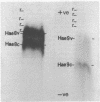
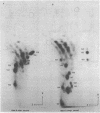
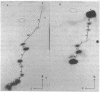
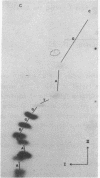
Abstract
The nucleotide sequence of a double stranded DNA fragment from the gene AB region of bacteriophage S13 DNA has been determined. The fragment was isolated as two adjacent shorter fragments by cleavage of S13 replicative form (RF) DNA with restriction endonuclease III from Hemophilus aegyptius. The strands of the fragments were separated electrophoretically and hydrolyzed with T4 endonuclease IV to yield short oligonucleotides which were then sequenced by partial exonuclease digestion. The complete nucleotide sequence of the restriction fragments was obtained by ordering the inter- and intrastrand overlapping oligonucleotide sequences. The adjacent fragments were 190 nucleotides in length. The sequences included a HindII site, an AluI site and two sequences which may be possible transcription initiation sequences, one with an adjacent sequence homologous to the canonical promoter site sequence T-A-T-Pu-A-T-Pu. Examination of the three possible reading frames for translation of the sequence revealed only one possible complete translation product. The postulated partial sequence of gene A protein has a highly positively charged arginine-rich area which may have importance in DNA binding.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker R., Tessman I. The circular genetic map of phage S13. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1438–1445. doi: 10.1073/pnas.58.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambara R., Jay E., Wu R. DNA sequence analysis: a formula to predict electrophoretic mobilities of oligonucleotides on cellulose acetate. Nucleic Acids Res. 1974 Nov;1(11):1503–1520. doi: 10.1093/nar/1.11.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow R. M., Hutchison C. A., Fabricant J. D., Sinsheimer R. L. Genetic Map of Bacteriophage phiX174. J Virol. 1971 May;7(5):549–558. doi: 10.1128/jvi.7.5.549-558.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi A., Maat J., de Waard A., Bernardi G. Preparation and specificity of endonuclease IV induced by bacteriophage T4. Eur J Biochem. 1976 Jun 15;66(1):175–179. doi: 10.1111/j.1432-1033.1976.tb10437.x. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- Delaney A. D., Spencer J. H. Nucleotide clusters in deoxyribonucleic acids. XIII. Sequence analysis of the longer unique pyrimidine oligonucleotides of bacteriophage S13 DNA by a method using unlabeled atarting oligonucleotides. Biochim Biophys Acta. 1976 Jul 2;435(3):269–281. doi: 10.1016/0005-2787(76)90108-8. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T., Marvin D. A. Altered coding in single stranded DNA viruses? Nature. 1969 Feb 22;221(5182):769–770. doi: 10.1038/221769a0. [DOI] [PubMed] [Google Scholar]
- GAREN A., LEVINTHAL C. A fine-structure genetic and chemical study of the enzyme alkaline phosphatase of E. coli. I. Purification and characterization of alkaline phosphatase. Biochim Biophys Acta. 1960 Mar 11;38:470–483. doi: 10.1016/0006-3002(60)91282-8. [DOI] [PubMed] [Google Scholar]
- Galibert F., Sedat J., Ziff E. Direct determination of DNA nucleotide sequences: structure of a fragment of bacteriophage phiX172 DNA. J Mol Biol. 1974 Aug 15;87(3):377–407. doi: 10.1016/0022-2836(74)90093-x. [DOI] [PubMed] [Google Scholar]
- Godson G. N. Evolution of phiX174 III. Restriction map of S13 and its alignment with that of phiX174. Virology. 1976 Dec;75(2):263–280. doi: 10.1016/0042-6822(76)90027-1. [DOI] [PubMed] [Google Scholar]
