Abstract
We report a capillary-based DNA sequencing read length of 100 bases in 16 min using end-labeled free-solution conjugate electrophoresis (FSCE) with a monodisperse poly-N-substituted glycine (polypeptoid) as a synthetic drag-tag. FSCE enabled rapid separation of single-stranded (ss) DNA sequencing fragments with single-base resolution without the need for a viscous DNA separation matrix. Protein-based drag-tags previously used for FSCE sequencing, for example, streptavidin, are heterogeneous in molar mass (polydisperse); the resultant band-broadening can make it difficult to obtain the single-base resolution necessary for DNA sequencing. In this study, we synthesized and HPLC-purified a 70mer poly-N-(methoxyethyl)glycine (NMEG) drag-tag with a molar mass of~11 kDa. The NMEG monomers that comprise this peptoid drag-tag are interesting for bioanalytical applications, because the methoxyethyl side chain's chemical structure is reminiscent of the basic monomer unit of polyethylene glycol, a highly biocompatible commercially available polymer, which, however, is not available in monodisperse preparation at an ~11 kDa molar mass. This is the first report of ssDNA separation and of four-color, base-by-base DNA sequencing by FSCE through the use of a chemically synthesized drag-tag. These results show that high-molar mass, chemically synthesized drag-tags based on the polyNMEG structure, if obtained in monodisperse preparation, would serve as ideal drag-tags and could help FSCE reach the commercially relevant read lengths of 100 bases or more.
Keywords: FSCE, DNA sequencing, peptoid, polyethylene glycol, sulfo-SMCC
INTRODUCTION
Capillary electrophoresis (CE) and microchip electrophoresis significantly advanced the technology of high-throughput DNA sequencing relative to original slab gel methods. Although in 2011, CE is not “cutting-edge” in comparison with newer next-generation DNA sequencing technologies, it is still ubiquitously used for small-scale DNA sequencing and for the sequencing of clinical or forensic DNA samples, because it offers the practical advantages of simplicity, high accuracy, and very low per-lane cost. This is the case even if only a few hundred bases are sequenced in total.1–5 Next-generation DNA sequencers are geared to larger-scale sequencing projects—often whole chromosomes, or whole genomes—and cannot be run or provide any data at all for less than ~$4000 (at present). CE separations of DNA, especially for DNA sequencing, require viscous separation networks (highly entangled polyacrylamide solutions), which are expensive and also quite difficult to load into narrow microchannels. This problem is intensified for microfluidic chips, which cannot withstand the high loading pressures that are usually required to force in the viscous DNA separation matrix.6,7
Free-solution capillary electrophoresis (FSCE) is a matrix-free separation technique that relies on appending a mobility modifier or “drag-tag” to each DNA fragment,8,9 resulting in size-based separation of the DNA in a simple aqueous buffer, with no added polymer. Although FSCE has tremendous potential, most of the current research and development setbacks have been related to inferior molecular characteristics of the drag-tag and the difficulty of obtaining an ideal drag-tag.7,10–12 The list of needed attributes for an ideal drag-tag is demanding and includes (i) good water-solubility and hydro-philicity and a very low tendency to “stick” to the glass capillary surface by hydrophobic interactions; (ii) large molecular size to provide sufficient drag for the separation of long DNA fragments; (iii) total monodispersity (size and structure homogeneity) to ensure clear and readable electropherograms in which each individual DNA size (e.g., 50 bases) has one, sharp peak; (iv) charge-neutrality (or near-neutrality) to avoid situations in which (if negatively charged) a drag-tag contributes its own, strong electrophoretic mobility in the same direction as the DNA, or (if positively charged) the drag-tag is a source of band-broadening via nonspecific ionic interactions with negatively charged glass microchannel walls.7,13–16
Both natural and engineered, bacterially expressed recombinant proteins have been demonstrated as drag-tags for FSCE DNA sequencing. Typically, the globular nature, charged residues, and polydispersity of these proteins have resulted in substantial band broadening and severe limitations on the single-base resolution of ssDNA sequencing fragments.11 As a recent exception, after rigorous and highly specialized protein purification procedures, systematically designed “protein polymers” have been shown to be able to provide high-resolution ssDNA sequencing by CE.10
The unavoidable polydispersity of synthetic molecules that might have a sufficient size and molar mass to be useful for ssDNA sequencing, such as polyethylene glycol (PEG), render them unsuitable as drag-tags. This is true even of so-called monodisperse PEG preparations that are commercially available at (for example) weight–average molar masses of 3, 5, or 12 kDa; they are not sufficiently monodisperse to be used. For instance, a monodisperse, 5 kDa PEG sample, appended end-on to a single, monodisperse ssDNA 20mer oligonucleotide, yields ~110 separate, resolved peaks by free-solution CE, where each peak differed in mass by one PEG monomer unit.17 This study did, however, demonstrate the extraordinarily high resolving power of bioconjugate CE methods.
