Abstract
The Notch signaling pathway controls patterning and cell fate decisions during development in metazoans, and is associated with human diseases such as cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) and certain cancers. Studies over the last several years have revealed sophisticated regulation of both the membrane-bound Notch receptor and its ligands by vesicle trafficking. This is perhaps most evident in neural progenitor cells in Drosophila, which divide asymmetrically to segregate Numb, an endocytic adaptor protein that acts as a Notch pathway inhibitor, to one daughter cell. Here, we discuss recent findings addressing how receptor and ligand trafficking to specific membrane compartments control activation of the Notch pathway in asymmetrically dividing cells and other tissues.
Introduction
In this review we will focus on the role of endocytosis- the process of internalization of membrane components, including phospholipids and integral membrane proteins, from the plasma membrane to intracellular compartments- in regulating trafficking of Notch signaling pathway components. The Notch signaling pathway is used in a variety of cellular contexts during embryonic development to establish distinct cell fate among individual cells. Importantly, Notch pathway activation is intimately associated with the membrane, as most core components of the pathway, with the exception of the nuclear factors, are membrane proteins. Genetic analysis in flies has highlighted the importance of vesicle trafficking events on Notch signaling during wing and eye development, and particularly in the development of the sensory organ precursor (SOP) cell lineage. The SOP cells arise from clusters of equipotent epithelial cells, termed proneural clusters, which use Notch signaling-mediated lateral inhibition to select a neural progenitor cell (Fig. 1). The progenitor cell retains its proneural cell fate while inhibiting the same in the rest of the cells in the cluster. Later, the neural precursor cells again use the Notch pathway to establish different cell fates following asymmetric cell division.
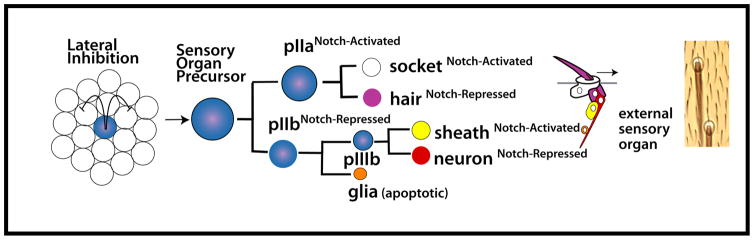
Figure 1.
Notch-mediated lateral inhibition selects a sensory organ precursor cell from a group of equipotent neurogenic epithelial cells. The progenitor divides four times giving rise to five cells that compose the external sensory organ. Notch signaling status determines cell fates in lineage.
The Notch signaling pathway
The canonical Notch signaling pathway is induced by binding of membrane-tethered ligands of the DSL family (Delta/Serrate/Lag2) in the signal-sending cell to the Notch transmembrane receptor protein in the signal-receiving cell. Prior to ligand binding, Notch signaling activation requires multiple rounds of glycosylation of receptor and processing by the endopeptidase Furin (S1 cleavage) in the Golgi. Notch is then trafficked as a heterodimer to the plasma membrane. Ligand binding is mediated through the EGF repeats of the Notch extracellular domain (NECD) and induces sequential cleavage of the Notch receptor at the plasma membrane by the ADAM metalloprotease (S2 cleavage) and the γ-secretase complex (S3 cleavage) [1–3]. The final S3 cleavage releases the Notch intracellular domain (NICD) from the membrane, which interacts with DNA binding protein CSL (CBF1/Suppressor of Hairless/Lag1) and Mastermind to regulate gene expression in the nucleus. In Drosophila, the requirement of endocytosis in both signal-sending and signal-receiving cells was first demonstrated by the clonal analysis of temperature-sensitive shibire mutants [4]. Shibire encodes the Drosophila homolog of dynamin [5], a protein which is required to pinch off endocytic vesicles from the plasma membrane. Subsequently, the endocytosis of the Notch ligand Delta in the signal-sending cell was shown to be required to activate Notch signaling in the signal-receiving cell[6••]. As both the Notch receptor and the DSL ligands are detected at the plasma membrane and in endocytic compartments in most cells, it is at present unclear whether ligand-induced Notch activation occurs exclusively at the plasma membrane and/or within endocytic membranes.
