The accumulated data on microRNA dysregulation in the different thyroid cancer types is reviewed, its diagnostic utility is critically assessed, and future study strategies are proposed.
Keywords: MircroRNA, Thyroid, Nodules, FNAB, Cancer, Review
Abstract
Thyroid cancer accounts for 1.5% of all malignancies in the U.S., and it is the most common endocrine malignancy. Detection of thyroid cancer mostly relies on evaluation of thyroid nodules, which are very common but only 5%–7% harbor malignancy. Fine-needle aspiration biopsy (FNAB) is currently the most important tool for the evaluation of thyroid nodules; however, it is limited in that it provides only a cytology assessment of the aspirated cells, and indeterminate diagnoses are present in up to 30% of FNAB results. This limitation can be overcome by the molecular analysis of FNAB, and more specifically with the use of microRNAs (miRs).
miRs constitute a class of endogenous small noncoding RNA fragments that regulate gene expression, and in vitro studies have shown that miRs play a significant role in cancer and regulate major processes, such as proliferation, differentiation, and cell death. Several studies have investigated the miR expression signature in different thyroid cancers, and data support its use as a diagnostic tool that is highly accurate for thyroid nodules. The purpose of this study is to review the accumulated data on miR dysregulation in the different thyroid cancer types, critically assess its diagnostic utility, and conclude with future study strategies.
Thyroid Cancer
Palpable nodules in the thyroid gland are very common, affecting up to 5% of the general population. The incidence of nonpalpable nodules within the general population is estimated to reach 50%. These nodules are identified mainly by neck ultrasound examinations and their incidence has been constantly increasing as a result of the increasing use of imaging techniques. Nevertheless, only 5%–7% of the thyroid nodules harbor malignancy and hence there is an obvious need to accurately characterize these nodules [1].
Thyroid cancer accounts for 1.5% of all malignancies in the U.S. and it is the most common endocrine malignancy, accounting for >95% of endocrine malignancies. Over 56,000 new cases are diagnosed annually in the U.S., 74% of these are in females, and it is the most rapidly increasing cancer in incidence. Despite its low malignant potential, every year, ∼1,740 patients die as a result of thyroid cancer in the U.S. Thyroid cancer is the fifth most common cancer in women; it is only surpassed by breast, lung, colon, and uterine cancer. The clinical spectrum of thyroid malignancy varies between highly differentiated incidental small cancers and undifferentiated, aggressive, and uniformly fatal tumors. Fortunately, >90% of thyroid tumors are well differentiated, with a concurrent good prognosis [2].
The most important tool in the diagnosis of thyroid cancer is fine-needle aspiration biopsy (FNAB), with an accuracy >95%. The inherent disadvantage of this test is that it supplies a cytopathological diagnosis based on a description of the cells, as opposed to a definitive, tissue-based, histopathological diagnosis. FNAB cannot provide information on tissue architecture and vascular or neural invasion, and thus it is very limited in diagnosing malignancy.
Currently, there are six different categories for FNAB results that are managed differently [3, 4].
(a) Nondiagnostic (5%–15%). In such biopsies, an insufficient number of cells has been aspirated and repeated FNAB is recommended.
(b) Benign (60%–70%). As previously stated, most nodules are benign and include adenoma, hyperplasia, Hashimoto's thyroiditis, and colloid cysts. The malignancy rate in such biopsies is 1%–3%, and therefore these patients can be referred to follow-up and, if the nodule increases in size or becomes symptomatic, repeated FNAB or surgical removal can be performed.
(c) Follicular lesion or neoplasm of unknown significance (10%). With this FNAB result, the expected malignancy rate is 5%–15% and thus patients are recommended to have a repeat FNAB [3].
(d) Follicular lesion or neoplasm (10%–20%). Follicular lesions are not classified as malignant or benign. This diagnosis includes Hürthle cell tumors. The rate of malignancy in such lesions varies in the range of 15%–30%. The recommended approach for these patients includes removal of the thyroid lobe or the entire thyroid gland [3, 4].
(e) Suspicious for malignancy (<10%). These aspirates are highly suspicious for malignancy but do not meet the malignancy criteria mentioned above. The malignancy rate found on final pathology of such results is 60%–75%. Total thyroidectomy is recommended for these patients.
(f) Malignant (5%–7%). This result has a very high positive predictive value and the malignancy rate is 97%–99%. Patients with malignant results should have their thyroid gland resected.
Categories (c), (d), and (e) include FNAB results that present both the patient and the surgeon with a challenging management decision [3, 4]. Patients who undergo thyroid lobectomy for an indeterminate nodule require another completion surgery if malignancy is identified on final pathology. In contrast, thyroid resections may prove to be unnecessary as a substantial portion of lesions with these results turn out to be benign on final pathology. This dilemma is encountered in up to 30% of the patients undergoing FNAB, and the need for an established better molecular diagnosis is clear.
