
4. Autophagy controls large events of the cell life and probably exerts also many other undefined functions
The main function of autophagy is to control protein turnover, cytosolic and organelles homeostasis also synonym of cytoplasmic remodeling. In light of its essential role for cell survival, microautophagy is usually considered as a constitutive mechanism and macroautophagy can be modulated in response to several conditions of stress [deprivation of nutrients (31, 32), oxydative stress (33), etc.]. Thus, macroautophagy not only prevents the accumulation of intracellular debris, but also allows to recycle cellular components. The best mean to stress the cell metabolism consists in nutrient starvation. In these environmental conditions, the deprivation of amino acids induces autophagy in vitro after a very short stress time (7–8 minutes), acting as a protective mechanism for the cells to guarantee the protein levels (31). Kuma et al demonstrated a similar phenomenon in vivo during the early neonatal starvation period (32••). More precisely, a cross-talk between amino acid signaling and G proteins in the regulation of macroautophagy has been described (34), strengthening the existence of a cell surface amino acid receptor (35).
Some evidences from in vivo experiments pointed out the potential implication of autophagy mechanisms in inflammation reactions (35, 36). Any tissue aggression by infectious agents (bacteria, virus, etc), excess of cell death or proliferation, mechanical stress, molecules or drugs, etc lead to the development of a strong organic reaction characterized by the chemoattraction and tissue infiltration of immune effectors (macrophages, lymphocytes, granulocytes) secreting a large panel of cytokines produced to limit this aggression. The main cells of innate immune reaction include dendritic cells and macrophages which recognize specific antigenic structures and function as antigen-presenting cells (APC). Indeed, antigens are processed by proteolysis into short peptides which are then presented by major histocompatibility complex (MHC) class I an II molecules to T cell receptors (TCR) expressed by T lymphocytes, the main effectors of the immune response. In addition, macrophages exert many other functions in host defense, in the maintenance of homeaostasis (i.e. removal of injured cells), killing infectious agents or malignant cells and thus mediate and control the inflammation. MCH class I molecules are thought to present endogenous antigens, MCH class II molecules to present peptides derived form exogenous materials (37) but the antigen processing for MCH class I or class II are quite different. Most endogenous antigens are synthesized on cytosolic ribosomes, processed by proteasomes, enter the endoplasmic reticulum where they are associated to MHC class I heavy chain, and thereafter traffic to the golgi and to the cell surface. Two pathways have been described for the antigen processing for class II MHC molecules. The most prominent processing used endocytosis (via receptor-mediated endocytosis such as Fc receptors or recycling via clathrin-coated pits of surface proteins) and a minor pathway includes transfer from the golgi to endosome and autophagy. Moreover, Nimmerjahn et al recently revealed that the endogenous presentation of an epitope derived from the cytosolic protein neomycin phosphotransferase II on MCH class II is mediated by autophagy (38••), opening new questions on the role of the link between cell protein turnover and the immune system. Unfortunatly, whereas autophagy has been clearly demonstrated to participate to the antigen processing, its impact on the control of immune response needs to be defined and is always controversial (39).
Lipocalins are a diverse family of proteins with a limited amino acid sequence similarity, but with a common tertiary structure (40). Cuervo et al (41) showed the accumulation in lysosomes of α2 microglobulin which belongs to the lipocalin superfamily via a chaperone mediated autophagy mechanism. The human homologuous of rat α2 microglobulin is the human neutrophil gelatinase-associated lipocalin (NGAL) (40). In light of its cellular induction in macrophages, LPS- or TNFα-activated hepatocytes and its intense synthesis in inflamed colon, NGAL appears as a modulator of inflammatory response. NGAL would be produced to limit the tissue damages inducible by an excessive inflammatory reaction. This hypothesis is strengthened by the inhibitory effect of the NGAL preincubation on neutrophils (40). The sequestration and degradation of lipocalins by autophagy clearly appears as modulation process of the inflammation reaction intensity (i.e. biological equilibrium between pro-inflammation reaction in response to pathogens and anti-inflammatory reaction to limit the tissue damages).
