Abstract
Both genetic and environmental factors contribute to the development and progression of systemic lupus erythematosus (SLE), a complex autoimmune disease. The disease exhibits a strong gender bias and develops predominantly in females. Additionally, most SLE patients exhibit increased serum levels of interferon-α (IFN-α) and the “IFN signature”. Studies using the mouse models of lupus have identified several lupus susceptibility loci, including the New Zealand Black (NZB)-derived autoimmunity 2 (Nba2) interval on the chromosome 1. The interval, which is syntenic to the human chromosome 1q region, harbors the FcγR family, SLAM/CD2-family, and the IFN-inducible Ifi200-family genes (encoding for the p200-family proteins). Studies involving the B6.Nba2 congenic mice revealed that the development of antinuclear autoantibodies (ANAs) depends on the age, gender, and activation of type I IFN-signaling. Interestingly, recent studies involving the generation of Nba2 subcongenic mouse lines and generation of mice deficient for the Fcgr2b or Aim2 gene within the interval have provided evidence that epistatic interactions among the Nba2 genes contribute to increased lupus susceptibility. Given that the expression of some of the p200-family proteins is differentially regulated by sex hormones and these proteins differentially regulate cytosolic DNA-induced production of type I IFN and proinflammatory cytokines (IL-1β and IL-18), the major known contributors of SLE-associated inflammation, we discuss the recent advancements in our understanding of the role of p200-family proteins in lupus susceptibility modification. An improved understanding of the role of p200-family proteins in the development of autoimmunity is likely to identify new approaches to treat SLE patients.
Keywords: Lupus susceptibility, Nba2 locus, Interferons, Sex hormones, Ifi200-family genes
1. Introduction
Systemic lupus erythematosus (SLE) is a complex autoimmune disease [1–6]. The disease is highly heterogeneous and has a potential to involve multiple organ systems. SLE in patients (and mouse models) is characterized by increased production of pathogenic anti-nuclear autoantibodies (ANAs), the presence of immune complexes containing nucleic acids that are bound to DNA-binding proteins, increased serum levels of interferon-α (IFN-α) and increased expression of a set of IFN-inducible genes (the “IFN-signature”), and increased serum levels of B cell activating factor (BAFF) [1–3].
The development of SLE involves immune dysregulation at the interface between the innate and adaptive immune systems [7, 8]. Studies have suggested that a defective clearance of cellular debris causes a loss of self-tolerance, autoantibody production, and the formation of immune complexes (ICs) [8]. ICs containing nucleic acids such as self DNA activate innate immune responses [8, 9]. Consequently, several clinical manifestations of SLE are thought to be the result of ICs deposition in tissues leading to secondary inflammatory responses and tissue damage [1, 2, 8, 9]. Therefore, ICs are thought to be the predominant cause of SLE-associated inflammation [1–3, 9].
SLE patients and certain mouse models exhibit a strong gender bias in the development of disease: develops at a female-to-male ratio of 9:1 [10–12]. Although, the etiology of human SLE remains unknown, there is considerable evidence that genetic, hormonal, and environmental factors contribute to the development and progression of lupus [1–3]. Correspondingly, multiple lupus susceptibility genes and pathways appear to be involved in lupus susceptibility [13, 14].
Genetic studies have identified polymorphisms within genes of the IFN pathway (and the IFN-regulated genes) that confer an increased risk for the development of SLE [15, 16]. Notably, the production of type IFNs in vivo is regulated by mechanisms that have the capacity to self-amplify upon recognition of IFN-inducing ligands (for example, ICs containing self DNA), thus, resulting in a feed-forward amplification loop of type I IFN production [17, 18]. Increased production of type I IFNs in SLE patients and certain mouse models is associated with organ and tissue damage [17–19].
Our overall understanding of the molecular mechanisms that contribute to the development of lupus disease and associated immunopathology has derived in part from studies involving mouse models [6, 20–22]. These models include the F1 hybrid of the New Zealand Black (NZB) and New Zealand White (NZW) strains, the MRL/lpr, and BXSB/Yaa strains, which spontaneously develop the disease. Additionally, there are models that are derived from the hybrid mice (for example, NZM2328 and NZM2410). Generation of B6.Nb2 congenic (congenic for the NZB-derived Nba2 interval on the C57BL/6 genetic background) mice indicated that female mice spontaneously develop splenomegaly (due to increases in the number of B and T cells) and increased levels of a variety of ANAs (anti-dsDNA, anti-chromatin, and anti-histone) much earlier than the age-matched male mice [23, 24]. The development of ANAs was age (~6–7-month) dependent. Additionally, the B6.Nba2 mice express increased levels of Ifi202 (a member of the IFN-inducible Ifi200-family of genes) mRNA and protein in splenic B cells and non-T/non-B cells [23]. Although, B6.Nba2 mice do not develop a kidney disease, however, (B6.Nba2 x NZW)F1 mice develop the disease like the (NZB x NZW)F1 mice, indicating a cooperation between the Nba2 interval genes and the NZW loci in the development of a kidney disease [23]. Interestingly, a deficiency of type I IFN-signaling in the B6.Nba2 mice reduces lupus-like disease [25]. Accordingly, treatment of mice with polyI:C, which induces the production of IFN-α/β, accelerates the development of ANA phenotype and lupus-like nephritis in the (B6.Nba2 x NZW)F1 mice [26], indicating a role for the IFN-signaling in modifying the development and progression of lupus disease. Moreover, TLR9-deficient B6.Nba2 mice, which express increased levels of TLR7, display increased production of ANAs and also develop lupus nephritis [27], suggesting opposing regulatory roles for the TLR7 and TLR9 in modifying the ANA phenotype and progression of lupus disease in these mice. Furthermore, overexpression of B cell activating factor (BAFF) in B6.Nba2.BAFF transgenic mice as compared to age-matched non-transgenic or B6.BAFF mice accelerated the development of a kidney disease [28]. Given that B6.Nba2 congenic mice exhibit a sex bias in the development of ANA phenotype [23, 29] and the development of the ANA phenotype requires the type I IFN-signaling [25, 26], this mouse model has emerged as a valuable model to investigate the potential role of sex hormones and IFN-signaling in the development of autoimmunity [12].
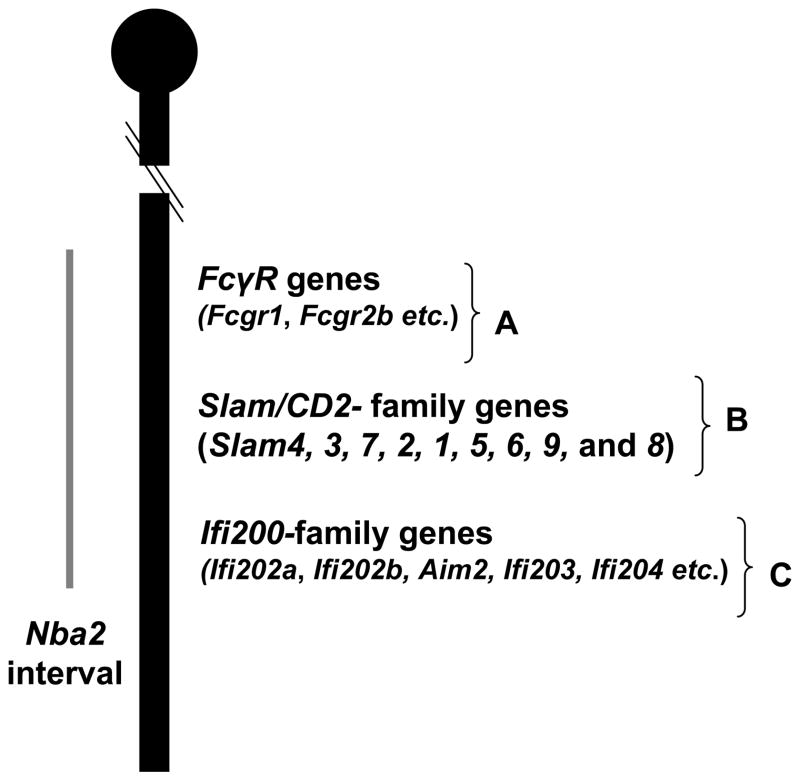
The Nba2 interval (~90–97 cM; see Fig. 1), which is located on the distal portion of the NZB chromosome 1, has emerged as a major genetic contribution to the disease susceptibility in the (NZB x NZW)F1 mice [20, 23, 30, 31]. The interval is syntenic to human lupus susceptibility region 1q and contains genes that encode for the Fcγ receptors (including the FcγRIIB receptor), Slam/CD2 family proteins, and p200-family proteins (encoded by the Ifi200-family genes) [30, 32, 33]. Interestingly, the generation of subcongenic mouse lines of the Nba2 interval [34, 35] and generation of mice deficient for the Fcgr2b (encoding for the inhibitory FcγRIIB receptor) [36, 37] or Aim2 (encoding for the Aim2 protein, a member of the p200-family) [38, 39] gene have provided important evidence that epistatic interactions among the Nba2 interval genes contribute to an increased lupus susceptibility. In this review, we discuss the recent advancements in our understanding of the role of Ifi200-genes as modifiers of lupus susceptibility.
