Abstract
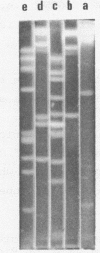
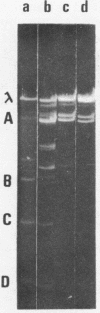
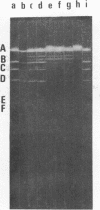
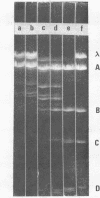
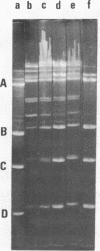
It is shown that three related antibiotics, distamycin A, propyl-distamycin and netropsin, can protect certain endo R.Hind III cleavage sites from attack by endonuclease, giving rise, after endo R.Hind III digestion, to larger DNA fragments. Bacteriophage lambda DNA has six recognition sites for Hind III enzyme. Three of these sites: shind III 2, 3 and 6 can be protected from nuclease action by all the antibiotics used. Propyl-distamycin protects partly shind III 5, too. Netropsin protects partly sites shind III 5 and 4, while distamycin A protects all the sites but shind III 1 so the Hind III digestion produces only two large fragments of lambda DNA.
Full text
PDF





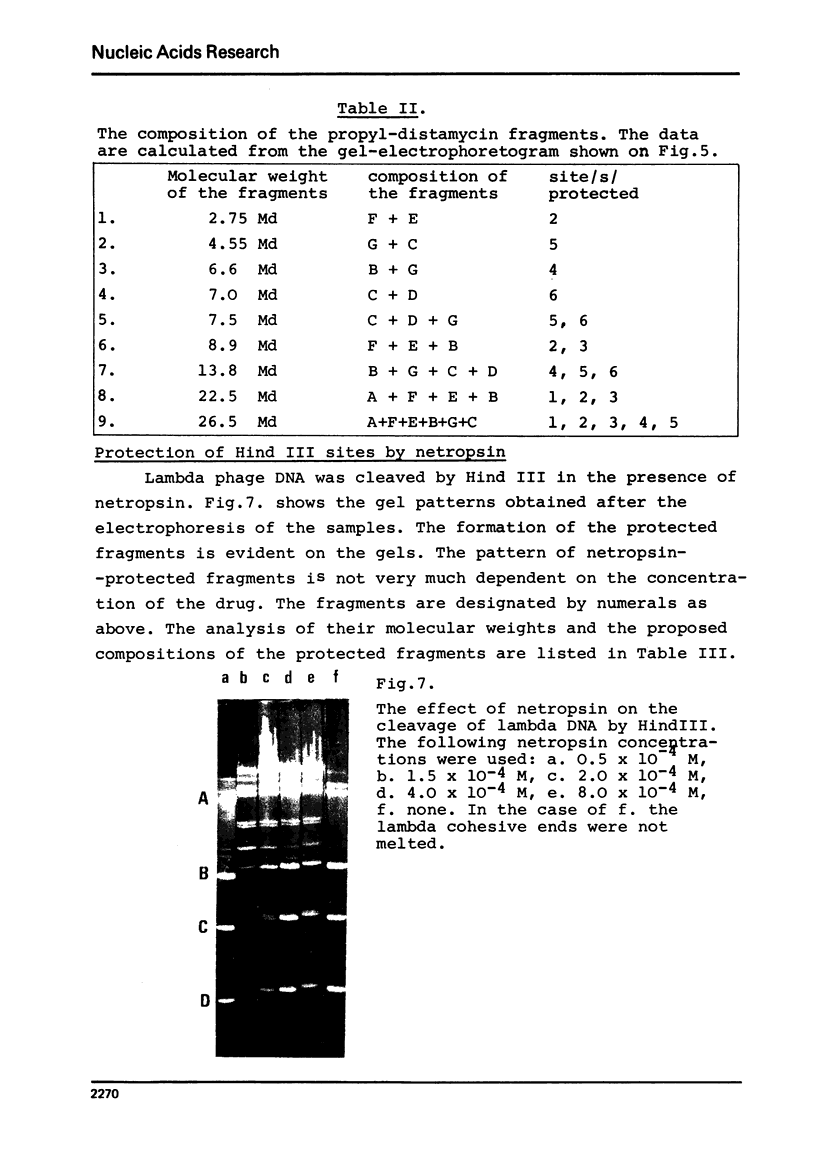



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Braga E. A., Nosikov V. V., Taniashin V. I., Zhuze A. L., Polianovskii O. L. Ogranichenie deistviia restriktsionnykh éndonukleaz antibiotikami, sviazyvaiushchimisia s DNK. Dokl Akad Nauk SSSR. 1975 Nov 21;225(3):707–710. [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries P., Gordon R. L., McConnell D. J., Connolly P. Endonuclease R. Hind fragments of T7 DNA. Virology. 1974 Mar;58(1):25–31. doi: 10.1016/0042-6822(74)90138-x. [DOI] [PubMed] [Google Scholar]
- KAISER A. D., HOGNESS D. S. The transformation of Escherichia coli with deoxyribonucleic acid isolated from bacteriophage lambda-dg. J Mol Biol. 1960 Dec;2:392–415. doi: 10.1016/s0022-2836(60)80050-2. [DOI] [PubMed] [Google Scholar]
- Lee A. S., Sinsheimer R. L. A cleavage map of bacteriophage phiX174 genome. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2882–2886. doi: 10.1073/pnas.71.7.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
- Nathans D., Smith H. O. Restriction endonucleases in the analysis and restructuring of dna molecules. Annu Rev Biochem. 1975;44:273–293. doi: 10.1146/annurev.bi.44.070175.001421. [DOI] [PubMed] [Google Scholar]
- Nosikov V. V., Braga E. A., Karlishev A. V., Zhuze A. L., Polyanovsky O. L. Protection of particular cleavage sites of restriction endonucleases by distamycin A and actinomycin D. Nucleic Acids Res. 1976 Sep;3(9):2293–2301. doi: 10.1093/nar/3.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Reinert K. E., Luck G., Wähnert U., Löber G., Thrum H. Interaction of the oligopeptide antibiotics netropsin and distamycin A with nucleic acids. J Mol Biol. 1971 May 28;58(1):329–348. doi: 10.1016/0022-2836(71)90250-6. [DOI] [PubMed] [Google Scholar]