Abstract
Phaeobacter gallaeciensis can antagonize fish-pathogenic bacteria in vitro, and the purpose of this study was to evaluate the organism as a probiont for marine fish larvae and their feed cultures. An in vivo mechanism of action of the antagonistic probiotic bacterium is suggested using a non-antagonistic mutant. P. gallaeciensis was readily established in axenic cultures of the two microalgae Tetraselmis suecica and Nannochloropsis oculata, and of the rotifer Brachionus plicatilis. P. gallaeciensis reached densities of 107 cfu/ml and did not adversely affect growth of algae or rotifers. Vibrio anguillarum was significantly reduced by wild-type P. gallaeciensis, when introduced into these cultures. A P. gallaeciensis mutant that did not produce the antibacterial compound tropodithietic acid (TDA) did not reduce V. anguillarum numbers, suggesting that production of the antibacterial compound is important for the antagonistic properties of P. gallaeciensis. The ability of P. gallaeciensis to protect fish larvae from vibriosis was determined in a bath challenge experiment using a multidish system with 1 larva per well. Unchallenged larvae reached 40% accumulated mortality which increased to 100% when infected with V. anguillarum. P. gallaeciensis reduced the mortality of challenged cod larvae (Gadus morhua) to 10%, significantly below the levels of both the challenged and the unchallenged larvae. The TDA mutant reduced mortality of the cod larvae in some of the replicates, although to a much lesser extent than the wild type. It is concluded that P. gallaeciensis is a promising probiont in marine larviculture and that TDA production likely contributes to its probiotic effect.
Introduction
One of the major challenges of marine aquaculture is the continuous and reliable production of juveniles. Severe losses in marine larviculture are caused by infection with opportunistic pathogenic bacteria, including several members of the Vibrionaceae family [1], [2], that accounts for approximately 1.5% of the bacterial community in the oceans [3]. Only some Vibrio species are pathogenic to organisms reared in marine aquaculture and one of the most prominent fish and shellfish pathogens is Vibrio (Listonella) anguillarum that causes serious disease and mortalities [2]. The main source of pathogenic bacteria in marine aquaculture is supply water [4], but also brood stock, humans, or starter cultures are possible sources of pathogens [5]. The majority of marine fish larvae are reared intensively in presence of microalgae (green water), which improves feeding, growth, and survival of the larvae [6]–[9]. The larvae require live feed, and rotifers (Brachionus plicatilis) are typically used as first feed. The rotifers themselves are fed or enriched with live microalgae, such as Tetraselmis suecica, Nannochloropsis oculata, and Isochrysis galbana. Opportunistic pathogens can proliferate in larval feed cultures of phytoplankton and invertebrates due to high concentrations of organic matter. Algae, rotifer and Artemia cultures can therefore harbor high concentrations of pathogenic bacteria [1], [5], [10], [11]. Prophylactic treatment of larvae or their feed cultures with antibiotics can reduce the pathogen load, but has to be avoided, since it leads to emergence of antibiotic-resistant pathogens, and since it impedes the establishment of a normal non-pathogenic microbiota [1], [12], [13].
The potential use of probiotic bacteria to limit outbreaks or effects of bacterial diseases in fish and invertebrate cultures has been investigated for more than two decades. Most studies have focused on the intestinal microbiota [14]–[19], although the use of probiotics is not confined to the intestinal tract of the cultured organisms [15], [20]. Biotic and abiotic surfaces, algal and fecal particles, and the nutrient-rich water serve as habitat and reservoir of opportunistic pathogenic bacteria in cultures of fish larvae or their food organisms [1], [5], [10], [11], [20], and it is hypothesized that competition by non-pathogenic bacteria that are superior in colonizing and persisting in these habitats could reduce the incidence of pathogenic bacteria.
Phaeobacter gallaeciensis (formerly Roseobacter gallaeciensis) is a Gram-negative α-proteobacterium from the Roseobacter-clade [21]. The bacterium produces the antibacterial compound tropodithietic acid (TDA) that is an efficient inhibitor of V. anguillarum and other fish-pathogenic bacteria [22]–[25]. Phaeobacter spp. are commonly isolated from larval cultures of marine fish and shellfish [26]–[28], and do not appear to adversely affect fish larvae [22], [29]. Ruegeria mobilis, a close relative to Phaeobacter also producing TDA, is a cosmopolitan marine bacterium that can be isolated from most ocean waters, apart from polar waters [30].
In a previous study, it was demonstrated that Phaeobacter and Ruegeria isolated from a turbot hatchery [26] could eliminate V. anguillarum in a seawater-based combined liquid-surface system [22]. It was demonstrated, using a TDA-negative mutant, that TDA production was likely a key factor in the pathogen inhibition. The purpose of the present study is to determine if P. gallaeciensis BS107 (DSM 17395) could antagonize V. anguillarum in fish larvae and cultures of their feed organisms. To the authors' knowledge, no study on antagonistic probiotic bacteria has yet elucidated the mechanism of action in vivo. Therefore a non-antagonistic TDA-negative mutant of P. gallaeciensis BS107 (DSM 17395) was created to investigate the in vivo mechanism of action, as emphasized by Tinh et al. [15]. The type strain P. gallaeciensis BS107 (DSM17395) [28] was chosen, since its inhibition of V. anguillarum in Tetraselmis cultures was more pronounced than that of other Phaeobacter and Ruegeria strains, as assessed in a preliminary experiment (data not shown). Gnotobiotic algae and rotifers were used for studying probiotic and nutritional effects of the introduced organism, as recommended [15], [31], [32].
Materials and Methods
Bacterial strains and media
All strains and plasmids are listed in Table 1. Phaeobacter (Roseobacter) gallaeciensis BS107 (DSM17395) was isolated from seawater in scallop (Pecten maximus) cultures [28]. Vibrio anguillarum serotype O1 strain NB10 was used in algal and rotifer experiments. It was isolated from the Gulf of Bothnia and has caused disease in rainbow trout [33], [34]. The strain has been tagged by insertion of plasmid pNQFlaC4-gfp27 (cat, gfp) into an intergenic region on the chromosome, and was kindly provided by D. Milton, University of Umeå [35]. V. anguillarum serotype O2α HI610 was used in challenge trials with cod larvae. The strain was isolated from diseased cod juveniles in the closed seawater basin Lake Parisvatn by the Institute of Marine Research (IMR), Norway, and has been used in challenge trials with cod [36]–[39]. It has been selected from a group of V. anguillarum strains of different serotypes, being the strain that caused the highest mortality in challenge trials with turbot, halibut and cod larvae [40].
Table 1. Bacterial strains and plasmids.
| Strain or plasmid | Genotype or relevant markers | Source or reference |
| Strains | ||
| P. gallaeciensis BS107 (DSM17395) | Wild type | Ruiz-Ponte et al. 1998 [28] |
| P. gallaeciensis BS107-Pda8 | CDS104961::EZ-Tn5, KanR | This study |
| P. gallaeciensis dsRed | MiniTn7(GmR)PA1/04/03 DsRedExpress-a | This study |
| V. anguillarum NB10 | Serotype O1, cmR, PA1/04/03-RBSII-gfpmut3*-T1 | Croxatto et al. 2007 [35] |
| V. anguillarum HI610 | Serotype O2α | Samuelsen & Bergh 2004 [36] |
| Plasmids |
Bacteria from frozen stock cultures (−80°C) were streaked on half-strength Marine Agar (½MA; 27.6 g Difco 212185 Marine Agar, 15 g Instant Ocean Sea Salts, 7.5 g Agar, 1 l deionized water). ½MA was also used for counting P. gallaeciensis. V. anguillarum was counted on Tryptone-Soy Agar (TSA; Oxoid CM0131) containing 6 mg/l chloramphenicol. The bacterial precultures for the algae and rotifer experiments were grown in 20 ml of ½YTSS (2 g Bacto Yeast extract, 1.25 g Bacto Tryptone, 20 g Sigma Sea Salts, 1 l deionized water) [41] at 25°C with aeration (200 rpm) until OD600 = 1.0. The cells were harvested at 5,000 x g, washed twice, and used as inoculum for algae and rotifer experiments. Bacteria were diluted and washed in artificial seawater (ASW; 2% Sigma Sea Salts). Axenicity of algae and rotifer cultures was controlled by plating 100 µl on ½MA and incubating for 7 days at 25°C.