The discovery of somatic mutations that are present in >40% and >60% of patients with papillary thyroid cancer (PTC) and follicular thyroid cancer (FTC), respectively, enabled molecular analysis to improve FNAB accuracy [5, 6]. In recent years, nearly all somatic mutations are tested for their applicability to FNAB diagnosis in different settings [7]. The same FNAB material can be used for cytological and molecular analyses, and it is possible to use a part of the total FNAB obtained or leftover cells in the needle bevel [8]. Studies that use an additional FNAB increase the likelihood of generating contradictory results. In an updated review by Ferraz et al. [6], 20 studies were reviewed and analyzed. Those studies used indeterminate FNAB categories and mutation detection was compared with the final histopathology results. Although most studies used mutation in one gene for analysis (e.g., BRAF or RET/PTC), four studies analyzed mutations in several genes (BRAF, RAS, RET/PTC, and PAX8/PPARg). This review of the studies identified a specificity of 95%–100% and a sensitivity of 38%–86% [9–12]. These results are promising and offer a clear clinical decision when mutation analysis is positive; however, the relatively low sensitivity indicates the need for other complementary molecular markers, such as microRNAs (miRs).
miRs
miRs constitute a class of endogenous small noncoding RNA fragments (18–24 nucleotides) that regulate gene expression. The first miR was discovered in the Caenorhabditis elegans worm as early as 1993, which was followed by identification of numerous miRs in plants, animals, and humans [13]. A search of updated databases revealed 1,921 unique mature human miRs (http://www.mirbase.org). Approximately half of all human miR genes are contained within the introns of protein-coding genes, whereas others reside apart from known genes or in the exons of untranslated genes [14, 15]. The biogenesis of miRs begins with transcription as long double-stranded primary transcripts that are subsequently converted into a precursor of ∼70 nucleotides, which is finally cleaved in the cytoplasm into the 22-nucleotide double-stranded miR. The duplex is then unwound, and one of the strands is incorporated into the RNA-induced silencing complex (RISC). miRs incorporated into the RISC are able to bind to the 3′ untranslated region (UTR) of the target mRNA, causing a block in translation or mRNA degradation depending on the level of complementarity [16]. The effect of miRs on their target genes is based on the degree of their sequence homology with their target genes [17]. Mature miRs target and inhibit translation or promote mRNA degradation by annealing to complementary sequences in the mRNA 3′ UTR. Because the specificity of miRs is dictated by six to seven nucleotides that bind to the 3′ UTR of their target mRNAs, a single miR can potentially target hundred of genes, and a single gene could be a potential target of many different miRs [18]. This diversity allows miRs to act as regulators of different cell mechanisms, and the extent of this regulation is yet to be elucidated. It is speculated that miRs altogether regulate as much as 30% of the human genome [19].
miRs play a significant role in cancer and regulate major processes such as proliferation, differentiation, and cell death. Lower expression of specific miRs can lead to upregulation of oncogenes whereas greater expression of other miRs may lead to downregulation of different tumor-suppressor genes, and thus both paths contribute to the development of malignancy [16, 20, 21]. To date, numerous cases of miR dysregulation have been identified as potential contributors to several malignancies. The first breakthrough was the finding that miR-15a and miR-16–1 are frequently deleted in B-cell chronic lymphocytic leukemia [22]. Many other malignancies followed: miR-21 is upregulated in glioblastoma [23], miR-143 and miR-145 are downregulated in colorectal cancer [24], and there is elevated expression of miR-205 and miR-155 in breast cancer [25]. miR dysregulation was also identified in many other malignancies including, but not limited to, acute myeloid leukemia, pancreatic cancer, lung cancer, melanoma, ovarian cancer, hepatoma, prostate cancer, bladder cancer, kidney cancer, gastric cancer, and thyroid cancer [26–35].
The exact target genes of each miR in every malignancy are not clear. The current focus of miR studies covers multiple oncologic aspects. The diagnostic utility of miR dysregulation in tissue and blood samples is being evaluated. Commercial protocols are available for RNA extraction and miR profile analysis for snap-frozen tissues, formalin-fixed paraffin-embedded (FFPE) samples, and even cells obtained with FNAB. Other studies focused on target gene mapping for every specific miR in an attempt to elucidate their effect on oncogenes and tumor-suppressor genes [36–38]. Lastly, miRs may serve as prognostic markers for certain cancers, and their therapeutic value is also being evaluated [26, 37].
The focus of this review is the role of miRs as a diagnostic tool for thyroid cancer (Table 1). Despite the importance of differentiating between benign and malignant thyroid nodules, several challenges are encountered while using miR for this model. Thyroid malignancies originate from different cells and can be divided into well-differentiated thyroid carcinoma and undifferentiated thyroid carcinoma. PTC and FTC constitute the well-differentiated thyroid cancer group whereas anaplastic thyroid cancer (ATC) is undifferentiated and medullary thyroid cancer (MTC) develops in C cells. As expected, different miRs are dysregulated in each tumor. Moreover, molecular changes that differentiate follicular adenoma (FA) from carcinoma and oncocytic adenoma from carcinoma are not clear. Therefore, it cannot be assumed that dysregulated miRs in thyroid malignancies will not be present in thyroid nodules that are considered benign. Lastly, critical evaluation of the existing literature should include special attention to the tissue used for analysis—fresh ex vivo thyroid tissue, FFPE slides, or FNAB samples—as well as what controls were used. Needless to say, in order to serve as a clinical tool, studies should focus on identifying malignancies in samples that mimic clinical scenarios such as FNAB.