Fig. 1.
Schematic map of the distal end of murine chromosome 1 encompassing the Nba2 interval (indicated by a gray line). The map includes the relative locations of the cluster of genes within the interval that encode for the Fcγreceptors, Slam/CD2 -familyreceptors, and p200-family proteins. Some of the genes that are discussed in this review are indicated.
2. The interferon-inducible Ifi200-family genes as immune regulators
The Ifi200-family genes encode for structurally and functionally-related p200-family proteins [12, 32, 40–43]. The family includes murine (for example, Ifi202a, Ifi202b, Ifi203, Ifi204, and Aim2) and human (for example, MNDA, IFIX, IFI16, and AIM2) genes [32]. The Ifi202a and Ifi202b are highly homolgous genes and encode for p202a and p202b proteins [44]. Therefore, in this review, we refer both p202a and p202b proteins as p202 protein.
The p200-family proteins share a partially conserved repeat of 200-amino acids residues (also called HIN-200 domain). The HIN-200 domain allows binding to double-stranded DNA (dsDNA) in a sequence independent manner [45, 46]. Interestingly, the crystal structures of both HIN-A and HIN-B domains of IFI16 protein (encoded by the IFI16 gene) revealed that each HIN domain is capable of enhancing p53-DNA complex formation and transcriptional activation [47]. However, both domains distinctly regulate the p53-mediated transcription: the HIN-A domain binds to the basic C terminus of p53 protein, whereas the HIN-B domain binds to the core DNA-binding region of p53. Notably, both protein-protein interactions are compatible with the DNA-bound state of p53. Except the p202a and p202b proteins, all p200-family proteins also share a homotypic protein-protein interaction pyrin domain (PYD) [39]. Most p200-family proteins (except the p202a and p202b proteins) contain both PYD and HIN-200 domain, therefore, these proteins are also called PYHIN-family proteins [48].
Most p200-family proteins (except the murine Aim2 and human AIM2 proteins), depending upon the cell type, are detected both in the cytoplasmic as well as in the nuclear fractions to some extents [39, 49]. Interestingly, activation of IFN-signaling in immune cells, which induces the expression of p200-family proteins, also potentiates the nuclear localization of certain proteins, including the p202 proteins [39, 50, 51].
Recent studies have indicated that certain p200-family proteins can sense cytosolic DNA in innate immune cells (macrophage and dendritic cells) [52–56]. Upon sensing cytosolic DNA in bone marrow-derived macrophages, the p202 protein (possibly both p202a and p202b) does not initiate an innate immune response [46]. However, PYHIN proteins such as p204 [52], Aim2 and AIM2 [53–55], and IFI16 [52] can initiate innate immune responses after sensing cytosolic DNA in monocytes or macrophages. Interestingly, Aim2 and AIM2 proteins form inflammasomes [46, 53–55], which through activation of caspase-1, increase the secretion of proinflammatory cytokines, such as IL-1β and IL-18. Additionally, the activation of an inflammasome also induces cell death by pyroptosis (caspase-1-dependent death) in macrophages [46]. In contrast to the Aim2/AIM2 proteins, upon sensing dsDNA, the p204 and IFI16 proteins were reported to recruit stimulator of interferon genes (STING) protein to stimulate the expression of IFN-β through the activation of interferon-regulated factor 3 (IRF3) and NF-κB [52]. Consistent with the above role for the p200-family proteins in innate immune responses, a recent study has noted that IFI16 protein may play an important role in sensing intracellular dsDNA in human monocyte-derived DCs as well as primary DCs [56]. Importantly, the study reported that human dsDNA-activated DCs, rather than LPS- or inflammatory cytokine mixture-activated DCs, represent the most potent inducers of naive CD4+ T cells to promote Th1-type cytokine production and generate CD4+ and CD8+ cytotoxic T cells. These studies clearly indicate an important role for the p200-family proteins in the initiation of innate immune responses upon sensing cytosolic DNA.
Overexpression of Ifi202 gene enhances the LPS-induced IL-12p40 and NF-κB promoter activities in a murine macrophage cell line (RAW 264.7) [57]. In addition, forced expression of Ifi202 gene enhances IL-12p40 mRNA induction in NZW bone marrow-derived dendritic cells (BMDCs) [57]. This observation raises the possibility that increased levels of the p202 protein in macrophages regulate the expression of IL-12p40, which is required for the production of IL-12 and IL-23. Increased expression of both IL-12 and IL-23 is important for TH1 and TH17 differentiation [58].
In addition, there is emerging evidence that p200-family proteins also participate in the regulation of adaptive immune responses through regulating the production of cytokines, including the type I IFN, which regulate cell proliferation, survival, and differentiation of immune cells, including B and T cells [23, 59–63]. Treatment of splenic B and T cells with IFN-α or their stimulation up-regulates the expression of p202 protein [62]. Increased levels of p202 protein in splenic B cells from lupus-prone B6.Nba2 female mice (see below) are associated with defects in the expression of target genes for E2F1 [64] and p53 [65] transcription factors that encode for pro-apoptotic proteins. Moreover, increased levels of the p202 protein in B cells are associated with defects in apoptosis after an in vitro ligation with anti-IgM [23].
p202 protein also inhibits the transcriptional activities of AP-1 [66] and c-Myc [67] in a variety of cell types. Moreover, p202 protein modulates the transcriptional activity of NF-κB, a potent regulator of immunoregulatory genes, in a cell-type dependent manner [57, 66, 68]. These observations raise the possibility that increased levels of the p202 protein in immune cells, including B and T cells, promote survival of autoreactive cells through transcriptional modulation of genes that promote cell survival.