Inflammatory process is also characterized by an extensive production of growth factors and cytokines which modulate vascular permeability, leukocyte infiltration and activation, and production of acute phase proteins. The control of autophagic cell death by growth factors (42–44) has been recently proved by Lum et al (45••). Indeed, using growth factor-dependent cells from Bax/Bak-deficient mice, these authors demonstrated that following growth factor withdrawal, Bax/Bak-deficient cells activate autophagy, undergo progressive atrophy, and ultimately succumb to death. Furthermore, cells respond to growth factor readdition by rapid restoration of their metabolism and subsequently recovery of their original size and proliferation rate. Such process explains in part the equilibrium between apoptosis and autophagy (46, 47). Whereas the most of apoptosis mechanisms involve caspase activation, autophagy is a caspase-independent mechanism process involving the lysosome pathway. Moreover, in both cases, the inhibition of caspases, proteasome or lysosome reduce cell death. In this context, autophagy is a strategy for survival in response to apoptosis which incurably leads to cell death. Thus, extracellular signals tightly regulate autophagy and cell death. Similar phenomenon takes place in inflammatory processes and participates to death control of injured cells or infected cells. Whereas cytokines can reduce autophagy, some of them induce it in defined circumstances. This is the case of the tumor necrosis factor-related apoptosis inducing ligand (TRAIL), one member of the TNF-α surperfamily strongly involved in inflammation (48). Similarly, IFN-γ induced cell death by autophagy through a mechanism dependent of Atg5 and its interaction to the Fas-Associated protein with Death Domain (FADD) (49). Autophagy is then sensitive to a great number of growth factors that transduce a variety of signalling molecules (ERK1/2, PI3K, GTPase, etc). In fine, the signals are transduced to a common target which occupies a privileged position in autophagic process. The kinase TOR (Target Of Rapamycin) plays this role in association with its downstream signalling molecules, the Atg kinases (50). In this context, Rapamycin which induces autophagy through the inhibition of TOR, can favor the clearance of aggregating proteins and then may be helpful in ameliorating the response to inflammation. α1-antitrypsin is the archetype of the Serpin supergene family and is the main inhibitor of destructive neutrophil protease (elastase, cathepsin G, proteinase 3). This glycoprotein, considered as an acute-phase reactant, is secreted by liver cells and its concentration increases during the host response to inflammation. Teckman and Perlmutter demonstrated that autophagosomes appear in response to α1-antitrypsin induction and its accumulation in the endoplasmic reticulum (51•). This study suggests that the autophagic response is induced to protect liver cells from the toxic effect of aggregated α1-antitrypsin retained in endoplasmic reticulum (52).
Recently, Shao et al showed that histone deacetylase inhibitors can induce both mitochondria mediated-apoptosis and caspase-independent autophagic cell death (53••). Moreover, Kiffin et al demonstrated the activation of chaperone-mediated autophagy during oxidative stress which clearly occurred during inflammation through i.e. the accumulation of oxidatively damaged proteins (54). Similarly, histone deacetylase inhibitors reduced NO production by peritoneal macrophages, IL-12 secretion by monocytes, without alterations of T-cell-receptor-stimulated IFN-γ production by peripheral blood mononuclear cells (55). These data strengthen the therapeutic potential of histone deacetylase inhibitors as potent contenders for anti-inflammatory drugs (56).
5. Autophagy in infectious diseases is induced by microorganisms but does not succeed in the eradication of the bacterial or viral infections
Microorganism infections lead to the development or local of systemic inflammatory processes. The degradation of undesirable invading microbes, essential for cell survival, involves autophagy. Autophagy appears as a defense mechanism inhibiting the survival of microorganisms (57, 58). Unfortunatly, intracellular bacteria and viruses must survive the antimicrobial responses of their hosts to replicate successfully. Thus, although microorganisms are sensitive to autophagic destruction, pathogens evolved strategies to avoid autophagy (56). After intracellular infection, they modulate the host phagosome to establish an intracellular niche for their survival and replication (59). In many cases, autophagy is dispensable for intracellular replication (60), inducing the cell death and the propagation of the pathogens (61). Other pathogens such as Shigella were found to escape to the autophagy process by secreting specific molecules (62). The identification of interactions occurring between microorganisms and eukaryotic cells is then the key point to develop novel drugs with antimicrobial activities (63).