Table 1.
Summary of micro-RNA (miR) dysregulation in thyroid malignancies
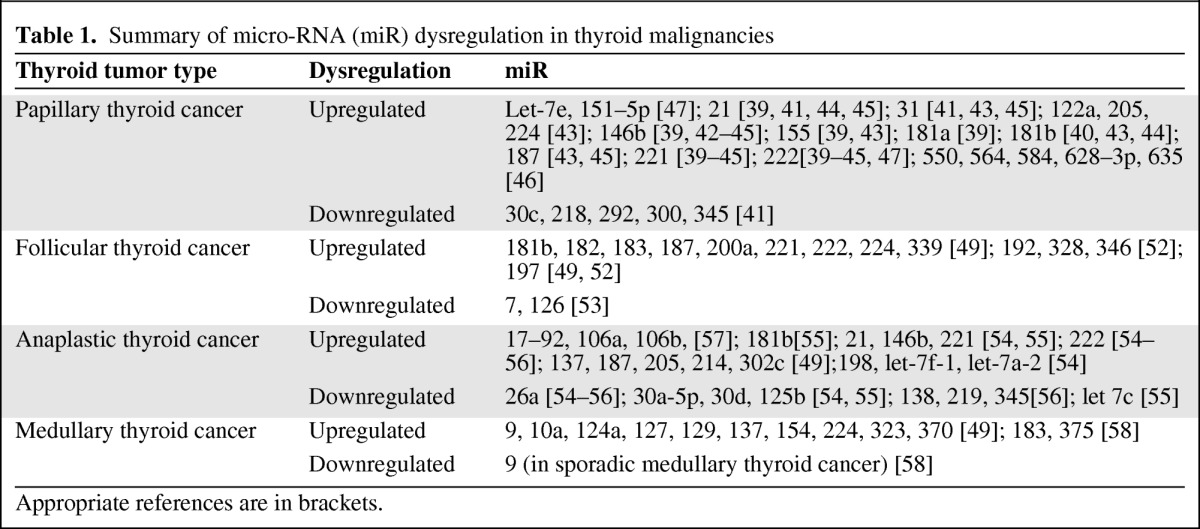
Appropriate references are in brackets.
miR Dysregulation in PTC
In 2005, He et al. [39] used pairwise significance analysis of microarray analysis of 30 tumor samples to identify dysregulation of 23 miRs and overexpression occurring in 17. Six miR genes were significantly overexpressed in PTC >1.5-fold. These miRs were miR-21, miR-146, miR-155, miR-181a, miR-221, and miR-222. The top three miRs (miR-146, miR-221, and miR-222) showed dramatic overexpression, with 11-fold to 19-fold higher level in PTC tumors than in adjacent unaffected thyroid tissue. These results were later repeated by Pallente et al. [40], in 2006, for miR-181b, miR-221, and miR-222 and by Tetzlaff et al. [41], in 2007, for miR-21, miR-31, miR-221, and miR-222. In the later study, FFPE tissue samples were used. In 2008, Chen et al. [42] identified significant upregulation of miR-146b, miR-221, and miR-222 in PTC samples obtained by both FFPE and ex vivo FNAB. It was concluded that miR-221 and miR-222 are upregulated in papillary and nonpapillary thyroid tumors and that miR-146b could potentially serve as a marker for diagnosing PTC using both FNAB and surgical pathology specimens. Nikiforova et al. [43] reported that miR-187 (on top of miR-146b, miR-181b, miR-221, and miR-222, all >10-fold) was upregulated in patients with PTC. miR-31, miR-122a, miR-155, miR-205, and miR-224 were upregulated in PTC to a lesser degree (by 5- to 10-fold). The analysis in that study included both fresh thyroid tissue and FNAB samples. Similar results were reported by Sheu et al. [44] (miR-21, miR-146b, miR-181, miR-221, and miR-222) and by Mazeh et al. [45] (miR-21, miR-31, miR-146b, miR-187, miR-221, and miR-222). In a recent study by Vriens et al. [46], five miRs were upregulated in thyroid cancer (miR-550, miR-564, miR-584, miR-628–3p, and miR-635). These miRs were not previously described and showed values 4- to 7.8-fold higher than normal; however, the specific thyroid cancer type (PTC versus others) was not clearly identified in the study. In a recent study by Yu et al. [47], miR-let-7e, miR-151–5p, and miR-222 were upregulated in the serum of PTC patients. Interestingly, the serum miR-151–5p and miR-222 levels decreased significantly following tumor excision. To date, this is the only study to evaluate circulating miRs in thyroid cancer patients. Upregulation of miR-21, miR-31, miR-146b, miR-155, miR-181b, miR-187, miR-221, and miR-222 was identified by more than one group (Table 1). Downregulation of some miRs has been described with varying results in relation to controls [39, 41, 46, 48]. The only miRs that have been reported on by more than a single group are miR-138 and miR-345.