For the challenge trials, V. anguillarum HI610 was grown in tryptone-soy broth with additional 0.5% NaCl at 20°C with shaking at 60 rpm to an OD600 of about 0.5. The P. gallaeciensis strains were grown in MB without shaking at 20°C until stationary phase was reached. All strains were harvested by centrifugation (1,825 x g), washed twice, and resuspended in aerated autoclaved 80% seawater. The bacterial concentrations in these suspensions were determined using a counting chamber for V. anguillarum, and for the P. gallaeciensis strains by measuring OD600 after centrifugation and dissolving in 0.1M NaOH.
Generation of a TDA-negative Phaeobacter mutant
A mutant library of P. gallaeciensis BS107 was created by random transposon insertion mutagenesis using the EZ-Tn5 <R6Kγori/KAN>Tnp Transposome Kit (Epicentre, Madison, WI), following the procedure of Geng et al. [42]. Ten non-pigmented mutants were selected, and absence of TDA production was confirmed by UHPLC-TOFMS and in an agar-diffusion test against V. anguillarum [27]. Growth rates of selected mutants were compared to the wild type in aerated (200 rpm) ½YTSS cultures at 30°C, and one of the strains with a growth rate comparable to the wild type, BS107-Pda8, was chosen for further experiments. Using rescue cloning as described in the transposome kit manual, the mutated locus was identified as CDS104961, which encodes for a “periplasmic component of a TRAP-type C4-dicarboxylate transport system”, as annotated on the BS107 genome on www.roseobase.org.
Fluorescence tagging of Phaeobacter
P. gallaeciensis BS107 was tagged chromosomally with a miniTn7(Gm)PA1/04/03 DsRedExpress-a cassette, using a mini-Tn7 tagging system [43], [44]. The delivery and helper plasmids were electroporated into P. gallaeciensis, followed by selection on ½MA containing 75 µg/ml gentamicin. pPDA11, a transcriptional fusion of the tdaC promoter to a promoterless gfp gene ligated to the broad-host range plasmid pRK415, was constructed in an analogous manner to pHG1011 as described in Geng et al. 2010 [45].
Phaeobacter antagonism in algae
Tetraselmis suecica CCAP 66/4 (Prasinophyceae) was obtained as axenic culture from the Culture Collection of Algae and Protozoa (Oban, UK). It was cultured in B-medium [46], a mineral algae medium, based on ASW. The 250-ml culture bottles were closed with cotton plugs and slowly aerated through a 0.2 µm syringe filter and a silicone tube, to prevent settling of particles. Light intensity on the bottles was 13,000 lux (daylight spectrum). Algal concentrations were assessed by measuring absorption at 665 nm, and calibrating with counts of axenic reference cultures in a Neubauer-improved counting chamber, using formaldehyde as fixative (0.5% final concentration). For each V. anguillarum inoculum level tested, eight bottles of 150 ml of B-medium were inoculated with 6.6×104 cells/ml axenic T. suecica. Two bottles were inoculated with approximately 107 cfu/ml washed P. gallaeciensis BS107, two bottles with the same level of washed mutant P. gallaeciensis BS107-Pda8 cells, and four bottles were left axenic. The cultures were grown for 2 days and axenicity was checked. All cultures, except two axenic negative controls, were inoculated with V. anguillarum NB10 to concentrations of 10, 100, 1000, or 104 cfu/ml. Inoculum levels were verified by plate-counting. Concentrations of algae and both bacterial species were observed until day 5 after inoculation of the pathogen. Two independent experiments were performed for every initial concentration of V. anguillarum. Nannochloropsis oculata CCMP525 (Eustigmatophyceae) was obtained as axenic culture from the Center for Culture of Marine Phytoplankton (West Boothbay Harbor, ME). Since it did not grow in ASW-based B-medium, it was cultured in f/2-medium [47] based on Atlantic Seawater obtained from CCMP. N. oculata cultures were not aerated. Antagonism experiments were done as in T. suecica, but only one initial V. anguillarum concentration (104 cfu/ml) was tested. Two independent experiments with two different initial densities of algae (lower density: 4×106 cells/ml, higher density: 2×107 cells/ml) were carried out.
TDA analysis
Samples of T. suecica – P. gallaeciensis co-cultures (20 ml) were extracted in 50-ml falcon tubes with 30 ml ethyl acetate (HPLC grade) containing 1% formic acid (HPLC grade) on a shaking table for 1 h. The samples were centrifuged at 8,000 x g, and 26 ml of the upper phase was transferred to a new Falcon tube and evaporated to dryness at 35°C with nitrogen flow. The samples were resuspended in 300 µl 85% acetonitrile, vortexed for 5 sec, placed in an ultrasonication bath for 10 min, vortexed again for 5 sec and filtered through a standard 0.22 µm PFTE syringe filter into a HPLC vial. Subsamples of 2 µl were then analyzed by UHPLC-TOFMS on a maXis G3 quadrupole time of flight mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with an electrospray (ESI) ion source which was connected to an Ultimate 3000 UHPLC system (Dionex, Sunnyvale, CA). Separation was performed at 40°C on a 100 mm×2.1 mm ID, 2.6 µm Kinetex C18 column (Phenomenex, Torrance, CA) equipped with Kinetex pre-column using a water-acetonitrile gradient (both buffered with 20 mM formic acid) at a flow of 0.4 ml min−1 starting at 10% acetonitrile and increased to 100% in 10 min, keeping this for 3 min. MS was operated in ESI+ with a data acquisition range of m/z 100–1000 at a resolution of 40,000 FMWH, the MS was calibrated using 20 mM sodium formate infused prior to each analytical run, providing a mass accuracy better than 1.5 ppm. TDA was detected and quantified from the extracted ion chromatograms of the [M+H]+ ions (± m/z 0.001).
For quantification, T. suecica cultures were spiked to a final concentration of 4800, 2400, 1600, 800, 320, 160, and 0 (blank) nM TDA (BioViotica, Dransfeld, Germany) by adding a maximum of 80 µl TDA-acetonitrile solution, and treated as described above. Spiked samples were left at room temperature for at least 1 h prior to extraction. The method was validated on 3 different days using spiked samples as described, and no false positives or negatives were recorded. Relative standard deviation was 30% and the limit of detection was estimated to be <50 nM (signal/noise 5∶1), based on the blank samples and lower calibration points. Sensitivity was greatly influenced by the age of the UHPLC column since TDA tailed (although a new pre-column was used) on columns which had been in use for only a few weeks. Samples from two individual biological experiments were analyzed independently.
Absence of TDA in static cultures of the TDA-negative mutant BS107-Pda8 was confirmed by UHPLC-TOFMS analysis.
Phaeobacter antagonism in rotifers, Brachionus plicatilis
A stock of the rotifer B. plicatilis (L-type) was obtained from Reed Mariculture (Campbell, CA). Axenic rotifers were attained by disinfecting approximately 50 amictic rotifer eggs in 1 ml of a strong antibiotic solution (150 µg/ml Tetracycline, 300 µg/ml Kanamycin, 60 µg/ml chloramphenicol, 1000 U/ml Penicillin in ASW) for 2 days. The hatched rotifers were filtered onto a sterile 50-µm polyamide mesh, rinsed with ASW and from then on fed with concentrated axenic T. suecica. No experiment with N. oculata as rotifer feed was conducted, since survival of V. anguillarum in presence of N. oculata was very variable (see Results section). Rotifer densities were determined by counting in a Sedgewick-Rafter counting chamber. Before counting, the cultures were thoroughly mixed, and 100 µl 5% formaldehyde added to a 1 ml sample. To set up co-culture experiments, axenic rotifer cultures were divided into eight 20-ml batches in 50-ml centrifuge tubes. The initial rotifer concentrations were 94 /ml (first replicate) and 30 /ml (second replicate). For each replicate, duplicate cultures were inoculated with washed wild type and mutant P. gallaeciensis at approximately 5×107 cfu/ml. On the next day, all cultures except the axenic controls were inoculated with 104 cfu/ml V. anguillarum. All rotifer cultures were fed daily with 10-fold concentrated T. suecica (1–2 ml depending on average rotifer density), so that the algae concentration was at approximately 106 cells/ml after feeding and did not drop below 2×105 cells/ml. The rotifers were counted daily, concentrations of V. anguillarum and P. gallaeciensis were determined, and axenicity of the negative controls was checked. The rotifer culture samples for enumeration of bacteria were homogenized by grinding and repeated pipetting through a 100-µl pipette tip. This was compared to homogenization with an Ultra-Turrax T25 (IKA, Germany) at 16,000 rpm and no significant differences in bacterial counts were found (P = 0.74).