6. New therapeutic approaches and conclusions
Whereas autophagy appears as a control mechanism of cell protein homeostasis, it also modulates numerous other activities. Particularly, it is likely that autophagy takes part in cell protection against toxicity caused by abnormal proteins or protein accumulations by helping their degradation (Figure 2d, 64), thereby allowing to adapt the immune and cellular responses to the inflammation stress (Figure 2e). Finally, autophagy may result in cell death or/and may correspond to a mechanism of cell survival in stress conditions. In this context the pharmacomodulators of apoptosis affect autophagy processes. Several drugs modulating the different types of cell death are being explored at the preclinical and clinical levels and are promising in the treatment of infectious, cancerous or degenerative pathologies associated with inflammatory process (65–68). Some of them are in clinical development or are approved by the FDA (66••). Thus, mTOR inhibitors induce autophagy and then inhibit the development of solid tumors (68). Similarly, histone deacetylase induce cell death through a mitochondria-mediated apoptosis and a caspase-independent autophagic cell death and then represent a very promising new drug class for the treatment of diseases associated with apoptotic defects (53). These agents act probably as true blockers of cell death and through their anti-inflammatory effects. The identification of molecules involved in autophagy will help a better understanding of this cellular mechanism and lead to the development of new therapeutic strategies.
Acknowledgments
I thank Dr Françoise Rédini and Dr Frédéric Blanchard for their critical comments on this manuscript and Dr Gilbert Pradal for his assistance on electron microscopy.
References
- 1.Baccino FM, Tessitore L, Bonelli G. Control of protein degradation and growth phase in normal and neoplastic cells. Toxicol Pathol. 1984;12:281–287. doi: 10.1177/019262338401200312. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3••.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. A key reference summarizing the works of A. Hershko, A. Ciechanover and I. Rose who received the 2004 Nobel Prize in Chemistry for the discovery of ubiquitin-mediated protein degradation. [DOI] [PubMed] [Google Scholar]
- 4.van Kerkhof P, Govers R, Alves dos Santos CM, Strous GJ. Endocytosis and degradation of the growth hormone receptor are proteasome-dependent. J Biol Chem. 2000;275:1575–1580. doi: 10.1074/jbc.275.3.1575. [DOI] [PubMed] [Google Scholar]
- 5.Li JG, Benovic JL, Liu-Chen LY. Mechanisms of agonist-induced down-regulation of the human k-opionic recetor: internalization is required for down-regulation. Mol Pharmacol. 2000;58:796–801. [PubMed] [Google Scholar]
- 6.Belouzard S, Rouillé Y. Ubiquitilation of leptin receptor OB-Ra regulates its clathrin-mediated endocytosis. Embo J. 2006 doi: 10.1038/sj.emboj.7600989. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols B. Caveosomes and endocytosis of lipid rafts. J Cell Sci. 2003;116:4707–4714. doi: 10.1242/jcs.00840. [DOI] [PubMed] [Google Scholar]
- 8.Nabi IR, Le PU. Caveolae/raft-dependent endocytosis. J Cell Biol. 2003;161 :673–677. doi: 10.1083/jcb.200302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tkachenko E, Lutgens E, Stan RV, Simons M. Fibroblast growth factor 2 endocytosis in endothelial cells proceed via syndecan-4-dependent of Rac1 and a Cdc42-dependent macropinocytic pathway. J Cell Sci. 2004;117:3189–3199. doi: 10.1242/jcs.01190. [DOI] [PubMed] [Google Scholar]
- 10.Fuki IV, Meyer ME, Williams KJ. Transmembrane and cytoplasmic domains of syndecan mediate a multi-step endocytic pathway involving detergent-insoluble membrane rafts. Biochem J. 2000;351:607–612. [PMC free article] [PubMed] [Google Scholar]
- 11.Llorente A, Prydk K, Sprangers M, Sktretting G, Kolset SO, Sandvig K. Proteoglycan synthesis is increased in cells with impaired clathrin-dependent endocytosis. J Cell Sci. 2001;114:335–343. doi: 10.1242/jcs.114.2.335. [DOI] [PubMed] [Google Scholar]
- 12•.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. Overview of the molecular mechanisms of autophagy. [DOI] [PubMed] [Google Scholar]
- 13.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 15.Muller O, Sattler T, Flotenmeyer M, Schwarz H, Plattner H, Mayer A. Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J Cell Biol. 2000;151:519–528. doi: 10.1083/jcb.151.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–2499. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 18.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. An unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 20.Juhasz G, Csikos G, Sinka R, Erdelyi M, Saas M. The drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543:154–158. doi: 10.1016/s0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 21.Melendez A, Talloczy Z, Seamans M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 22.Plomp PJA, Gordon PB, Meijer AJ, Hoyvik H, Seglen PO. Energy dependence of different steps in the autophagic-lysosomal pathway. J Biol Chem. 1989;264:6699–6704. [PubMed] [Google Scholar]