2.1 Identification of Ifi202 as a candidate lupus susceptibility gene within the Nba2 interval
Generation of B6.Nba2 congenic mice allowed screening for candidate lupus susceptibility genes within the Nba2 interval through gene expression analyses [23]. The analyses, which involved pair-wise comparisons of gene expression between C57BL/6 (B6) and B6.Nba2 female mice (age ~4-mo), identified two genes (Ifi202 and Ifi203), expression of which was altered significantly. Splenic cells from the congenic mice had 10–100-fold higher steady-state levels of Ifi202 mRNA and lower levels of Ifi203 mRNA [23]. However, the expression pattern was opposite in the B6 cells. Considering that the lupus phenotype in SLE patients and mouse models is associated with multiple genes [4, 5], identification of only two genes with differential expression between B6 and B6.Nba2 mice in the above gene expression analyses was consistent with the inclusion of limited (only ~11,000) number of genes in microarray gene expression analysis. Of note, several observations provided support for the potential role of the Ifi202 gene in the Nba2 interval-associated phenotype (ANA production). Firstly, we identified promoter polymorphisms in the Ifi202 gene between B6 and NZB strain of mice, which are associated with increased basal and induced expression of Ifi202 gene in the B6.Nba2 congenic mice [23, 24, 32]. Secondly, several studies involving other mouse models of SLE provided evidence for a role for the Ifi202 gene in the development of autoimmunity [32]. Thirdly, nuclear localization of p202 protein modulates the transcriptional activity of immunoregulatory factors, including the NF-κB and AP-1, in a cell type-dependent manner [66, 68]. Thus, raising the possibility that mouse strain-dependent increased nuclear levels of the p202 protein contribute to autoimmunity through modulating the transcription of immunomodulatory genes [24, 32]. Fourthly, expression of Ifi202 gene is differentially regulated by sex hormones: the female sex hormone estrogen (17β-estradiole) up-regulates the expression, whereas the male hormone androgen (dihydrotestosterone) suppresses the expression [69]. Interestingly, our observations that a deficiency of type I IFN-signaling in NZB female mice [70], which reduced lupus disease, did not decrease levels of the p202 protein raised the possibility that other cytokines could regulate the expression of Ifi202 gene in B6.Nba2 mice. Accordingly, we noted that IL-6 signaling through STAT3 activation can up-regulate the expression of Ifi202 gene in the B6.Nba2 mice [71]. Given that p202 protein’s nuclear localization (thus, its ability to modulate the expression of immunoregulatory genes) is potentiated by an activation of type I IFN-signaling [50, 51] and a deficiency in type I IFN-signaling in the B6.Nba2 mice reduced lupus disease [25], these observations make it likely that increased nuclear levels of the p202 protein in immune cells enhance the ANA phenotype in B6.Nba2 female mice in part through transcriptional modulation of the Nba2 interval genes.
2.2. p202 proteins and their regulation
Both p202a and p202b proteins are highly similar proteins [44]. Interestingly, Ifi202a-deficiency in mice and in embryonic fibroblasts increases levels of the p202b protein by a posttranscriptional mechanism [44]. Given that p202a and p202b proteins differ by only seven amino acid residues in the N-terminus, a compensatory increase in the levels of p202b protein in Ifi202a-deficient mice (and cells) could account for a lack of phenotype [44].
Steady-state levels of p202 protein are regulated by both transcriptional and post-transcriptional mechanisms [72]. These mechanisms include stabilization of Ifi202 mRNA in IFN-treated cells. Consistent with the above observations, the 3′-untraslated region in the Ifi202 mRNA contains three APyTGA-like regulatory elements, which are known to stabilize mRNAs after estrogen treatment of cells [73]. Importantly, a 50% increase (or decrease) in the levels of p202 protein in a variety of cultured cell lines and splenic B cells inhibits cell cycle progression and also modulates cell survival [32, 72].
The amino acid sequence of p202 protein contains a potential mitochondrial targeting sequence (MTS) in the N-terminus [50]. Consistent with the presence of an MTS in the p202 protein, a fraction of the p202 in the cytoplasm is detected in the mitochondria [50, 51]. Because p202 protein lacks a classical nuclear localization signal (NLS) and a nuclear export signal (NES) [72] and activation of IFN-signaling potentiates the nuclear localization in B6.Nba2 MEFs [50] and B cells [35], these observations support the idea that nucleo-cytoplasmic distribution of p202 protein in immune cells depends on the p202-binding proteins.
p202 is a phosphoprotein [45, 51] and dephosphorylation of p202 protein increases its ability to bind DNA in vitro [45]. Although, protein kinases that phosphorylate p202 protein remain unknown, the protein is predicted to be phosphorylated by several protein kinases. These kinases include the cyclin-dependent kinase-5 (Cdk-5) on Thr-46, Cdk-1 on Thr-46, PKC-epsilon on Ser-85 and Ser-436, PKC-delta on Ser-436, PKC-zeta on Ser-185 and Ser-275, DNA-PK on Ser-346, casein kinase 1 on Thr-168, and casein kinase 2 on Ser-344 and Ser-345. Because the unphosphorylated p202 protein binds to DNA tightly[45] and p202 protein also associates with the chromatin in vivo [51], these observations support the idea that phosphorylation of p202 protein regulates its nuclear levels.
Like other members of the p200-family proteins, p202 appears to homo- and heterodimerize [74]. Moreover, homodimerization of p202 protein depends on a motif MFHATVAT, which is conserved among the p200-family proteins. Notably, a substitution of His residue in the motif with a Phe residue abrogated the p202-mediated inhibition of the transcriptional activity of AP-1 [75]. Importantly, there are indications that p202 protein heterodimerizes with p204 protein [72, 76]. These observations are consistent with the idea that heterodimerization of p202 with other p200-family proteins in immune cells may regulate its nucleocytoplasmic distribution and functions.
3. The NZB-derived Ifi200-family genes are insufficient in the development of autoimmunity
To assess the relative genetic contributions of the Nba2 interval genes in ANA phenotype, subcongenic mice (B6.Nba2-A, B6.Nba2-B, B6.Nba2-A′B, B6.Nba2-BC, and B6.Nba2-C) have been generated [34]. Comparisons of these subcongenic strains of female mice with the parental strains (B6 and NZB) and congenic B6.Nba2 (mice indicated as the B6.Nba2-ABC) female mice with respect to the ANA phenotype and type I IFN production revealed that the B6.Nba2-A female mice (mice harboring the subinterval A that comprises the FcγRs genes; see Fig. 1) and B6.Nba2-B female mice (mice harboring the subinterval B that comprises the Slam-family genes) develop detectable levels of certain ANAs. However, levels of ANA were much higher in the B6.Nba2-A′B mice (mice harboring both FcγRs and Slam-familygenes), thus, indicating that the FcγR interval and Slam interval genes cooperate with each other to influence the autoantibody production [34]. Additionally, increased ANA levels in the B6.Nba2-A′B mice were associated with increased production of type I IFN. Moreover, the study revealed that B6.Nba2-C female mice (harboring subinterval C that comprises the Ifi200-family genes) do not develop ANAs and produce type I IFN. Together, these observations demonstrated that the Ifi200-family genes are not sufficient to break the tolerance in the B6.Nba2-C subcongenic mice [34]. Given that expression of most of Ifi200-family genes is up-regulated by type I IFNs [32], the lack of ANA phenotype in the B6.Nba2-C subcongenic mice is consistent with a reduced expression of these genes in the absence of type I IFN production (Table 1). Moreover, because type I IFN-induced signaling potentiates the nuclear localization (and nuclear functions) of the p202 protein [50, 51], the lack of type I IFN production in B6.Nba2-C subcongenic mice reduced the nuclear localization of the p202 protein in splenic B cells (Table 1). Interestingly, the study revealed that epistatic interactions between the Ifi200-family genes and Fcgr2b gene may down-regulate the expression of the Fcgr2b gene, resulting in an inhibition of the FcγRIIB-induced apoptosis [34].
Table-1.
Increased p202 protein levels and its nuclear localization are associated with the production of ANAs and IFN-α, and reduced levels of the FcγRIIB and Slamf1 receptors.
| Mouse strain | Nba2 genes | IFN-α levelsa | Protein levels FcγRIIB | Protein levels Slamf1 | Protein levels p202 | ANAsb |
|---|---|---|---|---|---|---|
| B6 | None | Not detected | +++ | +++ | + (C) | Not detected |
| B6.Nba2-ABC | FcγRs, Slam, and Ifi200 | Detectable | + | + | +++ (N) | Yes (+++) |
| B6.Nba2-A′B | FcγRs, Slam | Detectable | ? | ? | + (?) | Yes (++) |
| B6.Nba2-C | Ifi200 | Not detected | +++ | +++ | − | Not detected |
Serum levels of IFN-α at the age of 7–8 months;
ANAs against chromatin, histones, and dsDNA;
C, cytoplasmic; N, Nuclear.