Challenge trial
The protocol was adapted from [39], [40]. Cod (Gadus morhua) embryos were obtained from the commercial hatchery Havlandet AS, in Florø, Western Norway. Transport of the embryos in polystyrene containers at around 8°C took 4 to 5 hours in total by boat and car. Two independent replicates of the challenge trial were conducted. The embryos used in the first trial were disinfected with Buffodine (Evans Vanodine, Preston, UK), the embryos for the second trial were left untreated. Upon arrival, the embryos were randomly picked and distributed to the wells of 24-well dishes (Nunc, Roskilde, Denmark) filled with 2 ml 80% autoclaved, aerated seawater, placing one embryo in each well. In each trial three dishes for each treatment (72 embryos) were prepared and inoculated immediately. The six treatment groups are listed in Table 2. All inocula were prepared in a volume of 100 µl, and the strains were not mixed before inoculation. Initial bacterial concentrations were 1×106 cfu/ml for V. anguillarum HI610 and about 107 cfu/ml for the P. gallaeciensis strains. The plates were incubated in the dark at 7°C. The day when 50% of the larvae had hatched was defined as day 0, which was 6 days after the start of the experiment. Dead larvae were registered daily for 14 days.
Table 2. Group numbers and treatments in the challenge trial.
| Group number | Treatment |
| T1 | Negative control; no bacteria added |
| T2 | Positive control; V. anguillarum O2α HI610 106 cfu/ml |
| T3 | Wild type P. gallaeciensis BS107 (DSM17395) ∼107 cfu/ml |
| T4 | TDA-mutant P. gallaeciensis BS107-Pda8 ∼107 cfu/ml |
| T5 | V. anguillarum O2α HI610 106 cfu/ml and wild type P. gallaeciensis BS107 (DSM17395) ∼107 cfu/ml |
| T6 | V. anguillarum O2α HI610 106 cfu/ml and TDA-mutant P. gallaeciensis BS107-Pda8 ∼107 cfu/ml |
Statistics
Differences between concentrations of bacteria or algae were assessed using repeated measures ANOVA after log-transformation. Tukey's multiple comparison test was used for pairwise comparisons. To address the effects of P. gallaeciensis presence on concentrations of V. anguillarum, initial values (day 0) were omitted in the analysis and the experiments were analyzed separately. Rotifer numbers were not log-transformed before applying ANOVA, and initial values were omitted. Numbers of P. gallaeciensis and homogenization methods were compared using paired t-tests after log-transformation.
The cumulative mortalities in the challenge trials were compared at day 10, prior to the onset of starvation towards the end of the experiment. A chi-square test for 22 contingency tables was implemented, using the software R, version 2.13.1 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Antagonism in algae cultures
Both wild type and mutant P. gallaeciensis colonized the cultures of T. suecica and N. oculata. In T. suecica cultures, P. gallaeciensis reached 107 cfu/ml (Figure 1). In the dense cultures of N. oculata, P. gallaeciensis concentrations were approximately 5×106 cfu/ml, and in the less dense cultures approximately 8×105 cfu/ml (Figure S1). The wild type Phaeobacter reached slightly higher numbers in T. suecica than the TDA-negative mutant (P = 0.0211). This same slight difference was seen in one of the two Nannochloropsis experiments (P = 0.0335, P = 0.9259). P. gallaeciensis did not affect growth of the algae T. suecica (P = 0.9977) and N. oculata (P = 0.9919). Particles consisting of dead T. suecica and algal cell walls that were shed during cell division served as habitat for rosette forming P. gallaeciensis that formed dense biofilms on the particles (Figure 2 A–D).
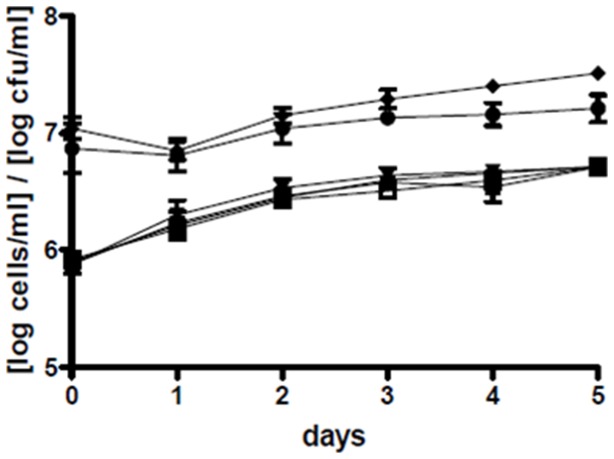
Figure 1. Concentrations of Tetraselmis suecica and Phaeobacter gallaeciensis in the co-cultures.
Means and standard deviations of eight experiments: colony-forming units of P. gallaeciensis wild type (♦) and the TDA-negative mutant (•), and concentrations of T. suecica with V. anguillarum (▾), T. suecica with P. gallaeciensis wild type (▪), T. suecica with P. gallaeciensis TDA-negative mutant (▴), and axenic T. suecica (□). The P. gallaeciensis strains were inoculated at 107 cfu/ml and remained as a steady population, while the algae went from late log into stationary phase.
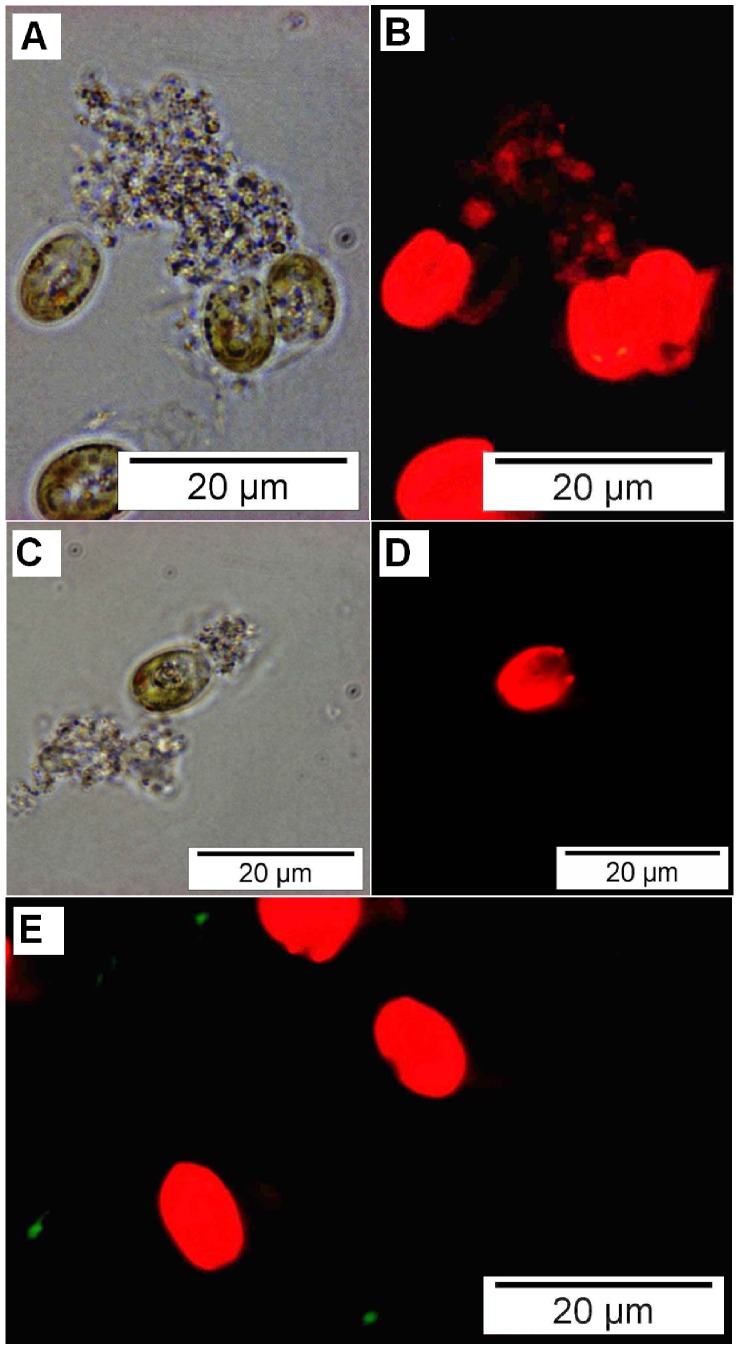
Figure 2. Localization of bacteria in cultures of Tetraselmis suecica.