- 23.Thumm M, Enger R, Koch B, Schlumberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 24•.Farré JC, Subramani S. Peroxisome turnover by microperoxyphagy : an autophagy- related process. Trends Cell Biol. 2004;14:515–523. doi: 10.1016/j.tcb.2004.07.014. Overview of the genes involved in autophagy. [DOI] [PubMed] [Google Scholar]
- 25.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABA-RAP and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 26.Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 27••.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. First visualization of autophagosome formation in real time using GFP-autophagy related protein (ATG5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role of Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;111:4837–4838. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 30.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescence autophagosome markers. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. Recent study demonstrating the involvement of autophagy in the maintenance of energy homeostasis after birth. [DOI] [PubMed] [Google Scholar]
- 33.Keller JN, Dimayuga E, Chen Q, Thrope J, Gee J, Ding Q. Autophagy, proteasome, lipofuschine, and oxidative stress in the aging brain. Int J Biochem Cell Biol. 2004;36:2376–2391. doi: 10.1016/j.biocel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon HT-29 cells. J Biol Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 35.Miotto G, Veberando R, khurana KK, Siliprandi N, Mortimore GE. Inhibition of macroautophagy and proteolysis in the isolated rat hepatocyte by a nontransportable derivative of the multiple antigen peptide Leu8-Lys4-Lys2-Lys-beta Ala. J Biol Chem. 1994;269:25348–25353. [PubMed] [Google Scholar]
- 36.Saadane A, Neveux N, Feldmann G, Lardeux B, Bleiderg-Daniel F. Inhibition of liver RNA breakdown during inflammation in the rat. Biochem J. 1996;317:907–912. doi: 10.1042/bj3170907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodgers JR, Rich R. Antigens and antigen presentation. In: Rich R, Fleisher T, Kotzin B, Schroeder H Jr, editors. Clinical Immunology: principles and practice. 2. St Louis: Mosby; 2001. p. 7.1. [Google Scholar]
- 38••.Mimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–1259. doi: 10.1002/eji.200323730. first identification of lysosome as the processing compartment for cytosolic antigens and link endogenous antigen presentation on MHC II with the process protein turnover by autophagy. [DOI] [PubMed] [Google Scholar]
- 39.Dissanayake SK, Tuera N, Ostrand-Rosenberg S. Presentation of endogenously synthesized MHC class II-restricted epitopes by MHC class II cancer vaccines is independent of transporter associated with Ag processing and the proteasome. J Immunol. 2005;174:1811–1819. doi: 10.4049/jimmunol.174.4.1811. [DOI] [PubMed] [Google Scholar]
- 40.Kjeldsen L, Cowland JB, Borregaard N. Human neutrophil gelatinase-associated lipocalin and homologous proteins in rat and mouse. Biochim Biophys Acta. 2000;1482:272–283. doi: 10.1016/s0167-4838(00)00152-7. [DOI] [PubMed] [Google Scholar]
- 41.Cuervo AM, Hildebrand H, Bomhard EM, Dice JF. Direct lysosomal uptake of alpha 2 microglobulin contributes to chemically induced nephropathy. Kidney Int. 1999;55:529–545. doi: 10.1046/j.1523-1755.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- 42.Cardenas-Aguayo MC, Santa-Olalla J, Baizabal JM, Salgado LM, Covarrubias L. Growth factor deprivation induces an alternative non-apoptotic death mechanism that is inhibited by Bcl2 in cells derived from neural precursor cells. J Hematother Stem Cell Res. 2003;12:735–748. doi: 10.1089/15258160360732759. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi H, Kanzawa T, Kondo Y, Kondo S. Inhibition of platelet-derived growth factor signalling induces autophagy in malignant glioma cells. Br J Cancer. 2004;90:1069–1075. doi: 10.1038/sj.bjc.6601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Y, Wang C, Cohen A. Effect of IGF-1 on the balance between autophagy of dysfunctional mitochondria and apoptosis. FEBS Lett. 2004;577:357–360. doi: 10.1016/j.febslet.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 45••.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. demonstration of autophagy control by extracellular signals due to growth factors. [DOI] [PubMed] [Google Scholar]