4. The Fcγr2b gene is a lupus susceptibility modifier gene within the Nba2 interval
As noted above, the Nba2 subinterval-A harbors genes that encode for Fc receptors for IgG [77, 78]. These receptors regulate innate and adaptive immune responses. Four Fcγ receptors have been reported in mice [77, 78]. Upon stimulation by immune complexes, the FcγRI and FcγRIII receptors initiate a stimulatory response through the FcRγ chain. The chain contains an intracellular immunoreceptor tyrosine-based activating motif (ITAM). In contrast to the stimulatory receptors, the FcγRIIB receptor (encoded by the Fcgr2b gene) transduces inhibitory signals via intracellular immunoreceptor tyrosine-based inhibitory motif (ITIMs). Because FcγRIIB receptor is the only Fcγ receptor, which is expressed by B cells, its expression is regulated tightly [78]. In humans and mice, isoforms of the inhibitory FcγRIIB receptor have been reported [79, 80]. The FcγRIIB1 isoform is predominantly expressed by B cells, whereas FcγRIIB2 isoform is predominantly expressed by myeloid-derived cells [78, 80]. Expression of FcγRIIB receptor in immune cells inhibits the functions of the activating Fcγ receptors [78]. These functions include phagocytosis and pro-inflammatory cytokine release. Consequently, the FcγRIIB receptor regulates several immune responses [78].
4.1 Epistatic interactions involving the Fcgr2b gene in the ANA phenotype
A deficiency of FcγRIIB receptor in mice of certain genetic background results in the development of autoimmunity [36, 81–83]. For example, Fcgr2b-deficient mice on the mixed (mice indicated as FcγRIIB129−/−; embryonic stem cells derived from the 129 strain of mice) genetic background develop autoimmunity [36, 81]. However, Fcgr2b-deficent mice on the B6 genetic background (mice indicated as FcγRIIBB6−/−; mice generated from the B6 embryonic stem cells) do not develop autoimmunity [36]. The development of autoimmunity in the FcγRIIB129−/− mice has been attributed to epistatic interactions between 129-derived Sle16 locus (the locus contains the Nba2 interval genes) and B6 genes [36]. Interestingly, splenic cells from the lupus-prone FcγRIIB129−/− female mice exhibit activation of an IFN-response and express increased levels of the p202 protein [37, 84]. Similarly, a blockade of the inhibitory FcγRIIB receptor in human innate immune cells (dendritic cells and monocytes) induces a type I IFN response, activates phosphorylation of STAT1, and induces expression of certain IFN-inducible genes, including the IFI16 and AIM2 p200-family proteins [85]. These observations provided evidence for epistatic interactions involving the Fcgr2b gene with other genes, including the Nba2 interval genes, in the modification of Nba2 interval-associated ANA phenotype through activation of an IFN response.
4.2 Epistatic interactions between the Ifi202 and Fcgr2b genes
Certain polymorphisms involving short deletions in the promoter and intronic regions of the murine Fcgr2b gene are associated with reduced steady-state levels of mRNA in germinal center B cells and plasma cells in certain lupus-prone strains of female mice (including the NZB mice) [78–80]. However, the observation that epistatic interactions between Fcγ receptor genes and p200-family genes may contribute to reduced expression of the Fcgr2b gene in the B6.Nba2 mice [34, 35] (independent of known polymorphisms) served as a basis to investigate whether IFNs or IFN-inducible p202 protein could down-regulate the expression of the Fcgr2b gene. The investigation revealed that activation of IFN-signaling in splenic cells by IFN-α or IFN-γ significantly decreases levels of the FcgRIIB mRNA and protein [84]. Furthermore, increased expression of p202 protein in cells, which increased the expression IFN-β and activated an IFN response, suppressed the expression of the Fcgr2b gene [84]. These observations provided further evidence for epistatic interactions between the Ifi202 and the Fcgr2b genes in increased production of ANAs in the B6.Nba2 mice.
5. Epistatic interactions between the Slam-family and Ifi200-family genes
Genetic studies have implicated the signaling lymphocytic activation molecule (SLAM) family of cell surface receptors in the modulation of immune cell functions [86–88]. The family consists of nine transmembrane proteins (for example, the SLAMF1 to SLAMF9) that are differentially expressed on lymphoid and myeloid cells [87, 88]. Interestingly, studies have indicated that increased expression of SLAMF3 and SLAMF6 in human SLE T cells promotes Th17 differentiation [89]. Similarly, increased expression of Ly108.1 (or Slamf6) in immature B cells from lupus-prone B6.Sle1z mice is associated with the development of lupus phenotype [90]. Additionally, the SLAMF6-driven co-stimulation of peripheral T cells was found to be defective in human SLE T cells [91].
SLAMF1 (or CD150) is a self-ligand cell surface glycoprotein (~75 kDa), which is expressed on T, B, macrophages, and dendritic cells [86, 87]. Slamf1-deficient T cells (CD4+) produce increased levels of interferon-γ [92]. Moreover, Slam-deficient mice on the C57BL/6 genetic background develop a lupus-like disease [93]. Cross-linking of the Slamf1 receptor on CD4+ T cells induces rapid serine phosphorylation of the Akt/PKB protein kinase [94]. Given that the AKT kinase inactivates MDM2 protein (a negative regulator of the p53) through phosphorylation [95] and the activation of the p53 represses the transcription of a number genes, including the Ifi202 gene [96], it is likely that the Slamf1 receptor regulates T-cell proliferation and survival through suppression of the Ifi202 gene. Accordingly, in a NOD mouse line, which is congenic for the B6-derived Nkt1 locus (which includes the Slam and the Ifi200-family genes), the increased expression of Slamf1 gene was inversely correlated with the Ifi202 gene [97]. Moreover, increased expression of Slamf1 gene was associated with an increased expression of the Ifi203 gene. These observations predict that epistatic interactions between the Ifi200-family genes and Slam-family genes could contribute to ANA phenotype in the B6.Nba2 mice. Therefore, further studies are needed to investigate the potential role of the interactions between Ifi200-family genes and Slam-family genes (Slamf1-9) in the Nba2 interval-associated phenotype.
6. Interactions among the Ifi200-family genes
Steady-state levels of Ifi202 mRNA in B6.Nba2 splenic cells are inversely correlated with levels of Ifi203 mRNA [23]. Furthermore, overexpression of p202 protein in RAW264.7 macrophage cells decreases levels of the p203 protein [98]. These observations suggested a mutual regulation of gene expression by the p200-family proteins.