In a separate study by one of the above groups, Visone et al. [49] demonstrated that miR-221 and miR-222 negatively regulate p27kip1, which inhibits G1–S phase cell-cycle progression and serves as a checkpoint for cell proliferation. The effect of miR-221 in a PTC cell line was also investigated by Kim et al. [50], who demonstrated more than twofold lower mRNA levels of HOXB5. In comparison with the previous studies, Jazdzewski et al. [51] demonstrated that the expression of miR-146a is dysregulated in patients with PTC and associated this dysregulation with the IRAK1, TRAF6, and PTC1 genes.
miR Dysregulation in FTC
The progression of FA to FTC is poorly defined. The differentiation of follicular carcinoma from a benign adenoma or a cytology result of a follicular lesion is based on capsular or vascular invasion. Several studies have tried to identify the molecular miR profile that distinguishes FTC from other benign thyroid lesions. Weber et al. [52], in 2006, investigated a set of 235 human miRs and identified that miR-192, miR-197, miR-328, and miR-346 were upregulated in FTC compared with FA 1.3- to 1.8-fold. An association with cell growth with knockdown resulting in growth suppression was demonstrated and a number of target genes were suggested (ACVR1, TSPAN3, and EFEMP2). Nikiforova et al. [43] demonstrated differential upregulation of miR-187 and miR-197 in FTC, but not all other miRs mentioned by Weber et al. [52] were investigated. In that study, the most highly upregulated miRs in conventional FTC cases were miR-181b, miR-182, miR-187, miR-200a, and miR-224, and those in oncocytic variant cases were miR-183, miR-187, miR-221, miR-222, and miR-339. There was an overlap between upregulated miRs in FA and FTC. The most highly upregulated miRs in conventional FAs were miR-181b, miR-200a, miR-200b, miR-224, and miR-339, and those in oncocytic variants were miR-31, miR-181b, miR-182, miR-183, and miR-339. In a recent carefully designed study by Kitano et al. [53], 34 miRs were differentially expressed in diverse groups of benign and malignant thyroid histologies. In contrast to the previous studies, miR-7 and miR-126 were differentially expressed between FTC and benign thyroid lesions (e.g., FA and Hürthle cell adenoma). Both these miRs were downregulated by 2.7-fold and 2.0-fold, respectively.
miR Dysregulation in ATC
A subtype of undifferentiated thyroid cancer, ATC is the most aggressive one and the one that carries the worst prognosis. Fortunately, it is the least common subtype. It is not surprising that several miRs were found to be dysregulated in ATC with suggested oncogenes as their target or miR tumor suppressor activity. Visone et al. [54] identified upregulation of four miRs in ATC (miR-198, miR-222, miR-let-7f-1, and miR-let-7a-2) and downregulation of miR-26a, miR-30a-5p, miR-30d, and miR-125b by more than threefold. Possible target genes were HMGA1 and HMGA2. These findings were confirmed by Schwertheim et al. [55], who identified downregulation of miR-26a, miR-30a-5p, miR-30d, miR-125b, and miR-let 7c in ATC. Surprisingly, these miRs were found to be upregulated in PTC and in contrast, four of the five miRs included in a PTC panel (miR-21, miR-146b, miR-181b, miR-221, and miR-222) were also upregulated in ATC. These interesting results were repeated by Mitomo et al. [56], who showed that miR-21, miR-146b, miR-221, and miR-222 were upregulated in both ATC and PTC. They suggested that miR-21 targets 17-β-estradiol factor (E2F) (involved in the cell cycle and apoptosis) and inhibits phosphatase and tensin homologue deleted on chromosome ten (PTEN). In that study, miR-miR-26a, miR-138, miR-219, and miR-345 were downregulated. A potential target of miR-138 is the gene encoding human telomerase reverse transcriptase (hTERT), whose overexpression has been associated with dedifferentiation, tumor stage, and more metastatic and invasive phenotypes. Nikiforova et al. [43] identified different miRs (miR-137, miR-187, miR-205, miR-214, and miR-302c) that were all upregulated in ATC by 16- to 114-fold. miR-17 to miR-92 is a cluster of seven miRs (miR-17–5p, miR-17–3p, miR-18a, miR-19a, miR-19b, miR-20a, and miR-91–1) and was shown to be overexpressed in ATC samples by Takakura et al. [57]. miR-19a and miR-19b have PTEN as a target, thus explaining the oncogenic role of miR-17 to miR-92. In the same study, miR-106a and miR-106b were also upregulated and suggested to target E2F-1.
miR Dysregulation in MTC
Less than 5% of cells within the thyroid gland are C cells that give rise to MTC. This rare malignancy constitutes 3%–4% of thyroid cancers. It can be sporadic or familial (25%), and adequate surgery is the mainstay of treatment. Nikiforova et al. [43] identified 10 different miRs to be very highly upregulated in MTC by 32- to 142-fold. These miRs are mostly unique to MTC and include miR-9, miR-10a, miR-124a, miR-127, miR-129, miR-137, miR-154, miR-224, miR-323, and miR-370. Abraham et al. [58], in an outstanding study, identified that miR-183 and miR-375 were overexpressed and miR-9 was underexpressed in sporadic MTC versus hereditary MTC. Furthermore, it was demonstrated that knockout of miR-183 expression resulted in decreased cell counts.