The sensory organ precursor cell: model for regulating Notch by asymmetric cell division
The SOP is an ideal model to study the regulatory mechanisms of Notch-mediated binary cell fate controlled by asymmetric cell division. Development of the adult peripheral nervous system is extremely rapid: the sensory organ precursor cell undergoes four rounds of mitosis within six hours to generate five cells[7]. One cell promptly undergoes apoptosis [8], while the remaining four cells differentiate into the external sensory organ (Fig. 1). The proliferative phase of the lineage is characterized by tightly regulated cell divisions [7,9,10]. The sensory organ precursor cell gives rise to two daughter cells, the pIIa and pIIb [11]. Notch is required for pIIa cell fate and overexpression of Notch causes the pIIb to switch to a pIIa cell, indicating that Notch is differentially regulated to distinguish pIIa and pIIb cell fates [12]. During SOP mitosis, two endocytic regulators, Numb and Neuralized, are asymmetrically localized and segregated to the pIIb daughter cell. While we will cover the functions of Numb and Neuralized below, the mechanisms controlling the asymmetric targeting of both proteins are complex and have been discussed extensively in excellent recent reviews [13–15], and therefore not covered here.
Notch trafficking and the role of Numb
The core elements canonical Notch pathway described above (ligand, receptor, proteases and nuclear cofactors) are required in most contexts, including in asymmetrically dividing progenitor cells, for productive Notch signaling. However, genetic screens revealed a number of genes that have a functional role in controlling Notch signaling in asymmetrically dividing cells. Numb functions to promote pIIb cell fate and encodes an adaptor protein that can bind the Notch receptor and interact with endocytic proteins such as α-adaptin and EPS15 [16–18]). Overexpression of Numb suppresses pIIa cell fate, indicating that it acts to antagonize Notch function in the pIIb cell. Importantly, Numb asymmetric localization and interaction with the Notch pathway do appear to be conserved in vertebrate systems, particularly in neural precursor cells [19–21].
The mechanism of Numb’s antagonism toward Notch has been elusive, and controversial. Numb’s interaction with endocytic proteins has lead to the hypothesis that Numb may regulate Notch by promoting removal of the Notch receptor from the plasma membrane into endosomes. It is clear from numerous studies that Notch receptor trafficking through the endocytic pathway is important for Notch signaling regulation [22]. However, there is little direct evidence to support the model that Numb regulates bulk endocytosis of Notch in vivo, and removal of Numb’s endocytic binding motifs do not appear to interfere with Numb’s ability of inhibit Notch [17]. An alternative hypothesis is that Numb may alter the path of the Notch receptor trafficking, thereby depleting Notch from a specific membrane domain. Recent live imaging studies, using a GFP-tagged Notch, have shown that Numb blocks the accumulation of Notch along the membrane interface between pIIa and pIIb cells at cytokinesis [23••]. Furthermore, a Golgi-associated protein ACBD3 has been shown to be a conserved Numb-interacting protein that regulates progenitor cell fate and Notch signaling in both the mammalian and fly nervous systems [24•]. In mammalian cells ACBD3 disassociates from the Golgi during mitosis, potentially restricting its interaction with cortically associated Numb to mitosis. However, the role of ACBD3 in regulation of Notch signaling remains undetermined.