6.1 Interactions between Ifi202 and Aim2 genes in the Nba2 interval-associated phenotype
Generation of Aim2-deficeint mice on the C57BL/6 genetic background (ES cells derived from the B6 mice; mice indicated as Aim2B6−/−) [99] and the mixed genetic background (ES cells from the 129 strain of mice; mice indicated as Aim2129−/−) [38] and their characterization indicated that Aim2 protein expression is not required for the production of type I IFN upon infections with certain bacteria or viruses. Interestingly, the deficiency of the Aim2 gene in mice (both Aim2B6−/− and Aim2129−/−) increased constitutive as well as induced (induced by infection by pathogens or transfection of DNA) levels of IFN-β [38, 98, 99]. These observations raised the possibility that Aim2 expression in immune cells suppresses the expression of IFN-β. Accordingly, we noted that Aim2-deficiency in the Aim2129−/− mice was associated with activation of type I IFN response: increased expression of IFN-β, activation of IFN-signaling, and an induction of the IFN-inducible proteins, including the p202 protein [98]. Moreover, levels of the inhibitory receptor FcγRIIB were decreased [84]. These observations, which indicated that expression of Aim2 protein in immune cells suppresses the type I IFN response and the expression of p202 protein, served as a basis to investigate whether increased levels of p202 protein in innate immune cells activate an IFN response. Surprisingly, increased expression of p202 protein in macrophage cell line RAW264.7 activated an IFN response and suppressed the expression of both Aim2 and FcγRIIB receptor [84]. Accordingly, increased levels of p202 protein in splenic cells from lupus-prone B6.Nba2 female mice as compared to age-matched B6 females were associated with activation of an IFN response and reduced levels of Aim and FcγRIIB receptor [84]. Given that expression of Aim2 protein is up-regulated by male sex hormone androgen in immune cells [62], the above observations suggest that an inverse expression correlation between the Aim2 and p202 proteins in the B6.Nba2 female mice contributes to increased production of autoantibodies, up-regulation of type I IFN production, and activation of an IFN response.
7. Distinct regulation of the Nba2 genes by IRF5, a positive regulator of IFN expression
Genome-wide association studies (GWAS) have identified human IRF5 gene (encoding for the IFN-regulatory IRF5 transcription factor) and its down stream target gene, PRDM1 (encoding for Blimp-1 transcriptional repressor) in lupus susceptibility [15]. Similarly, studies involving mouse models of SLE have provided evidence for murine Irf5 and Prdm1 genes in lupus susceptibility [100]. The murine IRF5, which is a member of the IRF family [101], is primarily expressed in the B220+ mature B cells and levels of IRF5 protein decrease in CD138+ plasma cells [102]. The activated IRF5 induces the transcription of a number of genes in cell type-dependent manner. These genes include the type I IFN and the Prdm1 gene [102]. Blimp-1 is a master regulator of the B cell differentiation [103]. The Irf5−/− female mice show reduced serum levels of type I IFNs and develop an age-related splenomegaly that is associated with an accumulation of CD19+ B220− B cells [102]. Moreover, splenic cells from Irf5−/− female mice exhibit a decrease in the number of plasma cells and down-regulation of Blimp-1 expression. IRF5 contributes to murine SLE-like disease through its direct regulation of class-switch recombination of the γ2a locus in B cells [104]. Because Nba2 interval includes the IFN-regulated genes such as Ifi202, Aim2 and Fcgr2b genes, we investigated whether the IRF5/Blimp-1 axis could regulate the expression of these genes. Our study revealed that IRF5 up-regulates the expression of p202 protein [63]. However, IRF5 expression down-regulates the expression of Aim2 and FcγRIIB receptor in immune cells. Given that expression of IRF5 is up-regulated by the female sex hormone estrogen in immune cells [105], the above observations provide further evidence for sex-dependent epistatic interactions among the Nba2 interval genes in increased production of autoantibodies in the female B6. Nba2 mice.
8. Interactions between IFI16 and AIM2 genes in autoimmunity
Studies have indicated a role for IFI16 protein in the development of autoimmune diseases [106–108]. Accordingly, increased levels of IFI16 mRNA have been reported in peripheral blood mononuclear cells isolated from SLE patients as compared to healthy donors [108]. Given that the expression of IFI16 and AIM2 proteins is inversely correlated in a variety of cells [109] and decreased levels of the AIM2 mRNA have been reported in SLE patients [110, 111], it is likely that increased levels of the IFI16 protein in innate immune cells contribute to increased production of type I IFN in SLE patients. Therefore, further studies are needed to understand the potential role of IFI16 and AIM2 proteins in innate and adaptive immune responses that are associated with the human SLE.
9. Conclusions
Recent studies have revealed sex hormone-dependent epistatic interactions among the Nba2 interval genes in the modification of autoantibody production and type I IFN production. These interactions involve the IFN-inducible Ifi200-family genes. Therefore, understanding of the molecular mechanisms through which IFN-inducible p200-family proteins regulate innate and adaptive innate immune responses in sex-dependent manner will serve as a basis to understand the molecular mechanisms that contribute to immune dysregulation in the development and progression of human SLE. Consequently, an improved understanding of the regulation and role of the p200-family proteins in the modification of SLE is needed to identify new approaches to effectively treat SLE patients.
Highlights.
Recent studies have revealed sex-dependent epistatic interactions among the Nba2 interval genes in modification of lupus susceptibility
The interferon-inducible Ifi200-family genes within Nba2 interval regulate innate and adaptive immune responses
Epistatic interactions between Ifi200-family genes and Fcgr2b gene contribute to increased IFN-α production
p202 protein increase autoantibody production in part by activating an interferon response that regulate survival of autoreactive B and T cells
p202 protein is a cell type-dependent transcriptional modulator for NF-κB and AP-1 transcription factors
Acknowledgments
Work in author’s laboratory has been supported by grants (R01 AI066261 and R56 AI089775) from the National Institutes of Health. The author apologizes to those investigators, whose work could not be cited directly due to the space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–21. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Liu Z, Davidson A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat Med. 2012;18:871–82. doi: 10.1038/nm.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358:929–39. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 4.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–92. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotzin BL. Systemic lupus erythematosus. Cell. 1996;85:303–6. doi: 10.1016/s0092-8674(00)81108-3. [DOI] [PubMed] [Google Scholar]
- 6.Morel L. Genetics of SLE: evidence from mouse models. Nat Rev Rheumatol. 2010;6:348–57. doi: 10.1038/nrrheum.2010.63. [DOI] [PubMed] [Google Scholar]
- 7.Rönnblom L, Pascual V. The innate immune system in SLE: type I interferons and dendritic cells. Lupus. 2008;17:394–9. doi: 10.1177/0961203308090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muñoz LE, Lauber K, Schiller M, Manfredi AA, Herrmann M. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nat Rev Rheumatol. 2010;6:280–9. doi: 10.1038/nrrheum.2010.46. [DOI] [PubMed] [Google Scholar]