- 46.Boya P, Gonzales-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimoru T, Codogno P, Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia L, Dourmashkin RR, Allen PD, Gray AB, Newland AC, Kelsey SM. Inhibition of autophagy abrogates tumor necrosis factor alpha induced apoptosis in human T-lymphoblastic leukaemia cells. Br J Haematol. 1997;98:673–685. doi: 10.1046/j.1365-2141.1997.2623081.x. [DOI] [PubMed] [Google Scholar]
- 48•.Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc Natl Acad Sci USA. 2004;101:3438–3443. doi: 10.1073/pnas.0400443101. Demonstration of a cytokine-dependent autophagic pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi BY, Lee H, Kim JH, Mizushima N, Oshumi Y, Jung YK. Essential roles of Atg5 and FADD in autophagic cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 50.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitor for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 51••.Teckman JH, Perlmutter DH. Retention of the mutant secretory protein α1-antitrypsin Z in endosplasmic reticulum induces autophagy. Am J Physiol. 2000;278:G39–G48. doi: 10.1152/ajpgi.2000.278.1.G39. evidence of the protective role of autophagy against toxic aggregated proteins. [DOI] [PubMed] [Google Scholar]
- 52.Permultter DH. Liver injury in α1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J Clin Invest. 2002;110:1579–1583. doi: 10.1172/JCI16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Shao Y, Gao Z, Marks PA, Jiang X. Apoptosis and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. Article open area to the development of novel drugs for the treatment of inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxydative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leoni F, Zaliani A, Bertolini G, Porro G, Pagani P, Pozzi P, Dona G, Fossati G, Sozzani S, Azam T, Bufler P, Fantuzzi G, Goncharov I, Kim SH, Pomerantz BJ, Reznikov LL, Siegmund B, Dinarello CA, Mascagni P. The antitumor histone deacetylase inhibitors suberoylanilide hydroxamic acid exhibits anti-inflammatory properties via suppression of cytokines. Proc Natl Acad Sci USA. 2002;99:2996–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanchard F, Chipoy C. Histone deacetylase inhibitors: new drugs for the treatment of inflammatory diseases? Drug Discov Today. 2005;10:197–204. doi: 10.1016/S1359-6446(04)03309-4. [DOI] [PubMed] [Google Scholar]
- 57.Guttierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 58.Dorn BR, Dunn WA, Progulske-Fox A. Bacterial interactions with the autophagic pathway. Cell Microbiol. 2002;4:1–10. doi: 10.1046/j.1462-5822.2002.00164.x. [DOI] [PubMed] [Google Scholar]
- 59.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nature Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Otto GP, Wu MY, Clarke M, Lu H, Anderson OR, Hilbi H, Shuman HA, Kessin RH. Macroautophagy is dispensable for intracellular replication of Legionella pneumophila in Dictyostelium discoideum. Mol Microbiol. 2004;51:63–72. doi: 10.1046/j.1365-2958.2003.03826.x. [DOI] [PubMed] [Google Scholar]
- 61.Hernandez LD, Pypaert M, Flavell RA, Galan JE. A Salmonella protein causes macrophage cell death by inducing autophagy. J Cell Biol. 2003;163:1123–1131. doi: 10.1083/jcb.200309161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 63.Bera A, Singh S, Nagaraj R, Vaidya T. Induction of autophagic cell death in Leishmania donovani by antimicrobial peptides. Mol Biochem Parasitol. 2003;127:23–35. doi: 10.1016/s0166-6851(02)00300-6. [DOI] [PubMed] [Google Scholar]
- 64.Benhamed MD, Heymann D, Berreur M, Cottrel M, Godard A, Daculsi G, Pradal G. Ultrastructural study of degradation of calcium phosphate ceramic by human monocytes and modulation of this activity by HILDA/LIF cytokine. J Histochem Cytochem. 1996;44:1131–1140. doi: 10.1177/44.10.8813078. [DOI] [PubMed] [Google Scholar]
- 65.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66••.Green DR, Kroemer G. Pharmacological manipulation of cell death: clinical applications in sight? J Clin Invest. 2005;115:2610–2617. doi: 10.1172/JCI26321. exhaustive review summarizing the main pharmacological inducers and repressors of cell death that are FDA approved or in clinical development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson DA, White E. Exploiting different ways to die. Gene Dev. 2004;18:1223–1226. doi: 10.1101/gad.1212404. [DOI] [PubMed] [Google Scholar]
- 68.Raymond E, Alexandre J, Faivre S, Vera K, Materman E, Boni J, Leister C, Korth-Bradley J, Hanauske A, Armand JP. Safety and pharmacokinetics of escalated doses of weekly intravenous infusion of CCI-779, a novel mTOR inhibitor, in patients with cancer. J Clin Oncol. 2004;22:2270–2272. doi: 10.1200/JCO.2004.08.116. [DOI] [PubMed] [Google Scholar]