- Grosveld F. G., Ojamaa K. M., Spencer J. H. Fragmentation of bacteriophage S13 replicative form DNA by restriction endonucleases from Hemophilus influenzae and Hemophilus aegyptius. Virology. 1976 May;71(1):312–324. doi: 10.1016/0042-6822(76)90115-x. [DOI] [PubMed] [Google Scholar]
- Harbers B., Delaney A. D., Harbers K., Spencer J. H. Nucleotide clusters in deoxyribonucleic acids. Comparison of the sequences of the large pyrimidine oligonucleotides of bacteriophages S13 and phiX174 deoxyribonucleic acids. Biochemistry. 1976 Jan 27;15(2):407–414. doi: 10.1021/bi00647a026. [DOI] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Fragment maps of phiX-174 replicative DNA produced by restriction enzymes from haemophilus aphirophilus and haemophilus influenzae H-I. J Virol. 1974 Nov;14(5):1142–1151. doi: 10.1128/jvi.14.5.1142-1151.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T. J., Knippers R. Isolation and function of the gene A initiator of bacteriophage phi-chi 174, a highly specific DNA endonuclease. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1549–1553. doi: 10.1073/pnas.71.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N. W., Gilham P. T. The analysis of polydeoxyribonucleotides by digestion with phosphatase and phosphodiesterases. Biochim Biophys Acta. 1973 Apr 21;308(7):53–58. doi: 10.1016/0005-2787(73)90121-4. [DOI] [PubMed] [Google Scholar]
- Jay E., Bambara R., Padmanabhan R., Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974 Mar;1(3):331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng Y., Gelfand D., Hayashi M., Shleser R., Tessman E. S. The eight genes of bacteriophages phi X174 and S13 and comparison of the phage-specified proteins. J Mol Biol. 1970 Apr 28;49(2):521–526. doi: 10.1016/0022-2836(70)90262-7. [DOI] [PubMed] [Google Scholar]
- Junowicz E., Spencer J. H. Nucleotide clusters in deoxyribonucleic acids. Separation of oligo-nucleotides released by deoxyribonuclease I. Biochemistry. 1970 Sep 1;9(18):3640–3648. doi: 10.1021/bi00820a022. [DOI] [PubMed] [Google Scholar]
- LEHMAN I. R., NUSSBAUM A. L. THE DEOXYRIBONUCLEASES OF ESCHERICHIA COLI. V. ON THE SPECIFICITY OF EXONUCLEASE I (PHOSPHODIESTERASE). J Biol Chem. 1964 Aug;239:2628–2636. [PubMed] [Google Scholar]
- Ling V. Fractionation and sequences of the large pyrimidine oligonucleotides from bacteriophage fd DNA. J Mol Biol. 1972 Feb 28;64(1):87–102. doi: 10.1016/0022-2836(72)90322-1. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Ptashne M., Backman K., Kield D., Flashman S., Jeffrey A., Maurer R. Recognition sequences of repressor and polymerase in the operators of bacteriophage lambda. Cell. 1975 Jun;5(2):109–113. doi: 10.1016/0092-8674(75)90018-5. [DOI] [PubMed] [Google Scholar]
- Middleton J. H., Edgell M. H., Hutchison C. A., 3rd Specific fragments of phi X174 deoxyribonucleic acid produced by a restriction enzyme from Haemophilus aegyptius, endonuclease Z. J Virol. 1972 Jul;10(1):42–50. doi: 10.1128/jvi.10.1.42-50.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J. N., Borst P., Grosveld F. G., Spencer J. H. The size of the repeating unit of the repetitive mitochondrial DNA from a "low-density" petite mutant of yeast. Biochim Biophys Acta. 1974 Dec 6;374(2):115–128. doi: 10.1016/0005-2787(74)90355-4. [DOI] [PubMed] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975 Mar;72(3):784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J. Nucleotide sequence from the coding region of rabbit beta-globin messenger RNA. Nucleic Acids Res. 1976 Jul;3(7):1811–1821. doi: 10.1093/nar/3.7.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Apr;3(4):863–877. doi: 10.1093/nar/3.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski P. D., Bakyta I. T4 endonuclease IV. Improved purification procedure and resolution from T4 endonuclease 3. J Biol Chem. 1972 Jan 25;247(2):405–412. [PubMed] [Google Scholar]