Diagnostic Strategies Using miRs
A flux of studies has shown dysregulation of miR expression in different thyroid cancers. Evaluating the data on the different miR functions and effects on intracellular pathways is beyond the scope of this review; however, there are accumulating data on the diagnostic utility of the miR profile for each thyroid cancer. The most studied diagnostic potential of miRs is in FNAB. Most studies have investigated miR expression in fresh thyroid tissues or FFPE samples; however, such samples are available only after thyroid resection and thus do not mimic clinical scenarios. In order for miR analysis to serve as a diagnostic tool, it should be performed on FNAB samples. The extraction of RNA from FNAB samples is challenging due to the low number of cells aspirated. Several studies have attempted to predict the risk for malignancy using FNAB samples with the use of specific miR panels. The first to describe miR analysis in FNAB samples were Pallente et al. [40] in 2006 in a study that demonstrated upregulation of miR-181b, miR-221, and miR-222 in eight PTC samples. Nikiforova et al. [43] used a panel of seven miRs (miR-146b, miR-155, miR-187, miR-197, miR-221, and miR-222) to distinguish between hyperplastic nodules and thyroid malignancy in FNAB samples. With at least one miR upregulated more than twofold, they reached an accuracy of 95%, and when three or more were upregulated the accuracy was 98%. These results were confirmed later that year by Chen et al. [42] in an analysis of 40 ex vivo FNAB samples; they concluded that only miR-146b and miR-222 persisted as distinguishing markers for PTC. Recently, a panel of six miRs was used (miR-21, miR-31, miR-146b, miR-187, miR-221, and miR-222) on a sample of 27 ex vivo FNABs, with an accuracy of 98% for predicting PTC [45]. The values for FTC are not as high, and Vriens et al. [46] used miR-100, miR-125b, miR-138, and miR-768–3p with an accuracy of 71% for follicular neoplasms and 98% for Hürthle cell neoplasms. In that study, miR-138 was upregulated in malignancy samples with an independent accuracy of 75%. One study failed to demonstrate the diagnostic utility of miR-21, miR-146b, miR-181b, miR-221, and miR-222 in individual FTC tumors. In that study, FFPE samples were used as opposed to FNAB samples [44]. Lastly, Shen et al. [59] used archived FNAB slides to extract RNA and evaluate the malignancy potential of those determined “follicular lesion of unknown significance” with a panel of seven miRs (miR-30d, miR-138, miR-146b, miR-187, miR-197, miR-221, miR-302c, and miR-346). The overall diagnostic accuracy for these challenging FNAB samples was 85%. Recently in a preliminary study, residual cells left in the FNAB needle were used to diagnose malignancy in intermediate cytology results with an accuracy of 90% [60]. Table 2 summarizes the tissue types and controls used in the different studies.
Table 2.
Summary of the tissue types and controls used in the different studies
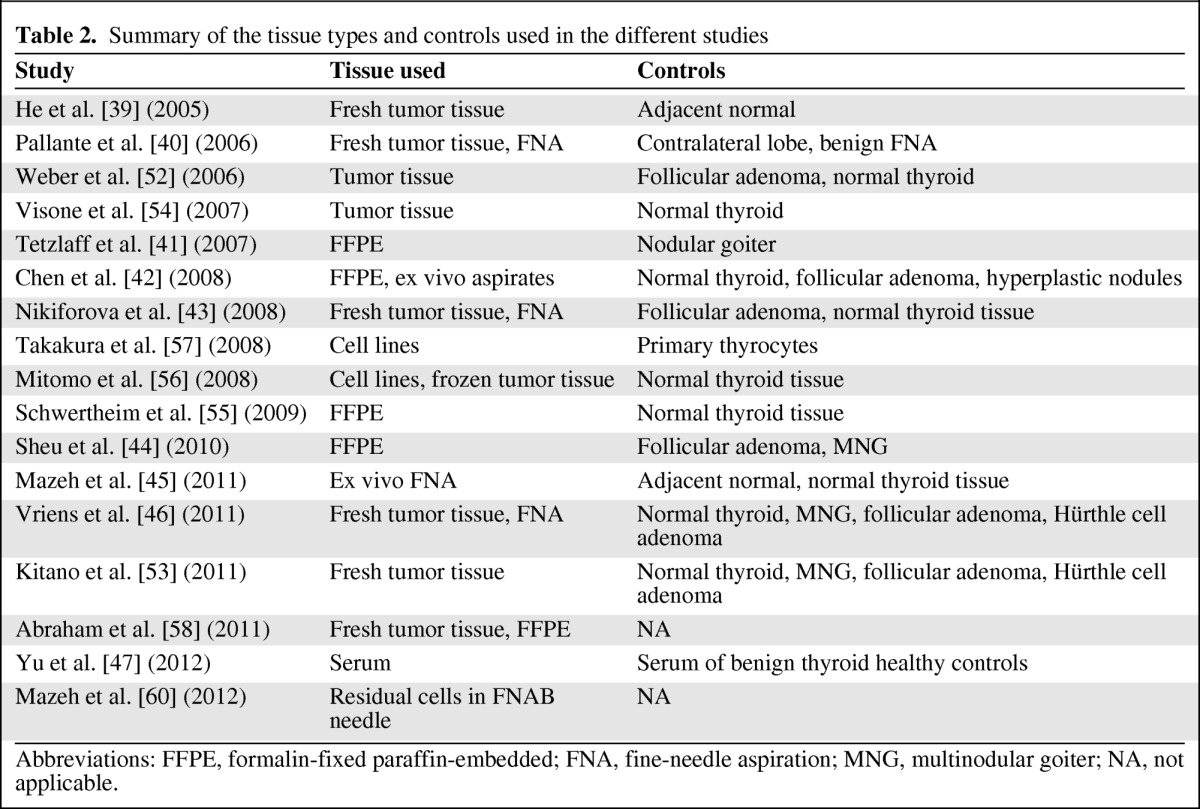
Abbreviations: FFPE, formalin-fixed paraffin-embedded; FNA, fine-needle aspiration; MNG, multinodular goiter; NA, not applicable.