In contrast to other contexts, such as in the eye or wing, Notch signaling in asymmetrically dividing cells requires the activity of Sanpodo, a type IIIa transmembrane protein whose expression is restricted to asymmetrically dividing cells in the CNS, PNS, and muscle lineages [25••]. Sanpodo is segregated to both the signal-sending and signal-receiving cells after SOP mitosis, and is targeted preferentially to early endosomes in the low-Notch pIIb cell in a Numb-dependent manner, and to the plasma membrane in pIIa cells [25••–29]. Sanpodo associates with both Numb and Notch, and co-localizes with Notch and Delta in endocytic vesicles. The role of Sanpodo in promoting Notch signaling is currently not known, but Sanpodo appears to deplete Notch from the apical membrane in daughter cells in a Numb-independent way [23••]. Interestingly, ectopic expression of Sanpodo suppresses Notch signaling in epithelial cells undergoing lateral inhibition and depletes the Notch receptor from the apical domain [23••,30]. In contrast, the AP-1 complex of endocytic adaptor proteins, which localizes to the trans-Golgi network and recycling endosome, act to suppress Notch signaling activation in the pIIb cell, perhaps by limiting accumulation of Notch and Sanpodo at the apical membrane region, proximal to adherens junctions components such as DE-cadherin [31]. Numb, Sanpodo, and AP-1 complex appear therefore to ensure trafficking of the Notch receptor, but not the Notch ligand Delta, to a specific membrane compartment shortly after completion of the asymmetric cell division (Fig. 2). However, what remains unclear is whether the activating Notch cleavage event occurs in a plasma membrane domain, such as apical region of the cell near the adherens junction, or whether Notch trafficking through an endosomal compartment is required for signaling in the pIIa cell.
Figure 2.
Trafficking of DSL ligands and the Notch receptor to and from the plasma membrane of both signal sending cells and signal-receiving cells control activation of the pathway. Genes involved in trafficking the receptor and/or ligand mentioned in the text are in blue, cytopslamic membrane compartments such as the trans-Golgi network (TGN), early endosome (EE), recycling endosome (RE), multi-vesicular body (MVB) and lysosome are represented. See text for details on roles of genes and compartments in control Notch receptor and ligand trafficking and signaling. Dashed arrows indicate potential site of Notch gamma-secretase cleavage and release of Notch intracellular domain.
Targeting Notch for degradation through the endocytic pathway is an additional control mechanism for Notch signaling. Mutations in the ESCRT complex genes vps25 and erupted, as well as the C2 domain containing protein lethal giant discs (lgd) result in accumulation of Notch in late endosomal compartments and Notch pathway activation in mutant cells [32–35] (Fig. 2). The E3 ubiquitin ligase Deltex appears to function with the HOPS complex and AP3 adaptor proteins to guide Notch to the lysome, determining whether it will be degraded in the lumen, or restricted to the limiting membrane, [36]. Taken together, these observations indicate that timely passage of the Notch receptor through the endosomal system limits release of the Notch intracellular domain, either by preventing accumulation of Notch and ligands in a permissive endosomal compartment, and/or processing by gamma-secretase within late endosomes.
Endocytic trafficking of DSL ligands
Two structurally unrelated RING-type E3 ubiquitin ligases, Neuralized and Mindbomb promote Notch ligand endocytosis by ubiquitylation [37–44]. Conserved intracellular domains/motifs that interact with the ubiquitin ligases and/or signal for endocytosis have been identified in the Drosophila Delta and Serrate by sequence comparison with their counterparts in other species. Both Neuralized and Mindbomb bind to the same Asn-based tripeptide stretch (NNL) on Serrate which is part of a longer motif conserved between invertebrate and vertebrate Serrate/Jagged proteins[45]. More recently, another motif (QNEEN) found adjacent to the NNL motif has been independently identified and shown to interact with Neuralized [46]. However in Delta, two discrete intracellular domains, ICD1 and ICD2 serve as interaction sites for Neuralized and Mindbomb respectively [47]. The motifs identified on Serrate have similarities with ICD1 (QNExN) as well as ICD2 (NNI/V) [47].