Poly-N-substituted glycines (peptoids) are non-natural oligomers, which have been developed and used for a number of different applications in biotechnology and medicine. We undertook to investigate polypeptoids as ssDNA sequencing drag-tags, because their inherent properties appeared to be suitable. Specifically, we considered their facile solid-phase synthesis and easy HPLC purification to be an advantage (PEGs, by contrast, require highly specialized, high-pressure chemical synthesis methods). We also considered the ease of controlling polypeptoid monomer sequence, given that they are made on solid phase, “monomer-by-monomer,” using an automated peptide synthesizer, providing high compound tunability and easy methods by which to modify the chain termini for chemoselective ligation to ssDNA molecules. Finally, we considered the good chemical stability of polyamides in aqueous buffer at pH 6–8, the relevant pH range for DNA separations.7,17–24 However, polypeptoids are limited in their accessible size by the solid-phase synthesis methods used to generate them. The direct, automated synthesis of peptoid chains longer than ~50 monomers (as needed for long read-length DNA sequencing) results in low overall product yield and challenging purifications.
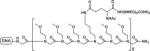
We previously investigated the effects of polypeptoid architecture on hydrodynamic drag, not by doing base-by-base DNA sequencing, but by separating a ladder of ssDNA molecules that we generated by PCR. In that study, we used CE with single-color laser-induced fluorescence (LIF) detection, whereas DNA sequencing requires an instrument that has a specialized detector that provides for four-color detection (one color for each of the four DNA bases). In that study, a comb-like, “branched” poly-N-(methoxyethyl)glycine (polyNMEG)-based 70mer was the highest molar mass compound used7 (the 70mer's chemical structure and properties are given in Table I). Smaller branched drag-tags were used as well. Importantly, the hydrodynamic drag generated by the branched polyNMEG drag-tags was found to scale linearly with drag-tag molar mass, indicating that a large amount of drag can be obtained by the creation of branched structures, without the tedious synthesis and purification of very long, totally monodisperse linear polymers.
Table I.
Polypeptoid Drag–Tag Structure and Properties, Including the Experimental Sequencing Results and Theoretical Prediction Using Eq. (2)
| Octamer-Branched 30mer Drag–Tag Structure | Mass (kDa) | Calculated α | Theoretical Read Length | Observed Read Length |
|---|---|---|---|---|
|
11.09 | 18.0 | 92 | 80–100 |
In the branched portion of the drag-tag chemical structure, N-(methoxyethyl)glycine peptoid residues are abbreviated as “NMEG”.
The amount of drag generated by a drag-tag is characterized in terms of an effective friction coefficient, α, which is the hydrodynamic drag quantified in units of “the hydrodynamic drag on a single ssDNA base.”8,25 The drag parameter α is estimated from knowledge of the electrophoretic mobility, μ, of a drag-tag-modified DNA fragment of Mc bases, relative to the free-solution mobility of DNA, μ0, as shown in Eq. (1):
| (1) |
The 70mer branched polyNMEG drag-tag was conjugated to 20 and 30mer DNA oligonucleotides as well as to single-stranded (ss) PCR products up to 150 bases in length; α for this drag-tag was found to be 17.2.7 The resolution R for diffusion-limited FSCE sequencing is given by Eq. (2),26 and the read length for a drag-tag with a friction of α can be predicted by solving this equation for Mc at a resolution of R = 1:
| (2) |
In this equation, R0 is an experimental parameter that is estimated to be 5.3 × 10−3 for experimental conditions. At the previously mentioned α of 17.2, Eq. (2) predicts a read length of 90 bases at an applied field of 313 V/cm. Therefore, we expected the synthetic 70mer drag-tag to produce a short but significant read length of 110–120 bases, on the same order of magnitude as obtained with streptavidin11 and our 127mer protein polymer.10
The synthesis and purification methods by which we obtained the 70mer NMEG-based peptoid drag-tag, which we use here for four-color, base-by-base ssDNA Sanger sequencing by CE, are described elsewhere.7 Briefly, a linear 30mer peptoid “backbone”—comprising not only NMEG monomers but also five evenly spaced, lysine-like monomers that are chemically reactive and hence easily modified—was synthesized by the solid-phase “submonomer” method using an ABI 433A automated peptide synthesizer and previously described methods.27–31 After full HPLC purification of this 70mer (away from shorter chains that resulted from incomplete monomer couplings), we grafted five octamer NMEG branches onto the lysine-like monomers' primary amine groups via solution-phase coupling. The product was purified by standard RP-HPLC to >99% homogeneity (assessed by analytical RP-HPLC), and the molar mass was confirmed to be both correct and monodisperse by MALDI-TOF/MS. The N-terminal primary amine of the drag-tag was then conjugated to a reduced, 5'-thiolated, M13mp18 (–40) ssDNA sequencing primer [5'-X1GTTTTCCCAGTCACGAC-3', where X1 is a C6-thiol modification] using the common heterobifunctional linker sulfo-SMCC.