Phase-contrast (A,C) and fluorescence (B,D,E) micrographs. Co-culture of Tetraselmis suecica with Phaeobacter gallaeciensis dsRed (A,B), axenic T. suecica (C,D), co-culture of T. suecica with V. anguillarum gfp (E). Panel A and B show two single (left) and one dividing algal cell (right side), and a marine snow-like particle consisting of algae-debris which is colonized by red-fluorescent P. gallaeciensis. Red fluorescence of algae is due to chlorophyll. Panels C and D show an algal cell and particles from an axenic culture, recorded using the same settings as for the panels above. Panel E shows red-fluorescent algae cells and green-fluorescent V. anguillarum, which do not colonize particles, but remain in suspension as single, motile cells.
V. anguillarum effectively colonized all Tetraselmis cultures that were not inoculated with P. gallaeciensis and numbers increased by up to 2.7 log units within the first day (Figure 3, Figure S2) and reached an average of 3×106 cfu/ml after 5 days. V. anguillarum did not colonize particles in the algae cultures, but remained in suspension (Figure 2 E). The numbers of V. anguillarum decreased markedly in presence of wild type P. gallaeciensis (Figure 3, Figure S2). Vibrio reductions were in the order of 3 log units, as compared to the monoxenic controls with only V. anguillarum, and complete elimination of the lowest Vibrio inoculum was achieved in 3 out of 4 replicates (Figure 3A). The effects of wild type P. gallaeciensis on V. anguillarum were highly significant in all Tetraselmis experiments, as compared to the controls (P<0.001) and to the mutant (P<0.001). Presence of the TDA-negative mutant did decrease concentrations of the pathogen by about one log unit, although this was only significant (α = 0.05) for two of the initial Vibrio concentrations (101: P = 0.0518, 102: P = 0.0011, 103: P = 0.0517, 104: P = 0.0008).
Figure 3. Reduction of V. anguillarum in cultures of Tetraselmis suecica by Phaeobacter gallaeciensis.
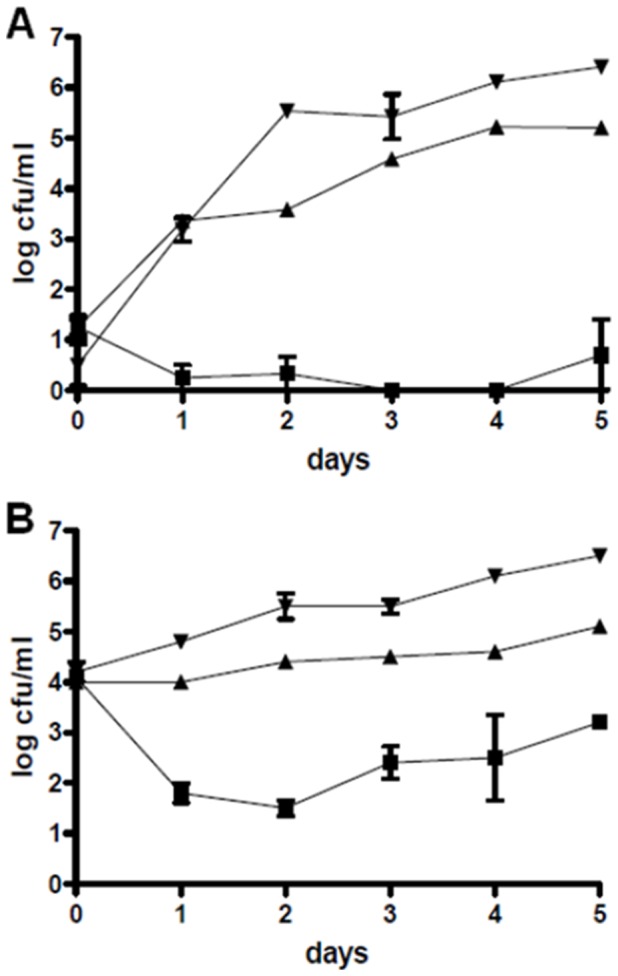
Colony-forming units of V. anguillarum inoculated at 101 cfu/ml (A) and at 104 cfu/ml (B) in presence of P. gallaeciensis wild type (▪), in presence of the P. gallaeciensis TDA-negative mutant (▴), and in the monoxenic control (▾).
The marked difference in V. anguillarum inhibition by the wild type P. gallaeciensis and the TDA negative mutant suggested that TDA was a major effector molecule. However, TDA was not detected by chemical analysis of the Phaeobacter–Tetraselmis co-cultures, where triplicate cultures were each analyzed in triplicates with a detection limit <50 nM TDA. The experiment and analysis were repeated with the same result. To determine if the wild type did indeed produce TDA in the algal cultures, a P. gallaeciensis carrying pPDA11 (tdaCp::gfp) was co-cultured with T. suecica. The tdaC promoter, indicative of TDA production, was induced when growing on particles in a T. suecica culture, as indicated by Gfp fluorescence (Figure 4). Adding pure TDA to Tetraselmis cultures inoculated with V. anguillarum caused a complete killing of the Vibrio population, but also affected survival of algae (50 µM TDA). A 50-fold lower concentration (1 µM) had no effect on the algae, but temporarily reduced V. anguillarum below 10 cfu/ml. A concentration of 50 nM TDA, which was the detection limit of the chemical analysis, did not have any effect (data not shown).
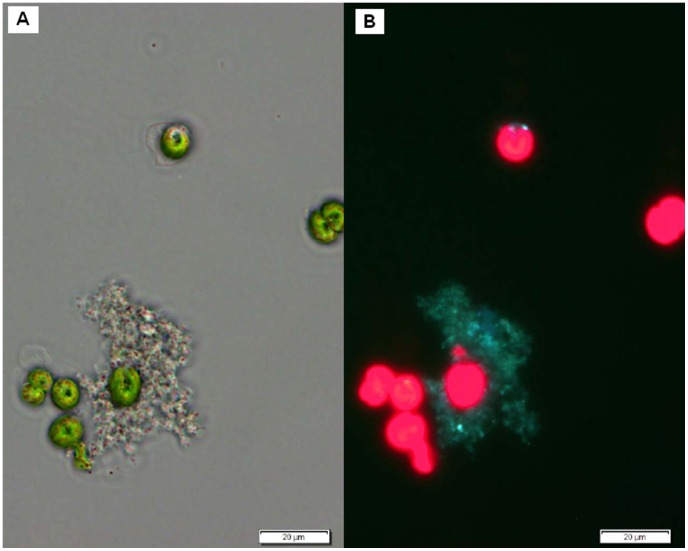
Figure 4. Expression of tdaC in co-culture with Tetraselmis suecica.
Phase contrast (A) and fluorescence (B) micrographs of P. gallaeciensis pPDA11 (tdaCp::gfp) in co-culture with T. suecica. The two panels show the same seven algal cells of which some are dividing, and a marine snow-like particle which is colonized by P. gallaeciensis carrying the promoter-fusion on a plasmid. The green fluorescence of P. gallaeciensis on the particle shows that the gfp gene is expressed from the tdaC promoter, indicating production of TDA.
V. anguillarum was completely eliminated in Nannochloropsis oculata cultures by wild type P. gallaeciensis within one or two days (Figure S3). However, V. anguillarum could only persist in dense cultures of N. oculata. In less dense N. oculata cultures V. anguillarum disappeared from the monoxenic control within 3 days. Consequently, the effect of the wild type P. gallaeciensis on V. anguillarum, as compared to the control, was significant in the experiment with high algae density (P = 0.001), but not in low density (P = 0.2106).
Antagonism in rotifer cultures
The concentrations of P. gallaeciensis and its mutant in the rotifer cultures were stable at about 106–107 cfu/ml, and no significant difference between the two strains was observed (P = 0.3689). The rotifers grew faster and reached higher densities in presence of P. gallaeciensis than in the axenic or monoxenic (V. anguillarum) controls (P<0.05) (Figure 5, Figure S4). Wild type P. gallaeciensis reduced V. anguillarum concentrations by 3 log units (P<0.01), in average from 6×105 to 9×102 cfu/ml (Figure 6). The effect of the TDA-negative mutant on the concentration of the pathogen was not significant (P>0.05).
Figure 5. Influence of bacterial strains on rotifer growth.