The E3 ubiquitin ligases add ubiquitin moieties to lysine residues on their target proteins. Two lysines close to the conserved motif for Neuralized and Mindbomb binding in Serrate have been shown to function as ubiquitin acceptor sites [45]. The K742 is the preferred lysine for Neuralized-mediated ubiquitylation of Drosophila Delta, while Mindbomb ubiquitylates different lysines without a specific preference [47].
The entry of the ubiquitylated Notch ligands into the endocytic machinery is assisted by the endocytic adaptor protein, Epsin, [48••,49]. Epsin has several motifs including the ubiquitin-interacting motifs (UIMs) which can interact with mono- or poly-ubiquitinated proteins [50–52] and an ENTH domain that interacts with PIP2 on the membrane and can induce membrane curvature[53]. Epsin also has motifs for interaction with AP2 and clathrin. In a recent study, one of the two UIMs, and the ENTH domain have been shown to be essential for Notch signaling [54]. In the same study, the clathrin binding motifs on the C-terminal region were dispensable although the extended region encompassing these and motifs for interaction with AP2 were necessary for Epsin function in Notch signaling. Clathrin involvement is further supported by the requirement of the endocytic protein Auxilin for Notch ligand endocytosis. Auxilin plays several roles during clathrin-mediated endocytosis including nucleating clathrin-coated pits and disassembly of clathrin coated vesicles along with its cofactor, Hsc70 [55]. Drosophila auxilin is required for Delta endocytosis and its function in clathrin dynamics is necessary for the internalization of Delta [56,57].
Mechanism of Notch activation by ligand endocytosis
Two major models have been proposed to explain the requirement of ligand internalization for Notch activation. The ligand-recycling model proposes that the ligand is internalized prior to its interaction with Notch in a ubiquitantion- and epsin-dependent manner and recycled back to the surface with a modification that renders the ligand to be signaling-competent[48••]. The requirement of Rab11, a GTPase on the recycling endosomes [58•] and Sec15, a component of the exocyst complex in Delta recycling in Drosophila SOPs support this model [59]. Recent studies on the Arp2/3 complex and Wiscott-Aldrich syndrome protein (WASp) as well as the Eps15 homology domain containing protein-binding protein 1 (dEHBP1) in Drosophila SOPs further support the requirement of Delta recycling for Notch activation. The dEHBP1 interacts with Rab11 and Sec15 and assists in the exocytosis and recycling of Delta in the SOPs [60]. The Arp2/3 and WASP are required for the trafficking of endocytosed Delta to the apical microvilli in the signal-sending cells [61] (Fig. 2).
According to the ‘pulling’ model, the endocytosis of the ligand bound to the Notch receptor induces a conformational change that allows access to the metalloproteases to the S2 cleavage site on the Notch receptor (6••). This model is supported by the observation of transendocytosis of NECD into the Delta-expressing central provein cells of Drosophila that do not express Notch [6••] and the localization of NECD in the vesicles of mammalian cells expressing Delta in a co-culture with Notch-expressing cells in the presence of ADAM protease inhibitors [62]. In further support of this model, Rab11 is not required for Notch signaling in Drosophila germline cells and the developing eye suggesting that Delta does not need to be recycled to activate Notch[63•]. Ligand endocytosis is still required for Notch activation in these contexts and it might provide the necessary force for the mechanical disruption of Notch [64]. However, in the germline cells, Delta is endocytosed in a dynamin- and epsin-dependent manner but does not require any other endocytic/recycling factors to promote signaling [65]. Whether dynamin and epsin are sufficient for NECD release or if lipid rafts are involved here as has been suggested in cultured mammalian cells for the ubiquitylation-dependent endocytosis of Dll3 and Dll1 [66,67] remains to be seen.