At this point, it is difficult to conclude which miRs should be included in a diagnostic panel due to the high variability in results. The most common denominator seems to include miR-146b, miR-221, and miR-222, which correctly identify PTC. The diagnostic utility of miRs for ATC and MTC can be criticized. Although the diagnosis of ATC is not always clear with FNAB, the clinical picture in these patients is significantly different than in patients with other thyroid cancers. Patients with ATC present with a neck mass and invasive symptoms are common. The diagnosis of MTC is clearer on FNABs and serum calcitonin levels confirm the diagnosis. It can be argued that miRs do not contribute to the diagnosis of these two thyroid malignancies.
Another diagnostic aspect that has been studied is the use of miRs to predict tumor aggressiveness. In contrast to the utility of FNAB for miR analysis, this aspect has been evaluated by fewer studies to date. Chou et al. [61] identified a significant association between the expression levels of specific miRs and extrathyroidal invasion (miR-146b, miR-221, and miR-222), high-risk disease (miR-146b, miR-221), and the presence of a BRAF mutation (miR-146b). These findings were corroborated by Yip et al. [62], who identified that upregulation of miR-146b, miR-221, miR-222, miR-155, and miR-31 and downregulation of miR-1, miR-34b, miR-130b, and miR-138 were correlated with aggressive PTC. Abraham et al. [58] identified that upregulation of miR-183 and miR-375 in MTC predicted lateral lymph node metastasis and was associated with residual disease, distant metastasis, and mortality. Preoperative aggressiveness prediction has clear clinical implications as it may assist in tailoring the appropriate surgery to each patient and offer high-risk patients a more extensive surgical procedure with central or lateral lymph node dissection. Postoperative aggressiveness prediction may be used to stratify patients for adjuvant therapies. Despite these promising results, larger prospective trials are needed before incorporating these results into clinical practice.
Conclusion and Future Strategies
In recent years, the number of publications on miRs has been exponentially rising, with >4,500 in the year 2011. The accumulated data also have been directed to investigate the role of miRs in thyroid cancer. Initial studies focused on determining the miR profile in benign and malignant thyroid lesions, and the purpose of this review was to summarize the existing data in an organized fashion that can be used to design future studies. It seems that different miR profiles should be used for identifying specific thyroid malignancies. A panel of four to six miRs may be used with very high accuracy to identify PTC, and similarly a different panel may be used for MTC as well. The less common scenario of ATC may also be accurately predicted using a separate miR panel. Currently, the accuracy of predicting FTC with miRs does not reach the prediction accuracy for other thyroid malignancies and the etiology for this has yet to be elucidated. The pure diagnostic utility of miRs for MTC and ATC may be challenged since these two thyroid malignancies do not present a diagnostic challenge. At present, the routine use of miRs is limited by their cost and by the lack of consistency in reporting the threshold defined for the diagnosis of malignancy in the different studies. Prospective clinical studies using miRs in FNAB specimens to determine surgical treatment are needed to ultimately prove the diagnostic utility of this molecular tool.
Recent and ongoing current studies focus on the effect of miRs on target genes; however, reviewing all the data is beyond the scope of this paper. The identification of target genes should lead to in vitro studies that may indicate possible therapeutic options. There are many more studies needed to elucidate miRs' effects.
The diagnostic utility of miRs can be further improved. miR analysis using peripheral blood samples may be useful in detecting ATC or other advanced thyroid cancers at earlier stages than currently possible. As mentioned above, the association of the miR expression profile with cancer aggressiveness needs further validation and may eventually impact surgical planning. The miR profile as well as the identification of specific mutations may lead to detection of a higher risk population as well as familial predisposition.
In conclusion, miRs have emerged as a robust diagnostic tool in the identification of malignancy in thyroid nodules. Current studies show promising results, with accuracy rates that far exceed the levels achieved using traditional cytology. Further studies are needed to translate these methods into routine clinical evaluations.
References
- 1.Dean DS, Gharib H. Epidemiology of thyroid nodules. Best Pract Res Clin Endocrinol Metab. 2008;22:901–911. doi: 10.1016/j.beem.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute. Surveillance Epidemiology and End Results. SEER Stat Fact Sheets: Thyroid. [accessed March 26, 2012]. Available at http://seer.cancer.gov/statfacts/html/thyro.html.
