Abstract
Objectives
To describe a novel MSH2 missense alteration co-segregating with pancreatic cancer.
Methods
Observational study of a kindred in which a novel MSH2 missense alteration was identified.
Results
We report a family in which a MSH2 P349L missense alteration is co-segregating with pancreatic cancers among three nonsmoking first degree relatives. Lynch syndrome-related tumors from individuals carrying this alteration consistently showed loss of immunohistochemical expression of MSH2 and in-silico analyses support interpretation of this DNA alteration as likely pathogenic.
Conclusions
The MSH2 P349L may increase the risk for pancreatic cancer beyond the usual mutations in DNA mismatch repair genes; however studies of additional families with the identical missense alteration are needed to confirm this initial impression.
Keywords: pancreatic cancer, HNPCC, Lynch Syndrome, hereditary, genetics
Introduction
Lynch syndrome (OMIM #s 120435, 609310) is an autosomal dominant cancer predisposition syndrome that underlies about 3–5% of all colorectal cancers.1-5 It is caused by germline mutations in one of several DNA mismatch repair genes, including MSH2, MLH1, MSH6, and PMS2. The genetic heterogeneity has made diagnostic testing a challenge, such that use of tumor assessment of either DNA mismatch repair deficiency (microsatellite instability) and/or expression of the four DNA mismatch repair gene products has been widely used to screen suspected cases. The Bethesda guidelines6 put forth recommendations based upon expert opinion for when tumor testing should be considered. Under that report, the cancers listed as Lynch Syndrome-associated included colorectal, endometrial, stomach, ovarian, pancreas, ureter and renal pelvis, biliary tract and brain (usually glioblastoma as seen in Turcot syndrome) tumors, sebaceous gland adenomas and keratoacanthomas in Muir-Torre syndrome, and carcinoma of the small bowel. Although pancreatic cancer is included in this list, risk usually appears to be only minimally increased, relative to the general population.
Materials and Methods
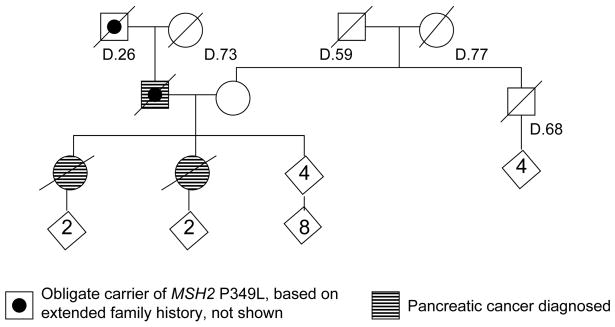
We have been following a family with a novel MSH2 missense alteration in which pancreatic cancer has been more commonly observed than colon or endometrial cancer (Figure 1). The ancestry is Northern European. None of the affected individuals smoked cigarettes nor had known exposure to unusual environmental agents. There is no family history of melanoma, early onset breast cancer, ovarian cancer, or pancreatitis. Table 1 lists the cancers of all relevant family members, and, where available, the results of tumor immunohistochemical expression of the DNA mismatch repair genes. All testing was done at Mayo Clinic using standard techniques.7–9 The MSH2 germline change, identified by sequencing, is in exon 6, c.1046C>T, (CCT>CTT), p.Pro349Leu, hereafter called P349L. This variant co-segregates with the development of pancreatic cancer and with the loss of MSH2 expression in tumor tissue in this family. The details of this family have not previously been published; however one aspect of this family’s laboratory results was included in a prior publication that reported on use of BRAF screening as a strategy to simplify HNPCC genetic testing.10, 11 No BRAF V600E somatic mutation was found in the MSI-high tumor tested in this family, consistent with this being a Lynch Syndrome family. The kindred is enrolled in an ongoing familial pancreatic cancer registry and an affected individual was studied and found to be negative for CFTR and CDKN2A mutations.
Figure 1.
Pedigree of family with MSH2 P349L missense substitution, showing those diagnosed with pancreatic cancer.
Table 1.
Results of germline and tumor molecular testing in family with three individuals with pancreatic cancer, co-segregating with an MSH2 P349L alteration.
| Tumor immunohistochemistry expression | ||||||
|---|---|---|---|---|---|---|
| Relation to proband, cancer, age at diagnosis | MSH2 P349L germline mutation | MSI | MLH1 | MSH2 | MSH6 | PMS2 |
| Paternal grandmother | ND | |||||
| Breast 73 | ND | ND | ND | ND | ND | |
| Father | ND (obligate carrier) | |||||
| Colon 43 | NA | NA | NA | NA | ||
| Pancreas 50 | Normal | Loss | Loss | normal | ||
| Mother | Negative | |||||
| Chronic lymphocytic leukemia | ND | ND | ND | ND | ND | |
| Proband | Positive | |||||
| Endometrial 39 | Normal | Loss | Loss | ND | ||
| Spindle Cell Sarcoma 39 | Normal | Normal | Normal | ND | ||
| Colon 50 | High | Normal | Loss | Loss | normal | |
| Papillary bladder-58 | Normal | Normal | Normal | ND | ||
| Pancreas 60 | Normal | Loss | Loss | normal | ||
| Sister | Positive | |||||
| Pancreas 54 | Normal | Loss | Loss | ND | ||
MSI-tumor microsatellite instability; ND-not done.