Recent data on transcytosis of Delta from the basolateral to the apical membrane in SOPs for Notch activation and in cultured MDCK cells suggest that the recycling of Delta localizes Delta in proximity with the Notch proteins located on the apical membrane and allows for ligand-receptor interaction[68•]. However this model does not preclude the pulling force model for the release of NECD after the recycled Delta interacts with Notch on the adjacent cells in which case a second ligand endocytic event would be required for transendocytosis of Notch [68•]. Since an activated form of Delta generated during its recycling has not been identified, the transcytosis of Delta is currently the most convincing mechanism for ligand recycling and Notch activation.
Asymmetry of endocytic components
An important unresolved question is how directionality of the ligand-receptor interaction is established and maintained. In asymmetrically dividing SOPs, Notch and Delta are present in both daughter cells. Numb and Neuralized are segregated to the low Notch daughter and contribute to biasing the signal toward Notch activation in the pIIa cell daughter. In addition, both the recycling endosomes and a subpopulation of early endosomes exhibit a remarkable asymmetry in SOP daughter cells (Fig. 3). Rab11 accumulates around the pIIb cell centrosome within minutes after completion of cytokinesis [58•], while a subpopulation of Rab5-positive early endosomes associated with Sara (Smad anchor for receptor activation) segregate into the pIIa cell at the completion of cytokinesis [69•]. The Rab11 asymmetry has been proposed to promote recycling and activation of Delta in signaling-sending cell, while the Sara-positive endosomes are a potential site of Notch activation as they contain Notch, Delta and Sanpodo. It remains to be determined how the asymmetry of endosomal compartments and the asymmetric targeting of Numb and Neuralized function together to ensure robust activation of Notch in one daughter cell, and complete suppression of the pathway in the other.
Figure 3.
The pIIa and pIIb daughter cells after asymmetric cell division of the sensory organ precursor cell. Numb and Neuralized are inherited by the pIIb cell daughter, and the recycling endsome accumulates around the pIIb cell centrosome. Sanpodo localizes to the apical membrane in the pIIa cell, as well as in a population of Sara-labeled early endosomes, which also contain Notch and Delta.
Conclusions
Although much progress has been made, our understanding of how membrane trafficking of Notch and its ligands can control activation or repression of the pathway, particularly in model organisms such as Drosophila, remains incomplete and some outstanding questions remain. In particular, how steady state levels of membrane Notch are established and maintained across a range of tissues is poorly understood, and may have important implications for controlling the output of the pathway. Although some studies have addressed the roles of conserved ubiquitin ligases that target Notch pathway components, such as mindbomb, the broader role of trafficking of Notch and its ligands in vertebrate development remains largely unexplored. Importantly, a better understanding of trafficking mechanisms that specifically regulate Notch signaling may provide new opportunities and therapeutic targets for treatment of human diseases, such as cancer, which involve inappropriate Notch activation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–38. [PubMed] [Google Scholar]
- 2.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 3.Ilagan MXG, Kopan R. SnapShot: notch signaling pathway. Cell. 2007;128:1246. doi: 10.1016/j.cell.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Seugnet L, Simpson P, Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev Biol. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- 5.van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 6••.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. In this study, the authors demonstrated transendocytosis of Notch in Delta-expressing cells and its colocalization with Delta. They suggested that endocytosis of the ligand promotes the dissociation of the Notch extracellular domain by unmasking the S2 cleavage sites, thus facilitating the S3 cleavage to release Notch intracellular domain. [DOI] [PubMed] [Google Scholar]