- Sadowski P. D., Hurwitz J. Enzymatic breakage of deoxyribonucleic acid. II. Purification and properties of endonuclease IV from T4 phage-infected Escherichia coli. J Biol Chem. 1969 Nov 25;244(22):6192–6198. [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sanger F. The Croonian Lecture, 1975. Nucleotide sequences in DNA. Proc R Soc Lond B Biol Sci. 1975 Dec 2;191(1104):317–333. doi: 10.1098/rspb.1975.0131. [DOI] [PubMed] [Google Scholar]
- Schekman R. W., Iwaya M., Bromstrup K., Denhardt D. T. The mechanism of replication of phi X174 single-stranded DNA. 3. An enzymic study of the structure of the replicative form II DNA. J Mol Biol. 1971 Apr 28;57(2):177–199. doi: 10.1016/0022-2836(71)90340-8. [DOI] [PubMed] [Google Scholar]
- Schendel P. F., Wells R. D. The synthesis and purification of (gamma-32P)-adenosine triphosphate with high specific activity. J Biol Chem. 1973 Dec 10;248(23):8319–8321. [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Smith L. H., Sinsheimer R. L. The in vitro transcription units of bacteriophage phiX174. II. In vitro initiation sites of phiX174 transcription. J Mol Biol. 1976 Jun 5;103(4):699–710. doi: 10.1016/0022-2836(76)90204-7. [DOI] [PubMed] [Google Scholar]
- Smith L. H., Sinsheimer R. L. The in vitro transcription units of bacteriophage phiX174. III. Initiation with specific 5' end oligonucleotides of in vitro phiX174 RNA. J Mol Biol. 1976 Jun 5;103(4):711–735. doi: 10.1016/0022-2836(76)90205-9. [DOI] [PubMed] [Google Scholar]
- TESSMAN E. S., SHLESER R. Genetic recombination between phages S13 and phi-X174. Virology. 1963 Feb;19:239–240. doi: 10.1016/0042-6822(63)90015-1. [DOI] [PubMed] [Google Scholar]
- Tessman I., Tessman E. S., Pollock T. H., Borrás M. T., Puga A., Baker R. Reinitiation mutants of gene B of bacteriophage S13 that mimic gene A mutants in blocking replicative form DNA synthesis. J Mol Biol. 1976 May 25;103(3):583–598. doi: 10.1016/0022-2836(76)90218-7. [DOI] [PubMed] [Google Scholar]
- Warner H. R., Lewis N. The synthesis of deoxycytidylate deaminase and dihydrofolate reductase and its control in Escherichia coli infected with bacteriophage T4 and T-4 amber mutants. Virology. 1966 May;29(1):172–175. doi: 10.1016/0042-6822(66)90208-x. [DOI] [PubMed] [Google Scholar]
- Weisbeek P. J., Vereijken J. M., Baas P. D., Jansz H. S., Van Arkel G. A. The genetic map of bacteriophage phiX174 constructed with restriction enzyme fragments. Virology. 1976 Jul 1;72(1):61–71. doi: 10.1016/0042-6822(76)90311-1. [DOI] [PubMed] [Google Scholar]
- Wu R., Bambara R., Jay E. Recent advances in DNA sequence analysis. CRC Crit Rev Biochem. 1975 Feb;2(4):455–512. doi: 10.3109/10409237509102550. [DOI] [PubMed] [Google Scholar]
- ZAHLER S. A. Some biological properties of bacteriophages S13 and theta X-174. J Bacteriol. 1958 Mar;75(3):310–315. doi: 10.1128/jb.75.3.310-315.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff E. B., Sedat J. W., Galibert F. Determination of the nucleotide sequence of a fragment of bacteriophage phiX 174 DNA. Nat New Biol. 1973 Jan 10;241(106):34–37. doi: 10.1038/newbio241034a0. [DOI] [PubMed] [Google Scholar]