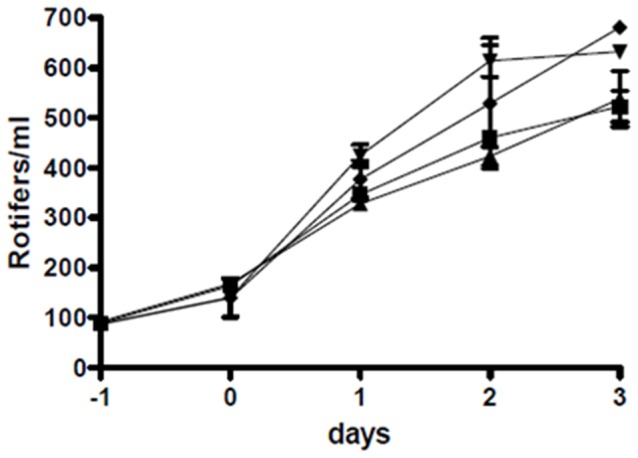
Rotifer numbers in co-culture with P. gallaeciensis wild type (▾), with the TDA-negative mutant of P. gallaeciensis (♦), with only V. anguillarum (▴), and axenic rotifers (▪), first experiment. All bacteria were inoculated at day 0. Both P. gallaeciensis strains promoted rotifer growth, whereas V. anguillarum had no influence.
Figure 6. Reduction of Vibrio anguillarum by Phaeobacter gallaeciensis in rotifer cultures.
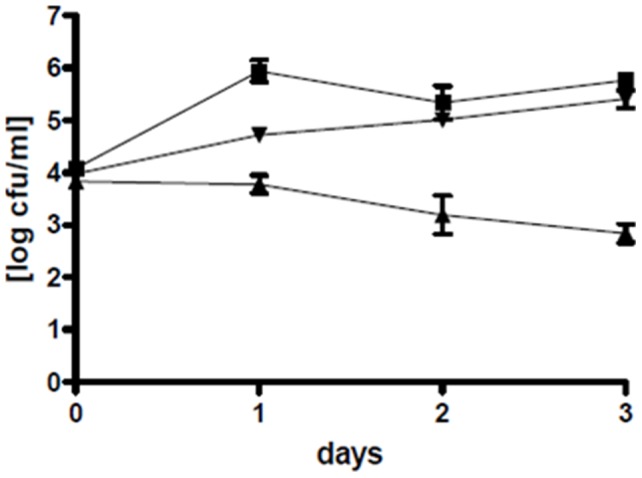
Mean values of two duplicate experiments: colony-forming units of V. anguillarum in co-culture with P. gallaeciensis wild type (▴), with the TDA-negative mutant of P. gallaeciensis (▾), and in the monoxenic control (▪).
Challenge trial
Six days after the arrival of the embryos and inoculation, more than 50% of the larvae had hatched. Total cumulative hatching success was 79.2% (first trial 79.6% and second trial 78.7%). The initial mortality was lower in the first trial (16.6%) than in the second (24.8%). In the non-challenged and non-treated control, 34.7%±9.8% (average ± standard deviation) of the larvae had died by day 1, yet after the initial mortality only 2.8%±0% of the larvae died between day 2 and day 10, reaching an accumulated mortality of 37.5%±9.8% at day 10. The larvae challenged with V. anguillarum HI610 died rapidly and reached 100%±0% accumulated mortality. Treating Vibrio-challenged larvae with wild type P. gallaeciensis caused a significant reduction in accumulated mortality by day 10 to 12.5%±2.0% which was not only lower than in the challenged larvae but also lower than in the non-challenged (37.5%). 96.1%±1.1% of the hatched larvae that had received wild type P. gallaeciensis survived until day 10, when starvation set in (Figure 7, Figure S5). The larvae exposed to only P. gallaeciensis wild type or mutant had a cumulative mortality of 12.1%±3.1% at day 10. The TDA-negative mutant of P. gallaeciensis did reduce accumulated mortality of the challenged larvae to 68.8%±30.4% (Figure 7, Figure S5), but was not nearly as efficient as the TDA-producing wild type.
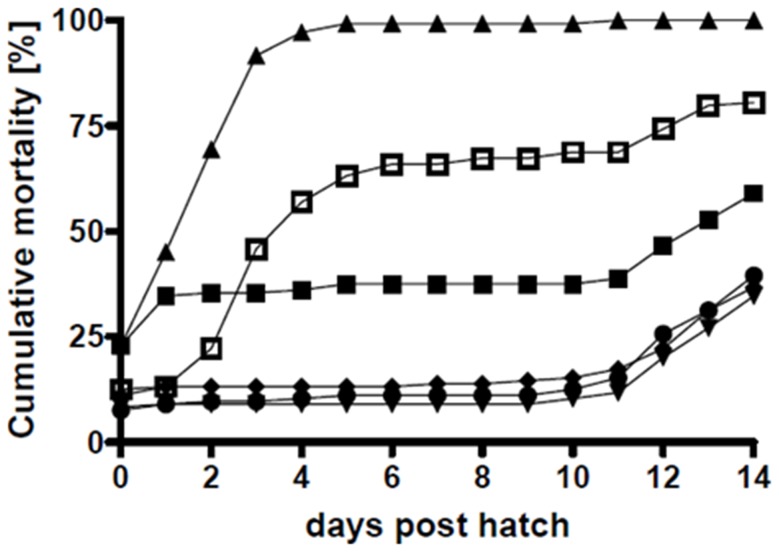
Figure 7. Mortality of cod larvae during the challenge trials.
Mean values of two independent triplicate experiments. The single-larvae cultures were simultaneously inoculated with P. gallaeciensis wild type and V. anguillarum (T5, •), or with the TDA-negative mutant of P. gallaeciensis and V. anguillarum (T6, □). Unexposed larvae and larvae exposed to single bacterial strains acted as controls: Negative Control (T1, ▪), only V. anguillarum (T2, ▴), only P. gallaeciensis wild type (T3, ▾), and only P. gallaeciensis TDA-negative mutant (T4, ♦).
Discussion
The present study demonstrates that Phaeobacter gallaeciensis is harmless and beneficial for the early life stages of cod. Equally important, P. gallaeciensis is highly efficient at preventing infections with V. anguillarum, and this probiotic effect can be achieved at the low temperature (7°C) used for the cod embryos and yolk sac larvae. It has previously been demonstrated that a Phaeobacter sp. can protect turbot larvae against vibriosis at higher temperatures (18°C) [29]. Non-infected larvae showed some level of initial mortality, which may have been due to opportunistic bacteria introduced with the embryos. Both challenged and unchallenged cod larvae exposed to P. gallaeciensis had a significantly lower initial mortality, indicating that the inherently occurring microbiota of the chorion may be controlled by the probiont.
A key question in the use of probiotics in aquaculture is how and where the probiont should be introduced to the system. Several studies have emphasized the potential role of feed organisms as a vehicle for probiotic bacteria [29], [48]–[51], or the potential of probiotic bacteria to control pathogenic bacteria in the feed cultures [51]–[53]. The majority of studies have focused on intestinal probiotic bacteria, and aimed at health-promoting effects within either the reared animal or the feed organism. In contrast, the present study takes a systems approach to preventing bacterial disease in aquaculture organisms, aiming at microbial control throughout the environment of the reared organism and the lower trophic levels of the production. Here it was found that cultures of two aquaculture-relevant algae and of the rotifer B. plicatilis can be colonized by P. gallaeciensis without compromising their growth, and that P. gallaeciensis in these cultures will strongly reduce, or eliminate fish-pathogenic V. anguillarum. Introducing P. gallaeciensis at this trophic level is very promising, since live feed is a common source of opportunistic pathogens [1], [5], [10], [11]. These findings corroborate the hypothesis from a previous study, that algae and rotifers in aquaculture can be cultured together with probiotic Roseobacters, and thus prevent proliferation of pathogens [22]. A reduction of a pathogenic Vibrio sp. by 3 log units, as it was achieved in the present study, is very promising in terms of larval health promotion, as only a one log reduction of the bacterial load in rotifers through UV radiation resulted in higher survival of turbot larvae [54]. Using probiotic bacteria, as compared to UV treatment, offers the advantage that nutrients are consumed, niches are occupied, and rapid re-growth of pathogens is prevented. It should be noted that the present study was done using gnothobiotic systems to rule out the influence of the inherent microbiota of algae and rotifer cultures. Thus, it cannot be determined, if or to what extent P. gallaeciensis would affect the inherent microbial communities of algae and rotifer cultures.