- 9.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–35. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2:777–80. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 11.Pennell LM, Galligan CL, Fish EN. Sex affects immunity. J Autoimmun. 2012;38:J282–91. doi: 10.1016/j.jaut.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Choubey D, Panchanathan R, Duan X, Liu H, Liu H. Emerging roles for the interferon-inducible p200-family proteins in sex bias in systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31:893–906. doi: 10.1089/jir.2011.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu T, Qin X, Kurepa Z, Kumar KR, Liu K, Kanta H, Zhou XJ, Satterthwaite AB, Davis LS, Mohan C. Shared signaling networks active in B cells isolated from genetically distinct mouse models of lupus. J Clin Invest. 2007;117:2186–96. doi: 10.1172/JCI30398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh RP, Waldron RT, Hahn BH. Genes, tolerance and systemic autoimmunity. Autoimmun Rev. 2012;11:664–9. doi: 10.1016/j.autrev.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–9. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salloum R, Niewold TB. Interferon regulatory factors in human lupus pathogenesis. Transl Res. 2011;157:326–31. doi: 10.1016/j.trsl.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crow MK. Interferon-alpha: a therapeutic target in systemic lupus erythematosus. Rheum Dis Clin North Am. 2010;36:173–86. doi: 10.1016/j.rdc.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–36. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 19.Niewold TB. Interferon alpha as a primary pathogenic factor in human lupus. J Interferon Cytokine Res. 2011;31:887–92. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vyse TJ, Kotzin BL. Genetic susceptibility to systemic lupus erythematosus. Annu Rev Immunol. 1998;16:261–92. doi: 10.1146/annurev.immunol.16.1.261. [DOI] [PubMed] [Google Scholar]
- 21.Kono DH, Theofilopoulos AN. Genetics of systemic autoimmunity in mouse models of lupus. Int Rev Immunol. 2000;19:367–87. doi: 10.3109/08830180009055504. [DOI] [PubMed] [Google Scholar]
- 22.Pathak S, Mohan C. Cellular and molecular pathogenesis of systemic lupus erythematosus: lessons from animal models. Arthritis Res Ther. 2011;13:241. doi: 10.1186/ar3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15:435–43. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- 24.Choubey D, Kotzin BL. Interferon-inducible p202 in the susceptibility to systemic lupus. Front Biosci. 2002;7:e252–62. doi: 10.2741/A921. [DOI] [PubMed] [Google Scholar]
- 25.Jørgensen TN, Roper E, Thurman JM, Marrack P, Kotzin BL. Type I interferon signaling is involved in the spontaneous development of lupus-like disease in B6.Nba2 and (B6. Nba2 x NZW)F1 mice. Genes Immun. 2007;8:653–62. doi: 10.1038/sj.gene.6364430. [DOI] [PubMed] [Google Scholar]
- 26.Jørgensen TN, Thurman J, Izui S, Falta MT, Metzger TE, Flannery SA, Kappler J, Marrack P, Kotzin BL. Genetic susceptibility to polyI:C-induced IFN-α/β-dependent accelerated disease in lupus-prone mice. Genes Immun. 2006;7:555–67. doi: 10.1038/sj.gene.6364329. [DOI] [PubMed] [Google Scholar]
- 27.Santiago-Raber ML, Kikuchi S, Borel P, Uematsu S, Akira S, Kotzin BL, Izui S. Evidence for genes in addition to Tlr7 in the Yaa translocation linked with acceleration of systemic lupus erythematosus. J Immunol. 2008;181:1556–62. doi: 10.4049/jimmunol.181.2.1556. [DOI] [PubMed] [Google Scholar]
- 28.Stohl W, Xu D, Kim KS, Koss MN, Jorgensen TN, Deocharan B, Metzger TE, Bixler SA, Hong YS, Ambrose CM, Mackay F, Morel L, Putterman C, Kotzin BL, Kalled SL. BAFF overexpression and accelerated glomerular disease in mice with an incomplete genetic predisposition to systemic lupus erythematosus. Arthritis Rheum. 2005;52:2080–91. doi: 10.1002/art.21138. [DOI] [PubMed] [Google Scholar]
- 29.Gubbels MR, Jørgensen TN, Metzger TE, Menze K, Steele H, Flannery SA, Rozzo SJ, Kotzin BL. Effects of MHC and gender on lupus-like autoimmunity in Nba2 congenic mice. J Immunol. 2005;175:6190–6. doi: 10.4049/jimmunol.175.9.6190. [DOI] [PubMed] [Google Scholar]
- 30.Vyse TJ, Rozzo SJ, Drake CG, Izui S, Kotzin BL. Control of multiple autoantibodies linked with a lupus nephritis susceptibility locus in New Zealand black mice. J Immunol. 1997;158:5566–74. [PubMed] [Google Scholar]
- 31.Kikuchi S, Fossati-Jimack L, Moll T, Amano H, Amano E, Ida A, Ibnou-Zekri N, Laporte C, Santiago-Raber ML, Rozzo SJ, Kotzin BL, Izui S. Differential role of three major New Zealand Black-derived loci linked with Yaa-induced murine lupus nephritis. J Immunol. 2005;174:1111–7. doi: 10.4049/jimmunol.174.2.1111. [DOI] [PubMed] [Google Scholar]
- 32.Choubey D, Panchanathan R. Interferon-inducible Ifi200-family genes in systemic lupus erythematosus. Immunol Lett. 2008;119:32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jørgensen TN, Gubbels MR, Kotzin BL. New insights into disease pathogenesis from mouse lupus genetics. Curr Opin Immunol. 2004;16:787–93. doi: 10.1016/j.coi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Jørgensen TN, Alfaro J, Enriquez HL, Jiang C, Loo WM, Atencio S, Bupp MR, Mailloux CM, Metzger T, Flannery S, Rozzo SJ, Kotzin BL, Rosemblatt M, Bono MR, Erickson LD. Development of murine lupus involves the combined genetic contribution of the SLAM and FcγR intervals within the Nba2 autoimmune susceptibility locus. J Immunol. 2010;184:775–86. doi: 10.4049/jimmunol.0901322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choubey D, Panchanathan R, Shen H, Duan X. Comment on “Development of murine lupus involves the combined genetic contribution of the SLAM and Fc gamma R intervals within the Nba2 autoimmune susceptibility locus”. J Immunol. 2010;184:4051–2. doi: 10.4049/jimmunol.1090015. [DOI] [PubMed] [Google Scholar]
- 36.Boross P, Arandhara VL, Martin-Ramirez J, Santiago-Raber ML, Carlucci F, Flierman R, van der Kaa J, Breukel C, Claassens JW, Camps M, Lubberts E, Salvatori D, Rastaldi MP, Ossendorp F, Daha MR, Cook HT, Izui S, Botto M, Verbeek JS. The inhibiting Fc receptor for IgG, FcγRIIB, is a modifier of autoimmune susceptibility. J Immunol. 2011;187:1304–13. doi: 10.4049/jimmunol.1101194. [DOI] [PubMed] [Google Scholar]
- 37.Choubey D, Panchanathan R, Liu H. Comment on “The inhibiting Fc receptor for IgG, FcgammaRIIB, is a modifier of autoimmune susceptibility”. J Immunol. 2011;187:3909. doi: 10.4049/jimmunol.1190055. [DOI] [PubMed] [Google Scholar]
- 38.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choubey D, Duan X, Dickerson E, Ponomareva L, Panchanathan R, Shen H, Srivastava R. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30:371–80. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnstone RW, Trapani JA. Transcription and growth regulatory functions of the HIN-200 family of proteins. Mol Cell Biol. 1999;19:5833–8. doi: 10.1128/mcb.19.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deschamps S, Meyer J, Chatterjee G, Wang H, Lengyel P, Roe BA. The mouse Ifi200-gene cluster: genomic sequence, analysis, and comparison with the human HIN-200 gene cluster. Genomics. 2003;82:34–46. doi: 10.1016/s0888-7543(03)00092-2. [DOI] [PubMed] [Google Scholar]