The ABI SNaPshot kit, intended for single-base extension genotyping, was used to obtain four-color sequencing samples, with small amounts of dNTPs added to ensure the generation of relatively small ssDNA sequencing fragments (<200 bases). An aliquot of 5 μl of the SNaPshot premix was mixed with 3 pmol of drag-tag-linked ssDNA primer, 0.12 μg of M13mp18 control template, and 200 nmol of each dNTP to a total volume of 10 μl. The mixture was subjected to a Sanger cycle sequencing protocol with 25 cycles of denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 30 s. Sequencing reactions were purified by gel filtration with a Centri-sep column (Princeton Separations) and denatured in formamide before analysis by CE.
The “drag-tagged” ssDNA sequencing reaction products were analyzed by FSCE with four-color LIF detection using an Applied Biosystems Prism 3100 capillary array electrophoresis instrument, with standard sequencing analysis protocols modified to allow for the loading of buffer rather than POP-5 polymer matrix into the capillary array. The capillary array had an effective length (length to the LIF detection window) of 36 cm and a total length of 47 cm. The running buffer was 50 mM Tris, 50 mM TAPS, and 2 mM EDTA, sometimes referred to as 0.53× TTE and 7M urea, pH 8.5, with a 3% (v/v) dilution of POP-5 polymer solution (as commercially obtained) added as a dynamic capillary wall coating agent to suppress electroosmotic flow (note that this low concentration of added polymer does not lead to the sieving of DNA molecules of any size). The separations were carried out at 14.7 kV total potential (313 V/cm) at 55°C.
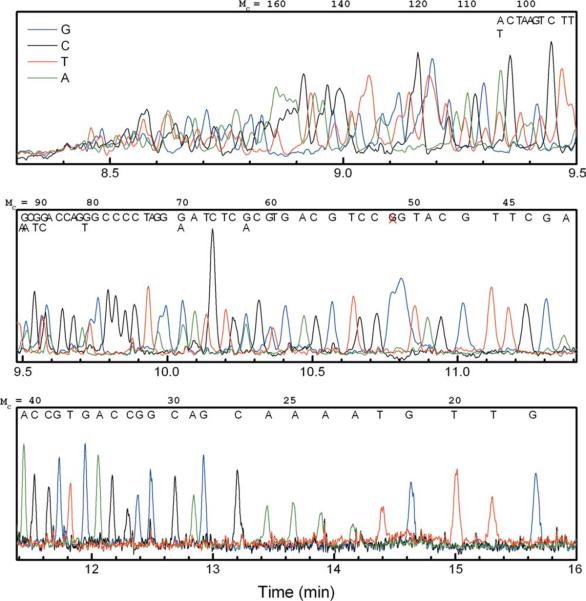
The resulting four-color sequencing electropherogram is shown in Figure 1. This is an essentially raw, unprocessed sequencing trace; that is, customary data processing typically used for matrix-based sequencing is absent. There were however two minor manipulations of the raw data shown in the figure: (1) for the sake of creating one plot, on a single set of axes, the “C” and “T” channel data were scaled by a constant factor of 2 or 3 respectively, to compensate for the less efficient excitation of the fluorescent dyes used on these base-specific terminators (when compared with “A” and “G”) by the 488 nm Ar-ion laser of the instrument; and (2) the data were smoothed with a Savitsky-Golay algorithm, implemented from within the plotting software (Origin™). This is a cosmetic change that compensated to some degree for a relatively noisy baseline, which could have been related to the fact that a relatively small amount of sample was injected, giving a lower signal-to-noise ratio. Neither manipulation affected our ability to interpret the sequence (nor indeed, the interpretation itself), and resolution was determined from the raw (unsmoothed) data. Manual alignment of the peaks to the known M13mp18 control sequence suggests a read length between 80 and 100 well-resolved ssDNA bases. As we will discuss in more detail below, this result is impressive when considering that we used a drag-tag comprising just 70 NMEG monomers. Note that each dye terminator has its own intrinsic electrophoretic mobility that is based on its chemical structure and thus gave a slightly different mobility shift in the raw data. Subsequently, the peaks for the given bases shifted out of their expected positions and caused overlap or position reversal of adjacent peaks. The ABI CE instrument has its own software package that applies a correction