The inhibition of V. anguillarum by a Phaeobacter sp. in a model aquaculture setting has been studied once before: Planas et al. [29] demonstrated that mortality in turbot larvae infected with V. anguillarum could be reduced by Phaeobacter sp. 27-4. A duplicate tank setup was used, and both the pathogen and the probiont were enclosed in rotifers and fed to the larvae. In spite of delivery with the feed, the probiont was only found in the lumen of the larval gut and did not colonize the intestinal epithelium. In contrast to this, the present study did not aim at a probiotic effect in the intestinal tract of the larvae, but assesses the potential of Phaeobacter to eliminate the pathogen in the environment of the larvae or embryos. It should be mentioned that, as larvae start to drink shortly after hatching [55], an intestinal presence of pathogens and probionts can occur. Phaeobacter sp. 27-4 is a TDA-producer, however, as opposed to P. gallaeciensis BS107, it produces TDA only in stagnant culture [26], suggesting that its TDA production may be more delimited and that BS107 could be more antagonistic in vivo.
In the challenge trial the TDA-negative mutant reduced the initial mortality as efficiently as the wild type, but could not prevent infection in the majority of the larvae. The probiotic effect of the mutant could be explained by competition for nutrients, space, or other resources or it could be attributed to a direct immunostimulatory effect on the larvae [56]–[59]. The mutated gene that renders P. gallaeciensis BS107-Pda8 unable to produce TDA belongs to an operon encoding the parts of a transport protein, which has not yet been reported to be involved in TDA production [42]. The role of this transmembrane protein in TDA production has not been investigated. It cannot be excluded that this mutation has pleiotropic phenotypic effects, and other functions than TDA production might be affected and could affect the antagonistic properties of Phaeobacter gallaeciensis. Nonetheless, the experiment using pure TDA indicated that this compound indeed has a major inhibitory effect against V. anguillarum in the algal system.
The difference in Vibrio-antagonism between the TDA-negative mutant and the wild type suggested that TDA production is the trait that enables P. gallaeciensis to antagonize V. anguillarum. However, TDA was not detectable by chemical analysis of the Phaeobacter-Tetraselmis co-cultures. Since a tdaC-promoter fusion (tdaCp::gfp), demonstrated that tdaC is expressed by P. gallaeciensis in particles in the algae cultures, the reason for the lack of chemical detection could be that the TDA concentration only reaches inhibitory concentrations in Phaeobacter-colonized particles. TDA is likely concentrated within and around the particles, adhering to organic mass of the particle, or being kept within the EPS produced by P. gallaeciensis. From an ecological point of view, for a particle-associated marine bacterium the production of an antagonistic compound would be more efficient if the compound was not dispersed, but kept in the close vicinity to fend off possible competitors.
Although TDA-producing Phaeobacter and Ruegeria spp. are likely to be already present in larviculture systems, their antagonistic properties, which may depend on growth conditions, are probably different from P. gallaeciensis BS107 [22], [26]. A preliminary experiment to this study showed that only a few of the tested Phaeobacter and Ruegeria strains were antagonistic in T. suecica cultures, whereas all of them did account for large inhibition zones in agar-based assays. Therefore, introduction of P. gallaeciensis BS107 in algae and rotifer cultures would likely enhance larval survival even though other Roseobacters are already present in the system. Its growth-promoting effect on rotifers may offer an unexpected additional advantage. Whether that is due to the nutritive value of the bacteria or to a potential role in the rotifer gut is not known. In the present study rotifer growth was not adversely affected by V. anguillarum. Nevertheless, a V. anguillarum strain was reported to cause pronounced growth inhibition of rotifers under suboptimal feeding schemes [60], which could possibly be remediated by P. gallaeciensis.
It cannot be predicted, if and how other pathogens in algae and rotifer cultures would be suppressed by P. gallaeciensis, however a range of fish pathogens are inhibited in vitro by P. gallaeciensis [22], [25] indicating that it likely could protect against other pathogens than V. anguillarum. Porsby et al. [61] have addressed the concern that resistance to TDA could develop, and found, using several experimental approaches, that no resistant mutants or variants could be isolated, neither from short-term selection cultures containing different concentrations of TDA nor from long-term adaptation cultures (>300 generations) containing increasing concentrations of TDA.
A recent study demonstrated that P. gallaeciensis, when incubated with p-coumaric acid, produced potent algicides, the roseobactins, which were effective against different microalgae, among them T. suecica [62]. P-coumaric acid is a degradation product of lignin, which is contained not only in terrestrial plants, but also in algae. Production of the algicides was only possible in concentrations of p-coumaric acid above 0.4 mM. The authors hypothesized that P. gallaeciensis contributes to algal health and growth by secreting TDA and phenylacetic acid, but will produce algicides in presence of p-coumarate, which is an indicator for algal senescence, in order to utilize the algal biomass for its own growth [62]. However, in the present study no negative effect of P. gallaeciensis on algal growth was observed. Possibly the levels of p-coumaric acid in the cultures of microalgae were too low for roseobacticide production. The environmental ecological niche of P. gallaeciensis has not yet been described, although studies of Rao et al. [63]–[65] indicate its preference for the surface of macroalgae, which during their decay may account for higher local concentrations of p-coumaric acid than microalgae. In an aquaculture farm, Phaeobacter spp. have been found to “naturally” occur on solid surfaces, whereas only Ruegeria spp. were inherently associated with algae cultures [26].
Based on the present findings, it is hypothesized that P. gallaeciensis can be used in marine larviculture, as a means of controlling the ambient, potentially harmful microbiota in cultures of rotifers and microalgae, and as a prophylaxis against vibriosis in fish larvae.
Supporting Information
Concentrations of Nannochloropsis oculata and Phaeobacter gallaeciensis in the co-cultures. Colony-forming units of P. gallaeciensis wild type (•) and the TDA-negative mutant (□), and concentrations of N. oculata with V. anguillarum (▴), N. oculata with P. gallaeciensis wild type (▾), N. oculata with P. gallaeciensis TDA-negative mutant (♦), and axenic N. oculata (▪) in the dense (A) and less dense (B) cultures.
(TIF)
Reduction of Vibrio anguillarum by Phaeobacter gallaeciensis in cultures of Tetraselmis suecica . Colony-forming units of V. anguillarum inoculated at 102 cfu/ml (A) and at 103 cfu/ml (B) in presence of P. gallaeciensis wild type (▪), in presence of the P. gallaeciensis TDA-negative mutant (▴), and in the monoxenic control (▾).
(TIF)
Reduction of Vibrio anguillarum by Phaeobacter gallaeciensis in cultures of Nannochloropsis oculata . Colony-forming units of V. anguillarum in presence of P. gallaeciensis wild type (▴), in presence of the P. gallaeciensis TDA-negative mutant (▾), and in the monoxenic control (▪), in dense (3×107 cells/ml; A) and less dense (1–7×106 cells/ml; B) cultures of N. oculata.
(TIF)
Influence of bacterial strains on rotifer growth. Rotifer numbers in co-culture with P. gallaeciensis wild type (▾), with the TDA-negative mutant of P. gallaeciensis (♦), with only V. anguillarum (▴), and axenic rotifers (▪), second experiment. All bacteria were inoculated at day 0. Both P. gallaeciensis strains promoted rotifer growth, whereas V. anguillarum had no influence.
(TIF)
Mortality of cod larvae during the challenge trials. Mean values of two independent triplicate experiments with error bars indicating standard deviations. The single-larvae cultures were simultaneously inoculated with P. gallaeciensis wild type and V. anguillarum (T5, •), or with the TDA-negative mutant of P. gallaeciensis and V. anguillarum (T6, □). Unexposed larvae and larvae exposed to single bacterial strains acted as controls: Negative Control (T1, ▪), only V. anguillarum (T2, ▴), only P. gallaeciensis wild type (T3, ▾), and only P. gallaeciensis TDA-negative mutant (T4, ♦).
(TIF)
Acknowledgments
We would like to thank Debra Milton, University of Umeå, for providing the chromosomally tagged Vibrio anguillarum NB10. We also thank Paul Henning Løvik for assistance with the challenge experiments.