- 3.Cibas ES, Ali SZ. The Bethesda System for reporting thyroid cytopathology. Thyroid. 2009;19:1159–1165. doi: 10.1089/thy.2009.0274. [DOI] [PubMed] [Google Scholar]
- 4.Cooper DS, Doherty GM, Haugen BR, et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009;19:1167–1214. doi: 10.1089/thy.2009.0110. [DOI] [PubMed] [Google Scholar]
- 5.Grogan RH, M E, Clark OH. The evolution of biomarkers in thyroid cancer—from mass screening to a personalized biosignature. Cancers (Basel) 2010;2:885–912. doi: 10.3390/cancers2020885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferraz C, Eszlinger M, Paschke R. Current state and future perspective of molecular diagnosis of fine-needle aspiration biopsy of thyroid nodules. J Clin Endocrinol Metab. 2011;96:2016–2026. doi: 10.1210/jc.2010-2567. [DOI] [PubMed] [Google Scholar]
- 7.Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7:569–580. doi: 10.1038/nrendo.2011.142. doi: 10.1038/nrendo.2011.142. [DOI] [PubMed] [Google Scholar]
- 8.Nikiforov YE, Ohori NP, Hodak SP, et al. Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: A prospective analysis of 1056 FNA samples. J Clin Endocrinol Metab. 2011;96:3390–3397. doi: 10.1210/jc.2011-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moses W, Weng J, Sansano I, et al. Molecular testing for somatic mutations improves the accuracy of thyroid fine-needle aspiration biopsy. World J Surg. 2010;34:2589–2594. doi: 10.1007/s00268-010-0720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantara S, Capezzone M, Marchisotta S, et al. Impact of proto-oncogene mutation detection in cytological specimens from thyroid nodules improves the diagnostic accuracy of cytology. J Clin Endocrinol Metab. 2010;95:1365–1369. doi: 10.1210/jc.2009-2103. [DOI] [PubMed] [Google Scholar]
- 11.Nikiforov YE, Steward DL, Robinson-Smith TM, et al. Molecular testing for mutations in improving the fine-needle aspiration diagnosis of thyroid nodules. J Clin Endocrinol Metab. 2009;94:2092–2098. doi: 10.1210/jc.2009-0247. [DOI] [PubMed] [Google Scholar]
- 12.Ohori NP, Nikiforova MN, Schoedel KE, et al. Contribution of molecular testing to thyroid fine-needle aspiration cytology of “follicular lesion of undetermined significance/atypia of undetermined significance.”. Cancer Cytopathol. 2010;118:17–23. doi: 10.1002/cncy.20063. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 14.Kim VN. Small RNAs: Classification, biogenesis, and function. Mol Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 15.Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 17.Boyd SD. Everything you wanted to know about small RNA but were afraid to ask. Lab Invest. 2008;88:569–578. doi: 10.1038/labinvest.2008.32. [DOI] [PubMed] [Google Scholar]
- 18.Santarpia L, Nicoloso M, Calin GA. MicroRNAs: A complex regulatory network drives the acquisition of malignant cell phenotype. Endocr Relat Cancer. 2010;17:F51–F75. doi: 10.1677/ERC-09-0222. [DOI] [PubMed] [Google Scholar]
- 19.de la Chapelle A, Jazdzewski K. MicroRNAs in thyroid cancer. J Clin Endocrinol Metab. 2011;96:3326–3336. doi: 10.1210/jc.2011-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 21.Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol. 2011;38:724–733. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 24.Akao Y, Nakagawa Y, Hirata I, et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 25.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 26.Sandhu S, Garzon R. Potential applications of microRNAs in cancer diagnosis, prognosis, and treatment. Semin Oncol. 2011;38:781–787. doi: 10.1053/j.seminoncol.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Marcucci G, Radmacher MD, Mrozek K, et al. MicroRNA expression in acute myeloid leukemia. Curr Hematol Malig Rep. 2009;4:83–88. doi: 10.1007/s11899-009-0012-7. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Sen S. MicroRNA functional network in pancreatic cancer: From biology to biomarkers of disease. J Biosci. 2011;36:481–491. doi: 10.1007/s12038-011-9083-4. [DOI] [PubMed] [Google Scholar]
- 29.Fanini F, Vannini I, Amadori D, et al. Clinical implications of microRNAs in lung cancer. Semin Oncol. 2011;38:776–780. doi: 10.1053/j.seminoncol.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Li SD, Zhang JR, Wang YQ, et al. The role of microRNAs in ovarian cancer initiation and progression. J Cell Mol Med. 2010;14:2240–2249. doi: 10.1111/j.1582-4934.2010.01058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu WK, Lee CW, Cho CH, et al. MicroRNA dysregulation in gastric cancer: A new player enters the game. Oncogene. 2010;29:5761–5771. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 32.Catto JW, Alcaraz A, Bjartell AS, et al. MicroRNA in prostate, bladder, and kidney cancer: A systematic review. Eur Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 33.Negrini M, Gramantieri L, Sabbioni S, et al. microRNA involvement in hepatocellular carcinoma. Anticancer Agents Med Chem. 2011;11:500–521. doi: 10.2174/187152011796011037. [DOI] [PubMed] [Google Scholar]
- 34.Menon MP, Khan A. Micro-RNAs in thyroid neoplasms: Molecular, diagnostic and therapeutic implications. J Clin Pathol. 2009;62:978–985. doi: 10.1136/jcp.2008.063909. [DOI] [PubMed] [Google Scholar]