to account for this (and which also smoothes the baseline), but because our DNA sequencing peaks elute “in the wrong order” (largest DNA first, smallest DNA last), this software package could not be used. Instead, to estimate the potential limitations on accurate read length, we had to look carefully at adjacent peaks within a given terminator track (e.g., just “G” or just “C”), which we knew, through knowledge of the sequence of the template DNA that we used, represented the electrophoretic zones of ssDNA molecules differing by just one base. We consider it likely that our estimate of where the readable resolution “stops,” that is, the upper limit in ssDNA size at which peak overlap would make even properly processed, smoothed, and corrected DNA sequence unreadable is reasonably accurate by the use of this approach.
FIGURE 1.
Four-color sequencing electropherogram obtained with 70mer branched drag-tag, with manual base calls of the M13mp18 sequence. The DNA fragment sizes (Mc) are shown at the top of each panel, and a red “X” around Mc = 52 bases marks an unexpected “G” peak. For the purposes of presentation, the “C” and “T” signals have been scaled by a factor of 2 to give peak heights comparable to the “G” and “C” traces, and the data has been smoothed to reduce the high-frequency noise attributed to a small bubble in the fluid path.
A peak-fitting routine (in the software package Origin) was used to estimate the peak widths of overlapping peaks, which in turn allowed the estimation of resolution for several runs of repeated bases. The resolution was lower for the G-terminated fragments than for the other terminators, for example, the GG pairs at 71–72 and 80–81 bases showed lower resolution than CCCC peaks at 75–78 bases. This may be a result of the interaction of the fluorescent dye on the G terminator with the capillary wall, which could cause additional band broadening; hence could well be the consequence of the particular dichlororhodamine dye terminator chemistry used in the SNaPshot kit. Interestingly, the G peak at 51 bases appeared to be split into two poorly resolved peaks, although there is only a single “G” in this position. Again, this could relate to the interference of the dye's interactions with the capillary wall (split peaks of this sort are sometimes seen in the free-solution CE separation of cationic or hydro-phobic proteins that “stick” to the glass capillary wall). Note that this shifting of peaks and decreased resolution for “G” terminated fragments was previously observed to occur in the FSCE sequencing with a protein polymer drag-tag.10 The CC pair at 84–85 bases is baseline-resolved. The overlapping TT pair at 95–96 bases was well resolved, whereas the AA pair at 100–101 bases was incompletely resolved. These results suggest that the upper limit in read length with this drag-tag is roughly 100 bases, in reasonable agreement with the prediction of Eq. (2) for diffusion-limited conditions. (A rigorous verification of diffusion-limited performance would require performing similar separations at multiple different electric field strengths and determining the extent of band-broadening as a function of the migration velocity or residence time of the different bioconjugates in the capillary.) Advantageously, the relatively bulky, branched polypeptoid molecule with a molar mass of 11 kDa, when attached to the sequencing primer during the Sanger cycle sequencing reaction, did not seem to interfere with the action of the DNA polymerase.