Results
In silico analyses. 17% of all mutations in MSH2 are missense mutations.12 The P349L variant is not listed in the Mismatch Repair Genes Variant Database from the Memorial University of Newfoundland (http://www.med.mun.ca/MMRvariants/search_results.aspx) nor is it included in the paper or supplemental materials in the MAPP-MMR database.13 It is also not reported in the MMR Gene Unclassified Variants Database (www.mmruv.info), although an MSH2 P349R mutation at the same site is reported by three in silico models, suggesting pathogenicity.13 The P349L variant has not been included in functional studies of pathogenicity of MSH2 missense variants.14, 15 However, the P349L variant is located in the lever domain of the MSH2 gene, a large domain that connects the ATP binding subunits to the clamp domains to mediate signals between the ATP- and the DNA-binding portions of the protein. Two of three missense substitutions studied functionally in the lever domain manifest lower stability and defective DNA mismatch repair and loss of expression in tumors, which is consistent with studies of homologous positions in yeast MSH2, in which half of missense alterations lead to inefficient expression of the gene.14, 15 The Uniprot database, referring to the Domingo report of this family,10 cites the Pro349Leu variant as possibly pathogenic (http://www.expasy.org/cgi-bin/variant_pages/get-sprot-variant.pl?VAR_043763). A BLOSUM score of -3 is reported in Uniprot.16 This is supported by in silico analyses using Align-GVGD, with Grantham Variation 0 and the Grantham Deviation 97.78 resulting in a C65 score for MSH2 P349L.17, 18 These findings together indicate that the residue is evolutionarily constrained, and predicts that this missense alteration is very likely to have functional consequences.
In order to derive a quantitative classification of pathogenicity, we performed Bayes factor analysis of variant segregation data using methods described previously.19 Calculations assumed age-specific relative risks for colorectal cancer, endometrial cancer, and other Lynch Syndrome-related cancers (including pancreatic cancer) as estimated in Quehenberger et al. (2005).20 This analysis also provided odds in favor of pathogenicity of 35.7:1, translating to a probability of pathogenicity of 0.97 for this variant. MSH2 P349L would thus be considered class 4 (likely pathogenic), based on the IARC 5 class classification system that is linked to posterior probability estimates.21
Discussion
Available data suggest that, in general, penetrance for pancreatic cancer in Lynch Syndrome is low. Prior to discovery of the genetic basis of the Lynch Syndrome, Watson and Lynch (1993) studied 1,424 at-risk persons from 23 large families (with 287 colorectal cancers) who were suspected of having this disorder.22 Six pancreatic cancers were recorded, compared with 4.1 expected, which was not statistically significantly different. In l999, Aarnio et al. had studied 360 gene carriers of 50 families with gene mutations (94% in MLH1 and 6% in MSH2), and found 3 pancreatic cancers, giving a Standardized Incidence Ratio of 4.5, but with a 95% CI of 1.0–14.23 In 2008, in a study that assessed extracolonic cancer risk among 6,041 members of 261 families with documented mutations in MLH1 (60%) or MSH2 (40%), cancers of the biliary tract, liver and pancreas combined accounted for 1.09% of the cancers in this study, giving a hazard ratio of 1.869 and a cumulative incidence of 4.1% to age 70 years.24 Most recently, risk of pancreatic cancer alone was addressed in 6,342 individuals from 147 families with MMR mutations (37.4% in MLH1, 55.1% in MSH2, and 7.55% in MSH6). A cumulative risk of pancreatic cancer was calculated as 1.31% (95% CI=0.31–2.32%) to age 50, and 3.68% (95% CI=1.45–5.88%) to age 70, which is an 8.6-fold (95% CI=4.7–15.7%) increase compared with the general population.25 In summary, we present a family in which a novel P349L missense substitution in MSH2 that co-segregates with disease in a Lynch Syndrome family appears particularly to be associated with a high risk for pancreatic cancer. All three cancer affected individuals carry the MSH2 P349L missense substitution, and there is loss of expression of MSH2/MSH6 in each of their pancreatic tumors. Together with in silico predictions, these data provide support that this alteration is the causative mutation. It would be of interest to learn of other families with the same missense alteration to determine if predisposition to pancreatic cancer is consistently associated with this MSH2 change.