- 42.Asefa B, Klarmann KD, Copeland NG, Gilbert DJ, Jenkins NA, Keller JR. The interferon-inducible p200 family of proteins: a perspective on their roles in cell cycle regulation and differentiation. Blood Cells Mol Dis. 2004;32:155–67. doi: 10.1016/j.bcmd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Mondini M, Costa S, Sponza S, Gugliesi F, Gariglio M, Landolfo S. The interferon-inducible HIN-200 gene family in apoptosis and inflammation: implication for autoimmunity. Autoimmunity. 2010;43:226–31. doi: 10.3109/08916930903510922. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Chatterjee G, Meyer JJ, Liu CJ, Manjunath NA, Bray-Ward P, Lengyel P. Characteristics of three homologous 202 genes (Ifi202a, Ifi202b, and Ifi202c) from the murine interferon-activatable gene 200 cluster. Genomics. 1999;60:281–94. doi: 10.1006/geno.1999.5923. [DOI] [PubMed] [Google Scholar]
- 45.Choubey D, Gutterman JU. The interferon-inducible growth-inhibitory p202 protein: DNA binding properties and identification of a DNA binding domain. Biochem Biophys Res Commun. 1996;221:396–401. doi: 10.1006/bbrc.1996.0607. [DOI] [PubMed] [Google Scholar]
- 46.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 47.Liao JC, Lam R, Brazda V, Duan S, Ravichandran M, Ma J, Xiao T, Tempel W, Zuo X, Wang YX, Chirgadze NY, Arrowsmith CH. Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure. 2011;19:418–29. doi: 10.1016/j.str.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–18. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 49.Veeranki S, Choubey D. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Mol Immunol. 2012;49:567–71. doi: 10.1016/j.molimm.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choubey D, Pramanik R, Xin H. Subcellular localization and mechanisms of nucleocytoplasmic distribution of p202, an interferon-inducible candidate for lupus susceptibility. FEBS Lett. 2003;553:245–9. doi: 10.1016/s0014-5793(03)01006-8. [DOI] [PubMed] [Google Scholar]
- 51.Choubey D, Lengyel P. Interferon action: cytoplasmic and nuclear localization of the interferon-inducible 52-kD protein that is encoded by the Ifi200 gene from the gene 200-cluster. J Interferon Res. 1993;13:43–52. doi: 10.1089/jir.1993.13.43. [DOI] [PubMed] [Google Scholar]
- 52.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 56.Kis-Toth K, Szanto A, Thai TH, Tsokos GC. Cytosolic DNA-activated human dendritic cells are potent activators of the adaptive immune response. J Immunol. 2011;187:1222–34. doi: 10.4049/jimmunol.1100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamauchi M, Hashimoto M, Ichiyama K, Yoshida R, Hanada T, Muta T, Komune S, Kobayashi T, Yoshimura A. Ifi202, an IFN-inducible candidate gene for lupus susceptibility in NZB/W F1 mice, is a positive regulator for NF-κB activation in dendritic cells. Int Immunol. 2007;19:935–42. doi: 10.1093/intimm/dxm054. [DOI] [PubMed] [Google Scholar]
- 58.Gee K, Guzzo C, Che Mat NF, Ma W, Kumar A. The IL-12 family of cytokines in infection, inflammation and autoimmune disorders. Inflamm Allergy Drug Targets. 2009;8:40–52. doi: 10.2174/187152809787582507. [DOI] [PubMed] [Google Scholar]
- 59.Ludlow LE, Purton LE, Klarmann K, Gough DJ, Hii LL, Trapani JA, Keller JR, Clarke CJ, Johnstone RW. The role of p202 in regulating hematopoietic cell proliferation and differentiation. J Interferon Cytokine Res. 2008;28:5–11. doi: 10.1089/jir.2007.0070. [DOI] [PubMed] [Google Scholar]
- 60.Chen J, Panchanathan R, Choubey D. Stimulation of T cells up-regulates expression of Ifi202, an interferon-inducible lupus susceptibility gene, through activation of JNK/c-Jun pathway. Immunol Lett. 2008;118:13–20. doi: 10.1016/j.imlet.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimmerman M, Yang D, Hu X, Liu F, Singh N, Browning D, Ganapathy V, Chandler P, Choubey D, Abrams SI, Liu K. IFN-γ upregulates survivin and Ifi202 expression to induce survival and proliferation of tumor-specific T cells. PLoS One. 2010;5:e14076. doi: 10.1371/journal.pone.0014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panchanathan R, Duan X, Arumugam M, Shen H, Liu H, Choubey D. Cell type and gender-dependent differential regulation of the p202 and Aim2 proteins: implications for the regulation of innate immune responses in SLE. Mol Immunol. 2011;49:273–80. doi: 10.1016/j.molimm.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panchanathan R, Liu H, Liu H, Fang CM, Erickson LD, Pitha PM, Choubey D. Distinct regulation of murine lupus susceptibility genes by the IRF5/Blimp-1 axis. J Immunol. 2012;188:270–8. doi: 10.4049/jimmunol.1102311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Panchanathan R, Xin H, Choubey D. Disruption of mutually negative regulatory feedback loop between interferon-inducible p202 protein and the E2F family of transcription factors in lupus-prone mice. J Immunol. 2008;180:5927–34. doi: 10.4049/jimmunol.180.9.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xin H, D’Souza S, Jørgensen TN, Vaughan AT, Lengyel P, Kotzin BL, Choubey D. Increased expression of Ifi202, an IFN-activatable gene, in B6. Nba2 lupus susceptible mice inhibits p53-mediated apoptosis. J Immunol. 2006;176:5863–70. doi: 10.4049/jimmunol.176.10.5863. [DOI] [PubMed] [Google Scholar]
- 66.Min W, Ghosh S, Lengyel P. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-kappa B, c-Fos, and c-Jun activities. Mol Cell Biol. 1996;16:359–68. doi: 10.1128/mcb.16.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H, Liu C, Lu Y, Chatterjee G, Ma XY, Eisenman RN, Lengyel P. The interferon- and differentiation-inducible p202a protein inhibits the transcriptional activity of c-Myc by blocking its association with Max. J Biol Chem. 2000;275:27377–85. doi: 10.1074/jbc.M003409200. [DOI] [PubMed] [Google Scholar]
- 68.Ma XY, Wang H, Ding B, Zhong H, Ghosh S, Lengyel P. The interferon-inducible p202a protein modulates NF-κB activity by inhibiting the binding to DNA of p50/p65 heterodimers and p65 homodimers while enhancing the binding of p50 homodimers. J Biol Chem. 2003;278:23008–19. doi: 10.1074/jbc.M302105200. [DOI] [PubMed] [Google Scholar]
- 69.Panchanathan R, Shen H, Bupp MG, Gould KA, Choubey D. Female and male sex hormones differentially regulate expression of Ifi202, an interferon-inducible lupus susceptibility gene within the Nba2 interval. J Immunol. 2009;183:7031–8. doi: 10.4049/jimmunol.0802665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santiago-Raber ML, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, Theofilopoulos AN. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197:777–88. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pramanik R, Jørgensen TN, Xin H, Kotzin BL, Choubey D. Interleukin-6 induces expression of Ifi202, an interferon-inducible candidate gene for lupus susceptibility. J Biol Chem. 2004;279:16121–7. doi: 10.1074/jbc.M313140200. [DOI] [PubMed] [Google Scholar]
- 72.Choubey D. p202: an interferon-inducible negative regulator of cell growth. J Biol Regul Homeost Agents. 2000;14:187–92. [PubMed] [Google Scholar]
- 73.Dodson RE, Shapiro DJ. An estrogen-inducible protein binds specifically to a sequence in the 3′ untranslated region of estrogen-stabilized vitellogenin mRNA. Mol Cell Biol. 1994;14:3130–8. doi: 10.1128/mcb.14.5.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koul D, Obeyesekere NU, Gutterman JU, Mills GB, Choubey D. p202 self-associates through a sequence conserved among the members of the 200-family proteins. FEBS Lett. 1998;438:21–4. doi: 10.1016/s0014-5793(98)01263-0. [DOI] [PubMed] [Google Scholar]
- 75.Datta B, Li B, Choubey D, Nallur G, Lengyel P. p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from the p53-binding protein 1 that binds to p202 overcomes this inhibition. J Biol Chem. 1996;271:27544–55. doi: 10.1074/jbc.271.44.27544. [DOI] [PubMed] [Google Scholar]