A linearized plot of electrophoretic mobility versus migration time is shown in Figure 2. The experimental data are fit well with the simple model represented by Eq. (1). In this case, we find that α = 18.0, which is close to the value of 17.2 observed and reported previously.7 The sequencing read length of 80–100 bases is also consistent with the prediction of ~90 bases from Eq. (2). Although this read length is short compared to common matrix-based sequencing reads (typically 650–800 bases), it is significant when accounting for the small size of the 70-monomer, 11-kDa peptoid drag-tag that was used. The much larger, >600 amino acid, 53-kDa streptavidin protein yielded only a slightly longer read length of ~110 bases.11 For branched polypeptoid drag-tags, it has been observed that α scales linearly with molar mass within the size range investigated so far, as opposed to a scaling with roughly the one-third power of molar mass for globular proteins like streptavidin. Thus, we expect that a branched drag-tag of a molar mass similar to that of streptavidin would provide dramatically better sequencing performance.
FIGURE 2.
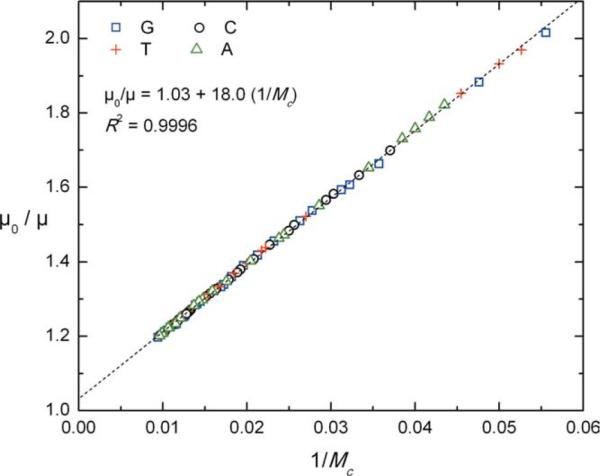
A fit of experimentally measured mobilities (μ0/μ) versus DNA fragment size 1/Mc, according to a rearranged version of Eq. (1), with a slope of α = 18.0 for the octamer-branched drag-tag. Different symbols are used to represent fragments with G, C, A, or T terminations, which reveal no terminator-specific deviation from model behavior.
In summary, there is great potential for long-read length FSCE DNA sequencing using synthetic polyNMEG-based drag-tags, provided that higher molar mass branched peptoids can be successfully prepared, while simultaneously adhering to FSCE's extremely strict purity and homogeneity requirements. The use of polypeptoids offers the advantages of facile sequence design and synthesis and the ability to readily incorporate chemically reactive monomer units at intervals via the submonomer synthesis method.31 Once this “backbone” polypeptoid structure has been purified, chemo-selective ligation chemistries afford the synthesis of comblike, branched polypeptoid structures that are reasonably large. Clearly, with some effort put into the design and chemical synthesis of these branched drag-tag molecules, substantially longer DNA sequencing read lengths are within reach.
Acknowledgments
Contract grant sponsor: National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH)
Contract grant number: 5 R01 HG002918-01
Contract grant sponsor: Northwestern University Henderson Fellowship
Footnotes
Russell D. Haynes and Robert J. Meagher contributed equally to this work.
REFERENCES
- 1.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. Nucleic Acids Res. 2008;36:D475–D479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J, Mitra RD, Varma C, Church GM. Nat Rev Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 3.Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 4.Fredlake CP, Hert DG, Kan CW, Chiesl TN, Root BE, Forster RE, Barron AE. Proc Natl Acad Sci USA. 2008;105:476–481. doi: 10.1073/pnas.0705093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kan CW, Fredlake CP, Doherty EAS, Barron AE. Electrophoresis. 2004;25:3564–3588. doi: 10.1002/elps.200406161. [DOI] [PubMed] [Google Scholar]
- 6.Meagher RJ, McCormick LC, Haynes RD, Won JI, Lin JS, Slater GW, Barron AE. Electrophoresis. 2006;27:1702–1712. doi: 10.1002/elps.200500554. [DOI] [PubMed] [Google Scholar]