Footnotes
Disclosure: Partial support provided by NIH Grant R01 CA97075 and NHMRC Project Grant 496616
Contributor Information
Noralane M. Lindor, Department of Medical Genetics, Mayo Clinic, Rochester, Minnesota.
Gloria M. Petersen, Department of Health Science Research, Mayo Clinic, Rochester, Minnesota.
Amanda B. Spurdle, Division of Genetics and Population Health, Queensland Institute of Medical Research, Brisbane, Australia.
Bryony Thompson, Division of Genetics and Population Health, Queensland Institute of Medical Research, Brisbane, Australia.
David E. Goldgar, Department of Dermatology, University of Utah School of Medicine, Salt Lake City, Utah.
Stephen N. Thibodeau, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota.
References
- 1.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005a;352(18):1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 2.Wijnen J, Vasen H, Kahn M, et al. Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N Engl J Med. 1998;339:511–518. doi: 10.1056/NEJM199808203390804. [DOI] [PubMed] [Google Scholar]
- 3.Vasen H, Watson P, Mecklin J, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999 Jun;116(6):1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 4.Lindor NM, Petersen GM, Hadley DW, et al. Recommendations for the Care of Individuals With an Inherited Predisposition to Lynch Syndrome. JAMA. 2006;296:1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- 5.Vasen H, Moslein G, Alonso A, et al. Guidelines for the clinical management of Lynch syndrome. J Med Genet. 2007;44(6):353–362. doi: 10.1136/jmg.2007.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Umar A, Boland C, Terdiman J, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004a Feb 18;96(4):261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002a Feb 15;20(4):1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham JM, Kim CY, Christensen ER, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69(4):780–790. doi: 10.1086/323658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baudhuin LM, Burgart LJ, Leontovich O, et al. Use of microsatellite instability and immunohistochemistry testing for the identification of individuals at risk for Lynch syndrome. Fam Cancer. 2005a;4(3):255–265. doi: 10.1007/s10689-004-1447-6. [DOI] [PubMed] [Google Scholar]
- 10.Domingo E, Laiho P, Ollikainen M, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41(9):664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Cunningham J, Winters J, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res. 2003a Sep 1;63(17):5209–5212. [PubMed] [Google Scholar]
- 12.Woods MO, Williams P, Careen A, et al. A new variant database for mismatch repair genes associated with Lynch syndrome. Hum Mutat. 2007;28:669–373. doi: 10.1002/humu.20502. [DOI] [PubMed] [Google Scholar]
- 13.Chao EC, Velasquez JL, Witherspoon MS, et al. Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR) Hum Mutat. 2008;29(6):852–860. doi: 10.1002/humu.20735. [DOI] [PubMed] [Google Scholar]
- 14.Gammie AE, Erdeniz N, Beaver J, et al. Functional characterization of pathogenic human MSH2 missense mutations in Saccharomyces cerevisiae. Genetics. 2007;177(2):707–721. doi: 10.1534/genetics.107.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ollila S, Dermadi Bebek D, Jiricny J, et al. Mechanisms of Pathogenicity in Human MSH2 Missense Mutants. Hum Mutat. 2008;29(11):1355–1363. doi: 10.1002/humu.20893. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S, Henikoff JG. Amino Acid Substitution Matrices from Protein Blocks. PNAS. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavtigian S, Deffenbaugh A, Yin L, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43(4):295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathe E, Olivier M, Kato S, et al. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 2006;34(5):1317–1325. doi: 10.1093/nar/gkj518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold S, Buchanan DD, Barker M, et al. Classifying MLH1 and MSH2 variants using bioinformatic prediction, splicing assays, segregation, and tumor characteristics. Hum Genet. 2009;30(5):757. doi: 10.1002/humu.20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quehenberger F, Vasen H, van Houwelingen H. Risk of colorectal and endometrial cancer for carriers of mutations of the hMLH1 and hMSH2 gene: correction for ascertainment. J Med Genet. 2005;42:491–496. doi: 10.1136/jmg.2004.024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plon S, Eccles D, Easton D, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29(11):1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson P, Lynch H. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer. 1993 Feb 1;71(3):677–685. doi: 10.1002/1097-0142(19930201)71:3<677::aid-cncr2820710305>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 23.Aarnio M, Sankila R, Pukkala E, et al. Cancer risk in mutation carriers of DNA- mismatch-repair genes. Int J Cancer. 1999 Apr;81(2):214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 24.Watson P, Vasen HF, Mecklin JP, et al. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. International Journal of Cancer. 2008;123(2):444–449. doi: 10.1002/ijc.23508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kastrinos F, Mukherjee B, Tayob N, et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009;302(16):1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]