- 35.Nikiforova MN, Chiosea SI, Nikiforov YE. MicroRNA expression profiles in thyroid tumors. Endocr Pathol. 2009;20:85–91. doi: 10.1007/s12022-009-9069-z. [DOI] [PubMed] [Google Scholar]
- 36.Tie J, Fan D. Big roles of microRNAs in tumorigenesis and tumor development. Histol Histopathol. 2011;26:1353–1361. doi: 10.14670/HH-26.1353. [DOI] [PubMed] [Google Scholar]
- 37.Gambari R, Fabbri E, Borgatti M, et al. Targeting microRNAs involved in human diseases: A novel approach for modification of gene expression and drug development. Biochem Pharmacol. 2011;82:1416–1429. doi: 10.1016/j.bcp.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Subramanyam D, Blelloch R. From microRNAs to targets: Pathway discovery in cell fate transitions. Curr Opin Genet Dev. 2011;21:498–503. doi: 10.1016/j.gde.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 41.Tetzlaff MT, Liu A, Xu X, et al. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18:163–173. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- 42.Chen YT, Kitabayashi N, Zhou XK, et al. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol. 2008;21:1139–1146. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- 43.Nikiforova MN, Tseng GC, Steward D, et al. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheu SY, Grabellus F, Schwertheim S, et al. Differential miRNA expression profiles in variants of papillary thyroid carcinoma and encapsulated follicular thyroid tumours. Br J Cancer. 2010;102:376–382. doi: 10.1038/sj.bjc.6605493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazeh H, Mizrahi I, Halle D, et al. Development of a microRNA-based molecular assay for the detection of papillary thyroid carcinoma in aspiration biopsy samples. Thyroid. 2011;21:111–118. doi: 10.1089/thy.2010.0356. [DOI] [PubMed] [Google Scholar]
- 46.Vriens MR, Weng J, Suh I, et al. MicroRNA expression profiling is a potential diagnostic tool for thyroid cancer. Cancer. 2011 Oct 17; doi: 10.1002/cncr.26587. [Epub ahead of print]. doi: 10.1002/cncr.26587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu S, Liu Y, Wang J, et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab. 2012 Apr 3; doi: 10.1210/jc.2011-3059. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 48.Leone V, D'Angelo D, Rubio I, et al. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1α. J Clin Endocrinol Metab. 2011;96:E1388–E1398. doi: 10.1210/jc.2011-0345. [DOI] [PubMed] [Google Scholar]
- 49.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 50.Kim HJ, Kim YH, Lee DS, et al. In vivo imaging of functional targeting of miR-221 in papillary thyroid carcinoma. J Nucl Med. 2008;49:1686–1693. doi: 10.2967/jnumed.108.052894. [DOI] [PubMed] [Google Scholar]
- 51.Jazdzewski K, Liyanarachchi S, Swierniak M, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci U S A. 2009;106:1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber F, Teresi RE, Broelsch CE, et al. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab. 2006;91:3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- 53.Kitano M, Rahbari R, Patterson EE, et al. Expression profiling of difficult-to-diagnose thyroid histologic subtypes shows distinct expression profiles and identify candidate diagnostic microRNAs. Ann Surg Oncol. 2011;18:3443–3452. doi: 10.1245/s10434-011-1766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visone R, Pallante P, Vecchione A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 55.Schwertheim S, Sheu SY, Worm K, et al. Analysis of deregulated miRNAs is helpful to distinguish poorly differentiated thyroid carcinoma from papillary thyroid carcinoma. Horm Metab Res. 2009;41:475–481. doi: 10.1055/s-0029-1215593. [DOI] [PubMed] [Google Scholar]
- 56.Mitomo S, Maesawa C, Ogasawara S, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takakura S, Mitsutake N, Nakashima M, et al. Oncogenic role of miR-17–92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–1154. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abraham D, Jackson N, Gundara JS, et al. MicroRNA profiling of sporadic and hereditary medullary thyroid cancer identifies predictors of nodal metastasis, prognosis, and potential therapeutic targets. Clin Cancer Res. 2011;17:4772–4781. doi: 10.1158/1078-0432.CCR-11-0242. [DOI] [PubMed] [Google Scholar]
- 59.Shen R, Liyanarachchi S, Li W, et al. MicroRNA signature in thyroid fine needle aspiration cytology applied to “atypia of undetermined significance” cases. Thyroid. 2012;22:9–16. doi: 10.1089/thy.2011.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazeh H, Levy Y, Mizrahi I, et al. Differentiating benign from malignant thyroid nodules using micro ribonucleic acid amplification in residual cells obtained by fine needle aspiration biopsy. J Surg Res. 2012 May 14; doi: 10.1016/j.jss.2012.04.051. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 61.Chou CK, Chen RF, Chou FF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20:489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 62.Yip L, Kelly L, Shuai Y, et al. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol. 2011;18:2035–2041. doi: 10.1245/s10434-011-1733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