- 7.Haynes RD, Meagher RJ, Won JI, Bogdan FM, Barron AE. Bioconjugate Chem. 2005;16:929–938. doi: 10.1021/bc0496915. [DOI] [PubMed] [Google Scholar]
- 8.Mayer P, Slater GW, Drouin G. Anal Chem. 1994;66:1777–1780. [Google Scholar]
- 9.Noolandi J. Electrophoresis. 1992;13:394–395. doi: 10.1002/elps.1150130180. [DOI] [PubMed] [Google Scholar]
- 10.Meagher RJ, Won JI, Coyne JA, Lin J, Barron AE. Anal Chem. 2008;80:2842–2848. doi: 10.1021/ac702591t. [DOI] [PubMed] [Google Scholar]
- 11.Ren H, Karger AE, Oaks F, Menchen S, Slater GW, Drouin G. Electrophoresis. 1999;20:2501–2509. doi: 10.1002/(SICI)1522-2683(19990801)20:12<2501::AID-ELPS2501>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 12.Won JI, Meagher RJ, Barron AE. Biomacromolecules. 2004;5:618–627. doi: 10.1021/bm034442p. [DOI] [PubMed] [Google Scholar]
- 13.Long D, Ajdari A. Electrophoresis. 1996;17:1161–1166. doi: 10.1002/elps.1150170628. [DOI] [PubMed] [Google Scholar]
- 14.Long D, Dobrynin AV, Rubinstein M, Ajdari A. J Chem Phys. 1998;108:1234–1244. [Google Scholar]
- 15.McCormick LC, Slater GW, Karger AE, Vreeland WN, Barron AE, Desruisseaux C, Drouin G. J Chromatogr A. 2001;924:43–52. doi: 10.1016/s0021-9673(01)00990-6. [DOI] [PubMed] [Google Scholar]
- 16.Nedelcu S, Meagher RJ, Barron AE, Slater GW. J Chem Phys. 2007;126 doi: 10.1063/1.2730799. [DOI] [PubMed] [Google Scholar]
- 17.Vreeland WN, Desruisseaux C, Karger AE, Drouin G, Slater GW, Barron AE. Anal Chem. 2001;73:1795–1803. doi: 10.1021/ac001380+. [DOI] [PubMed] [Google Scholar]
- 18.Meagher RJ, Coyne JA, Hestekin CN, Chiesl TN, Haynes RD, Won JI, Barron AE. Anal Chem. 2007;79:1848–1854. doi: 10.1021/ac061903z. [DOI] [PubMed] [Google Scholar]
- 19.Vreeland WN, Slater GW, Barron AE. Bioconjugate Chem. 2002;13:663–670. doi: 10.1021/bc0155871. [DOI] [PubMed] [Google Scholar]
- 20.Yoo B, Kirshenbaum K. Curr Opin Chem Biol. 2008;12:714–721. doi: 10.1016/j.cbpa.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Zuckermann RN, Kodadek T. Curr Opin Mol Ther. 2009;11:299–307. [PubMed] [Google Scholar]
- 22.Fowler SA, Blackwell HE. Org Biomol Chem. 2009;7:1508–1524. doi: 10.1039/b817980h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butterfoss GL, Renfrew PD, Kuhlman B, Kirshenbaum K, Bonneau R. J Am Chem Soc. 2009;131:16798–16807. doi: 10.1021/ja905267k. [DOI] [PubMed] [Google Scholar]
- 24.Culf AS, Ouellette RJ. Molecules. 2010;15:5282–5335. doi: 10.3390/molecules15085282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desruisseaux C, Long D, Drouin G, Slater GW. Macromolecules. 2001;34:44–52. [Google Scholar]
- 26.Meagher RJ, Won JI, McCormick LC, Nedelcu S, Bertrand MM, Bertram JL, Drouin G, Barron AE, Slater GW. Electrophoresis. 2005;26:331–350. doi: 10.1002/elps.200410219. [DOI] [PubMed] [Google Scholar]
- 27.Burkoth TS, Fafarman AT, Charych DH, Connolly MD, Zuckermann RN. J Am Chem Soc. 2003;125:8841–8845. doi: 10.1021/ja0352101. [DOI] [PubMed] [Google Scholar]
- 28.Horn T, Lee BC, Dill KA, Zuckermann RN. Bioconjugate Chem. 2004;15:428–435. doi: 10.1021/bc0341831. [DOI] [PubMed] [Google Scholar]
- 29.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, Spellmeyer DC, Tan RY, Frankel AD, Santi DV, Cohen FE, Bartlett PA. Proc Natl Acad Sci USA. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uno T, Beausoleil E, Goldsmith RA, Levine BH, Zuckermann RN. Tetrahedron Lett. 1999;40:1475–1478. [Google Scholar]
- 31.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J Am Chem Soc. 1992;114:10646–10647. [Google Scholar]