Funding Statement
This study was funded by the Danish Research Council for Technology and Production to the project 09-066524 (Bioactive bacterial biofilm surfaces in aquaculture – disease prevention without antibiotics). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Olafsen JA (2001) Interactions between fish larvae and bacteria in marine aquaculture. Aquacult 200: 223–247. [Google Scholar]
- 2. Toranzo AE, Magarinos B, Romalde JL (2005) A review of the main bacterial fish diseases 3. in mariculture systems. Aquacult 246: 37–61. [Google Scholar]
- 3. Wietz M, Gram L, Jorgensen B, Schramm A (2010) Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat Microb Ecol 61: 179–189. [Google Scholar]
- 4. Douillet PA, Pickering PL (1999) Seawater treatment for larval culture of the fish Sciaenops ocellatus Linnaeus (red drum). Aquacult 170: 113–126. [Google Scholar]
- 5. Eddy SD, Jones SH (2002) Microbiology of summer flounder Paralichthys dentatus fingerling production at a marine fish hatchery. Aquacult 211: 9–28. [Google Scholar]
- 6. Salvesen I, Reitan KI, Skjermo J, Oie G (2000) Microbial environments in marine larviculture: Impacts of algal growth rates on the bacterial load in six microalgae. Aquacult Int 8: 275–287. [Google Scholar]
- 7. Conceicao LEC, Yufera M, Makridis P, Morais S, Dinis MT (2010) Live feeds for early stages of fish rearing. Aquaculture Research 41: 613–640. [Google Scholar]
- 8. Naas KE, Naess T, Harboe T (1992) Enhanced 1St Feeding of Halibut Larvae (Hippoglossus hippoglossus L) in Green Water. Aquacult 105: 143–156. [Google Scholar]
- 9. Skjermo J, Vadstein O (1999) Techniques for microbial control in the intensive rearing of marine larvae. Aquacult 177: 333–343. [Google Scholar]
- 10. Munro PD, Barbour A, Birkbeck TH (1995) Comparison of the Growth and Survival of Larval Turbot in the Absence of Culturable Bacteria with Those in the Presence of Vibrio anguillarum, Vibrio alginolyticus, Or A Marine Aeromonas Sp. Appl Environ Microbiol 61: 4425–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reid HI, Treasurer JW, Adam B, Birkbeck TH (2009) Analysis of bacterial populations in the gut of developing cod larvae and identification of Vibrio logei, Vibrio anguillarum and Vibrio splendidus as pathogens of cod larvae. Aquacult 288: 36–43. [Google Scholar]
- 12. Cabello FC (2006) Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Env Microbiol 8: 1137–1144. [DOI] [PubMed] [Google Scholar]
- 13. Skjermo J, Salvesen I, Oie G, Olsen Y, Vadstein O (1997) Microbially matured water: A technique for selection of a non-opportunistic bacterial flora in water that may improve performance of marine larvae. Aquacult Int 5: 13–28. [Google Scholar]
- 14. Vine NG, Leukes WD, Kaiser H (2006) Probiotics in marine larviculture. FEMS Microb Rev 30: 404–427. [DOI] [PubMed] [Google Scholar]
- 15. Tinh NTN, Dierckens K, Sorgeloos P, Bossier P (2008) A review of the functionality of probiotics in the larviculture food chain. Mar Biotechnol 10: 1–12. [DOI] [PubMed] [Google Scholar]
- 16. Wang YB, Li JR, Lin JD (2008) Probiotics in aquaculture: Challenges and outlook. Aquacult 281: 1–4. [Google Scholar]
- 17. Kesarcodi-Watson A, Kaspar H, Lategan MJ, Gibson L (2008) Probiotics in aquaculture: The need, principles and mechanisms of action and screening processes. Aquacult 274: 1–14. [Google Scholar]
- 18. Irianto A, Austin B (2002) Probiotics in aquaculture. J? Fish Dis 25: 633–642. [Google Scholar]
- 19. Balcazar JL, de Blas I, Ruiz-Zarzuela I, Cunningham D, Vendrell D, et al. (2006) The role of probiotics in aquaculture. Veterinary Microbiology 114: 173–186. [DOI] [PubMed] [Google Scholar]
- 20. Verschuere L, Rombaut G, Sorgeloos P, Verstraete W (2000) Probiotic bacteria as biological control agents in aquaculture. Microbiol Mol Biol Rev 64: 655–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martens T, Heidorn T, Pukall R, Simon M, Tindall BJ, et al. (2006) Reclassification of Roseobacter gallaeciensis Ruiz-Ponte, et al. 1998 as Phaeobacter gallaeciensis gen. nov., comb. nov., description of Phaeobacter inhibens sp nov., reclassification of Ruegeria algicola (Lafay, et al. 1995) Uchino, et al 1999 as Marinovum algicola gen. nov., comb. nov., and emended descriptions of the genera Roseobacter, Ruegeria and Leisingera . International Journal of Systematic and Evolutionary Microbiology 56: 1293–1304. [DOI] [PubMed] [Google Scholar]
- 22. D'Alvise PW, Melchiorsen J, Porsby CH, Nielsen KF, Gram L (2010) Inactivation of Vibrio anguillarum by Attached and Planktonic Roseobacter Cells. Appl Environ Microbiol 76: 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bruhn JB, Nielsen KF, Hjelm M, Hansen M, Bresciani J, et al. (2005) Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade. Appl Environ Microbiol 71: 7263–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruhn JB, Gram L, Belas R (2007) Production of antibacterial compounds and biofilm formation by Roseobacter species are influenced by culture conditions. Appl Environ Microbiol 73: 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prado S, Montes J, Romalde JL, Barja JL (2009) Inhibitory activity of Phaeobacter strains against aquaculture pathogenic bacteria. Int Microbiol 12: 107–114. [PubMed] [Google Scholar]
- 26. Porsby CH, Nielsen KF, Gram L (2008) Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl Environ Microbiol 74: 7356–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hjelm M, Bergh O, Riaza A, Nielsen J, Melchiorsen J, et al. (2004) Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst Appl Microbiol 27: 360–371. [DOI] [PubMed] [Google Scholar]
- 28. Ruiz-Ponte C, Cilia V, Lambert C, Nicolas JL (1998) Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus . Int? J? System Bacteriol 48: 537–542. [DOI] [PubMed] [Google Scholar]
- 29. Planas M, Perez-Lorenzo M, Hjelm M, Gram L, Fiksdal IU, et al. (2006) Probiotic effect in vivo of Roseobacter strain 27-4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquacult 255: 323–333. [Google Scholar]
- 30. Gram L, Melchiorsen J, Bruhn JB (2010) Antibacterial Activity of Marine Culturable Bacteria Collected from a Global Sampling of Ocean Surface Waters and Surface Swabs of Marine Organisms. Mar Biotechnol 12: 439–451. [DOI] [PubMed] [Google Scholar]
- 31. Tinh NTN, Phuoc NN, Dierckens K, Sorgeloos P, Bossier P (2006) Gnotobiotically grown rotifer Brachionus plicatilis sensu strictu as a tool for evaluation of microbial functions and nutritional value of different food types. Aquacult 253: 421–432. [Google Scholar]
- 32. Martinez-Diaz SF, Varez-Gonzalez CA, Legorreta MM, Vazquez-Juarez R, Barrios-Gonzalez J (2003) Elimination of the associated microbial community and bioencapsulation of bacteria in the rotifer Brachionus plicatilis . Aquacult Int 11: 95–108. [Google Scholar]
- 33. Norqvist A, Norrman B, Wolfwatz H (1990) Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum . Infect Immun 58: 3731–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Norqvist A, Hagstrom A, Wolfwatz H (1989) Protection of rainbow trout against vibriosis and furunculosis by the use of attenuated strains of Vibrio anguillarum . Appl Environ Microbiol 55: 1400–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Croxatto A, Lauritz J, Chen C, Milton DL (2007) Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Env Microbiol 9: 370–382. [DOI] [PubMed] [Google Scholar]
- 36. Samuelsen OB, Bergh O (2004) Efficacy of orally administered florfenicol and oxolinic acid for the treatment of vibriosis in cod (Gadus morhua). Aquacult 235: 27–35. [Google Scholar]
- 37. Vik-Mo FT, Bergh O, Samuelsen OB (2005) Efficacy of orally administered flumequine in the treatment of vibriosis caused by Listonella anguillarum in Atlantic cod Gadus morhua. Dis Aquat Org 67: 87–92. [DOI] [PubMed] [Google Scholar]
- 38. Seljestokken B, Bergh O, Melingen GO, Rudra H, Olsen RH, et al. (2006) Treating experimentally induced vibriosis (Listonella anguillarum) in cod, Gadus morhua L., with florfenicol. Journal of Fish Diseases 29: 737–742. [DOI] [PubMed] [Google Scholar]