- 76.Liu C, Wang H, Zhao Z, Yu S, Lu YB, Meyer J, Chatterjee G, Deschamps S, Roe BA, Lengyel P. MyoD-dependent induction during myoblast differentiation of p204, a protein also inducible by interferon. Mol Cell Biol. 2000;20:7024–36. doi: 10.1128/mcb.20.18.7024-7036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 78.Smith KG, Clatworthy MR. FcgammaRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat Rev Immunol. 2010;10:328–43. doi: 10.1038/nri2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang Y, Hirose S, Sanokawa-Akakura R, Abe M, Mi X, Li N, Miura Y, Shirai J, Zhang D, Hamano Y, Shirai T. Genetically determined aberrant down-regulation of FcgammaRIIB1 in germinal center B cells associated with hyper-IgG and IgG autoantibodies in murine systemic lupus erythematosus. Int Immunol. 1999;11:1685–91. doi: 10.1093/intimm/11.10.1685. [DOI] [PubMed] [Google Scholar]
- 80.Jiang Y, Hirose S, Abe M, Sanokawa-Akakura R, Ohtsuji M, Mi X, Li N, Xiu Y, Zhang D, Shirai J, Hamano Y, Fujii H, Shirai T. Polymorphisms in IgG Fc receptor IIB regulatory regions associated with autoimmune susceptibility. Immunogenetics. 2000;51:429–35. doi: 10.1007/s002510050641. [DOI] [PubMed] [Google Scholar]
- 81.Bolland S, Ravetch JV. Spontaneous autoimmune disease in Fc(gamma)RIIB-deficient mice results from strain-specific epistasis. Immunity. 2000;13:277–85. doi: 10.1016/s1074-7613(00)00027-3. [DOI] [PubMed] [Google Scholar]
- 82.Bolland S, Yim YS, Tus K, Wakeland EK, Ravetch JV. Genetic modifiers of systemic lupus erythematosus in FcgammaRIIB(−/−) mice. J Exp Med. 2002;195:1167–74. doi: 10.1084/jem.20020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McGaha TL, Karlsson MC, Ravetch JV. FcgammaRIIB deficiency leads to autoimmunity and a defective response to apoptosis in Mrl-MpJ mice. J Immunol. 2008;180:5670–9. doi: 10.4049/jimmunol.180.8.5670. [DOI] [PubMed] [Google Scholar]
- 84.Panchanathan R, Shen H, Duan X, Rathinam VA, Erickson LD, Fitzgerald KA, Choubey D. Aim2 deficiency in mice suppresses the expression of the inhibitory Fcgamma receptor (FcgammaRIIB) through the induction of the IFN-inducible p202, a lupus susceptibility protein. J Immunol. 2011;186:6762–70. doi: 10.4049/jimmunol.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dhodapkar KM, Banerjee D, Connolly J, Kukreja A, Matayeva E, Veri MC, Ravetch JV, Steinman RM, Dhodapkar MV. Selective blockade of the inhibitory Fcgamma receptor (FcgammaRIIB) in human dendritic cells and monocytes induces a type I interferon response program. J Exp Med. 2007;204:1359–69. doi: 10.1084/jem.20062545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang A, Batteux F, Wakeland EK. The role of SLAM/CD2 polymorphisms in systemic autoimmunity. Curr Opin Immunol. 2010;22:706–14. doi: 10.1016/j.coi.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 87.Detre C, Keszei M, Romero X, Tsokos GC, Terhorst C. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin Immunopathol. 2010;32:157–71. doi: 10.1007/s00281-009-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 89.Chatterjee M, Rauen T, Kis-Toth K, Kyttaris VC, Hedrich CM, Terhorst C, Tsokos GC. Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J Immunol. 2012;188:1206–12. doi: 10.4049/jimmunol.1102773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312:1665–9. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- 91.Chatterjee M, Kis-Toth K, Thai TH, Terhorst C, Tsokos GC. SLAMF6-driven co-stimulation of human peripheral T cells is defective in SLE T cells. Autoimmunity. 2011;44:211–8. doi: 10.3109/08916934.2010.530627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004;199:1255–64. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keszei M, Latchman YE, Vanguri VK, Brown DR, Detre C, Morra M, Arancibia-Carcamo CV, Paul E, Calpe S, Castro W, Wang N, Terhorst C, Sharpe AH. Auto-antibody production and glomerulonephritis in congenic Slamf1−/− and Slamf2−/− [B6.129] but not in Slamf1−/− and Slamf2−/− [BALB/c. 129] mice. Int Immunol. 2011;23:149–58. doi: 10.1093/intimm/dxq465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Howie D, Okamoto S, Rietdijk S, Clarke K, Wang N, Gullo C, Bruggeman JP, Manning S, Coyle AJ, Greenfield E, Kuchroo V, Terhorst C. The role of SAP in murine CD150 (SLAM)-mediated T-cell proliferation and interferon gamma production. Blood. 2002;100:2899–907. doi: 10.1182/blood-2002-02-0445. [DOI] [PubMed] [Google Scholar]
- 95.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 96.D’Souza S, Xin H, Walter S, Choubey D. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J Biol Chem. 2001;276:298–305. doi: 10.1074/jbc.M007155200. [DOI] [PubMed] [Google Scholar]
- 97.Jordan MA, Fletcher JM, Jose R, Chowdhury S, Gerlach N, Allison J, Baxter AG. Role of SLAM in NKT cell development revealed by transgenic complementation in NOD mice. J Immunol. 2011;186:3953–65. doi: 10.4049/jimmunol.1003305. [DOI] [PubMed] [Google Scholar]
- 98.Panchanathan R, Duan X, Shen H, Rathinam VA, Erickson LD, Fitzgerald KA, Choubey D. Aim2 deficiency stimulates the expression of IFN-inducible Ifi202, a lupus susceptibility murine gene within the Nba2 autoimmune susceptibility locus. J Immunol. 2010;185:7385–93. doi: 10.4049/jimmunol.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, Dixit VM, Monack DM. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–6. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kozyrev SV, Alarcon-Riquelme ME. The genetics and biology of Irf5-mediated signaling in lupus. Autoimmunity. 2007;40:591–601. doi: 10.1080/08916930701510905. [DOI] [PubMed] [Google Scholar]
- 101.Barnes B, Lubyova B, Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res. 2002;22:59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- 102.Lien C, Fang CM, Huso D, Livak F, Lu R, Pitha PM. Critical role of IRF-5 in regulation of B-cell differentiation. Proc Natl Acad Sci U S A. 2010;107:4664–8. doi: 10.1073/pnas.0911193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–69. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 104.Savitsky DA, Yanai H, Tamura T, Taniguchi T, Honda K. Contribution of IRF5 in B cells to the development of murine SLE-like disease through its transcriptional control of the IgG2a locus. Proc Natl Acad Sci U S A. 2010;107:10154–9. doi: 10.1073/pnas.1005599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shen H, Panchanathan R, Rajavelu P, Duan X, Gould KA, Choubey D. Gender-dependent expression of murine Irf5 gene: implications for sex bias in autoimmunity. J Mol Cell Biol. 2010;2:284–90. doi: 10.1093/jmcb/mjq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mondini M, Vidali M, Airò P, De Andrea M, Riboldi P, Meroni PL, Gariglio M, Landolfo S. Role of the interferon-inducible gene IFI16 in the etiopathogenesis of systemic autoimmune disorders. Ann N Y Acad Sci. 2007;1110:47–56. doi: 10.1196/annals.1423.006. [DOI] [PubMed] [Google Scholar]
- 107.Choubey D, Deka R, Ho SM. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci. 2008;13:598–608. doi: 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- 108.Kimkong I, Avihingsanon Y, Hirankarn N. Expression profile of HIN200 in leukocytes and renal biopsy of SLE patients by real-time RT-PCR. Lupus. 2009;18:1066–72. doi: 10.1177/0961203309106699. [DOI] [PubMed] [Google Scholar]
- 109.Duan X, Ponomareva L, Veeranki S, Panchanathan R, Dickerson E, Choubey D. Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Mol Cancer Res. 2011;9:589–602. doi: 10.1158/1541-7786.MCR-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Flinn LJ. Genomic analysis of a human interferon-inducible gene family and systemic lupus erythematosus. J Interferon Cytokine Res. 2007;27:828. [Google Scholar]
- 111.Choubey D. DNA-responsive inflammasomes and their regulators in autoimmunity. Clin Immunol. 2012;142:223–31. doi: 10.1016/j.clim.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]