- 39. Sandlund N, Bergh O (2008) Screening and characterisation of potentially pathogenic bacteria associated with Atlantic cod Gadus morhua larvae: bath challenge trials using a multidish system. Dis Aquat Org 81: 203–217. [DOI] [PubMed] [Google Scholar]
- 40. Sandlund N, Rodseth OM, Knappskog DH, Fiksdal IU, Bergh O (2010) Comparative susceptibility of turbot, halibut, and cod yolk-sac larvae to challenge with Vibrio spp. Dis Aquat Org 89: 29–37. [DOI] [PubMed] [Google Scholar]
- 41. Sobecky PA, Mincer TJ, Chang MC, Helinski DR (1997) Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol 63: 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geng HF, Bruhn JB, Nielsen KF, Gram L, Belas R (2008) Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl Environ Microbiol 74: 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lambertsen L, Sternberg C, Molin S (2004) Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Env Microbiol 6: 726–732. [DOI] [PubMed] [Google Scholar]
- 44. Bao Y, Lies DP, Fu H, Roberts GP (1991) An Improved Tn7-Based System for the Single-Copy Insertion of Cloned Genes Into Chromosomes of Gram-Negative Bacteria. Gene 109: 167–168. [DOI] [PubMed] [Google Scholar]
- 45. Geng HF, Belas R (2010) Expression of Tropodithietic Acid Biosynthesis Is Controlled by a Novel Autoinducer. Journal of Bacteriology 192: 4377–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hansen PJ (1989) The Red Tide Dinoflagellate Alexandrium tamarense - Effects on Behavior and Growth of a Tintinnid Ciliate. Marine Ecology-Progress Series 53: 105–116. [Google Scholar]
- 47.Guillard RR, Ryther JH (1962) Studies of Marine Planktonic Diatoms.1. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Canadian Journal of Microbiology 8: : 229–&. [DOI] [PubMed] [Google Scholar]
- 48. Gatesoupe FJ (2002) Probiotic and formaldehyde treatments of Artemia nauplii as food for larval pollack, Pollachius pollachius . Aquacult 212: 347–360. [Google Scholar]
- 49. Prol MJ, Bruhn JB, Pintado J, Gram L (2009) Real-time PCR detection and quantification of fish probiotic Phaeobacter strain 27-4 and fish pathogenic Vibrio in microalgae, rotifer, Artemia and first feeding turbot (Psetta maxima) larvae. J? Appl Microbiol 106: 1292–1303. [DOI] [PubMed] [Google Scholar]
- 50. Carnevali O, Zamponi MC, Sulpizio R, Rollo A, Nardi M, et al. (2004) Administration of probiotic strain to improve sea bream wellness during development. Aquacult Int 12: 377–386. [Google Scholar]
- 51. Planas M, Vazquez JA, Marques J, Perez-Lomba R, Gonzalez MP, et al. (2004) Enhancement of rotifer (Brachionus plicatilis) growth by using terrestrial lactic acid bacteria. Aquacult 240: 313–329. [Google Scholar]
- 52. Marques A, Dinh T, Ioakeimidis C, Huys G, Swings J, et al. (2005) Effects of bacteria on Artemia franciscana cultured in different gnotobiotic environments. Appl Environ Microbiol 71: 4307–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marques A, Thanh TH, Sorgeloos P, Bossier P (2006) Use of microalgae and bacteria to enhance protection of gnotobiotic Artemia against different pathogens. Aquacult 258: 116–126. [Google Scholar]
- 54. Munro PD, Henderson RJ, Barbour A, Birbeck TH (1999) Partial decontamination of rotifers with ultraviolet radiation: the effect of changes in the bacterial load and flora of rotifers on mortalities in start-feeding larval turbot. Aquacult 170: 229–244. [Google Scholar]
- 55. Mangor-Jensen A, Adoff GR (1987) Drinking Activity of the Newly Hatched Larvae of Cod Gadus morhua L. Fish Physiology and Biochemistry. 3: 99–103. [DOI] [PubMed] [Google Scholar]
- 56. Picchietti S, Mazzini M, Taddei AR, Renna R, Fausto AM, et al. (2007) Effects of administration of probiotic strains on GALT of larval gilthead seabream: Immunohistochemical and ultrastructural studies. Fish & Shellfish Immunology 22: 57–67. [DOI] [PubMed] [Google Scholar]
- 57. Dimitroglou A, Merrifield DL, Carnevali O, Picchietti S, Avella M, et al. (2011) Microbial manipulations to improve fish health and production - A Mediterranean perspective. Fish & Shellfish Immunology 30: 1–16. [DOI] [PubMed] [Google Scholar]
- 58. Rollo A, Sulpizio R, Nardi M, Silvi S, Orpianesi C, et al. (2006) Live microbial feed supplement in aquaculture for improvement of stress tolerance. Fish Physiology and Biochemistry 32: 167–177. [Google Scholar]
- 59. Perez-Sanchez T, Balcazar JL, Merrifield DL, Carnevali O, Gioacchini G, et al. (2011) Expression of immune-related genes in rainbow trout (Oncorhynchus mykiss) induced by probiotic bacteria during Lactococcus garvieae infection. Fish & Shellfish Immunology 31: 196–201. [DOI] [PubMed] [Google Scholar]
- 60. Harzevili ARS, VanDuffel H, Defoort T, Dhert P, Sorgeloos P, et al. (1997) The influence of a selected bacterial strain Vibrio anguillarum TR 27 on the growth rate of rotifers in different culture conditions. Aquacult Int 5: 183–188. [Google Scholar]
- 61. Porsby CH, Webber MA, Nielsen KF, Piddock LJV, Gram L (2011) Resistance and Tolerance to Tropodithietic Acid, an Antimicrobial in Aquaculture, Is Hard To Select. Antimicrobial Agents and Chemotherapy 55: 1332–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Seyedsayamdost MR, Case RJ, Kolter R, Clardy J (2011) The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis . Nature Chemistry 3: 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rao D, Webb JS, Kjelleberg S (2005) Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata . Appl Environ Microbiol 71: 1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rao D, Webb JS, Kjelleberg S (2006) Microbial colonization and competition on the marine alga Ulva australis . Appl Environ Microbiol 72: 5547–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rao D, Webb JS, Holmstrom C, Case R, Low A, et al. (2007) Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl Environ Microbiol 73: 7844–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Concentrations of Nannochloropsis oculata and Phaeobacter gallaeciensis in the co-cultures. Colony-forming units of P. gallaeciensis wild type (•) and the TDA-negative mutant (□), and concentrations of N. oculata with V. anguillarum (▴), N. oculata with P. gallaeciensis wild type (▾), N. oculata with P. gallaeciensis TDA-negative mutant (♦), and axenic N. oculata (▪) in the dense (A) and less dense (B) cultures.
(TIF)
Reduction of Vibrio anguillarum by Phaeobacter gallaeciensis in cultures of Tetraselmis suecica . Colony-forming units of V. anguillarum inoculated at 102 cfu/ml (A) and at 103 cfu/ml (B) in presence of P. gallaeciensis wild type (▪), in presence of the P. gallaeciensis TDA-negative mutant (▴), and in the monoxenic control (▾).
(TIF)
Reduction of Vibrio anguillarum by Phaeobacter gallaeciensis in cultures of Nannochloropsis oculata . Colony-forming units of V. anguillarum in presence of P. gallaeciensis wild type (▴), in presence of the P. gallaeciensis TDA-negative mutant (▾), and in the monoxenic control (▪), in dense (3×107 cells/ml; A) and less dense (1–7×106 cells/ml; B) cultures of N. oculata.
(TIF)
Influence of bacterial strains on rotifer growth. Rotifer numbers in co-culture with P. gallaeciensis wild type (▾), with the TDA-negative mutant of P. gallaeciensis (♦), with only V. anguillarum (▴), and axenic rotifers (▪), second experiment. All bacteria were inoculated at day 0. Both P. gallaeciensis strains promoted rotifer growth, whereas V. anguillarum had no influence.
(TIF)
Mortality of cod larvae during the challenge trials. Mean values of two independent triplicate experiments with error bars indicating standard deviations. The single-larvae cultures were simultaneously inoculated with P. gallaeciensis wild type and V. anguillarum (T5, •), or with the TDA-negative mutant of P. gallaeciensis and V. anguillarum (T6, □). Unexposed larvae and larvae exposed to single bacterial strains acted as controls: Negative Control (T1, ▪), only V. anguillarum (T2, ▴), only P. gallaeciensis wild type (T3, ▾), and only P. gallaeciensis TDA-negative mutant (T4, ♦).
(TIF)