- 7.Gho M, Bellaïche Y, Schweisguth F. Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development. 1999;126:3573–3584. doi: 10.1242/dev.126.16.3573. [DOI] [PubMed] [Google Scholar]
- 8.Fichelson P, Gho M. The glial cell undergoes apoptosis in the microchaete lineage of Drosophila. Development. 2003;130:123–133. doi: 10.1242/dev.00198. [DOI] [PubMed] [Google Scholar]
- 9.Roegiers F, Younger-Shepherd S, Jan LY, Jan YN. Two types of asymmetric divisions in the Drosophila sensory organ precursor cell lineage. Nat Cell Biol. 2001;3:58–67. doi: 10.1038/35050568. [DOI] [PubMed] [Google Scholar]
- 10.Bellaïche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O’Kane CJ, Bryant PJ, Schweisguth F. The Partner of Inscuteable/Discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell. 2001;106:355–366. doi: 10.1016/s0092-8674(01)00444-5. [DOI] [PubMed] [Google Scholar]
- 11.Hartenstein V, Posakony JW. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- 12.Hartenstein V, Posakony JW. A dual function of the Notch gene in Drosophila sensillum development. Dev Biol. 1990;142:13–30. doi: 10.1016/0012-1606(90)90147-b. [DOI] [PubMed] [Google Scholar]
- 13.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roegiers F, Jan YN. Asymmetric cell division. Curr Opin Cell Biol. 2004;16:195–205. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Segalen M, Bellaïche Y. Cell division orientation and planar cell polarity pathways. Seminars in Cell & Developmental Biology. 2009;20:972–977. doi: 10.1016/j.semcdb.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, Di Fiore PP. Numb is an endocytic protein. The Journal of Cell Biology. 2000;151:1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang H, Rompani SB, Atkins JB, Zhou Y, Osterwalder T, Zhong W. Numb proteins specify asymmetric cell fates via an endocytosis- and proteasome-independent pathway. Molecular and Cellular Biology. 2005;25:2899–2909. doi: 10.1128/MCB.25.8.2899-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berdnik D, Török T, González-Gaitán M, Knoblich JA. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Developmental Cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhong W, Jiang MM, Schonemann MD, Meneses JJ, Pedersen RA, Jan LY, Jan YN. Mouse numb is an essential gene involved in cortical neurogenesis. Proc Natl Acad Sci US A. 2000;97:6844–6849. doi: 10.1073/pnas.97.12.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- 22.Fortini ME, Bilder D. Endocytic regulation of Notch signaling. Current Opinion in Genetics & Development. 2009;19:323–328. doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Couturier L, Vodovar N, Schweisguth F. Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat Cell Biol. 2012;14:131–139. doi: 10.1038/ncb2419. Using a GFP-tagged Notch, the authors show that Notch intracellular domain can be detected in the pIIa but not the pIIb cell nucleus, and confirming that both Numb and Sanpodo independently promote Notch endocytosis, with Numb blocking Notch trafficking to an apical domain in vivo. [DOI] [PubMed] [Google Scholar]
- 24•.Zhou Y, Atkins JB, Rompani SB, Bancescu DL, Petersen PH, Tang H, Zou K, Stewart SB, Zhong W. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 2007;129:163–178. doi: 10.1016/j.cell.2007.02.037. Describes the association between the Golgi protein ACBD3 and Numb. ACBD3 disassociates from the Golgi during mitosis, allowing it to potentially bind Numb at the plasma membrane. Overexpression ACBD3 alters cell fate assignments in the SOP lineage, supporting a role for this protein in regulation of Notch signaling. [DOI] [PubMed] [Google Scholar]
- 25••.O’Connor-Giles KM, Skeath JB. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Developmental Cell. 2003;5:231–243. doi: 10.1016/s1534-5807(03)00226-0. Identified Sanpodo as a four pass transmembrane protein that interacts with Numb and Notch and is required for Notch activation in pIIb cells. The authors proposed a model by which Sanpodo membrane association is controlled by Numb to regulate Notch activation. [DOI] [PubMed] [Google Scholar]
- 26.Hutterer A, Knoblich JA. Numb and alpha-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 2005;6:836–842. doi: 10.1038/sj.embor.7400500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langevin J, Le Borgne R, Rosenfeld F, Gho M, Schweisguth F, Bellaïche Y. Lethal giant larvae controls the localization of notch-signaling regulators numb, neuralized, and Sanpodo in Drosophila sensory-organ precursor cells. Curr Biol. 2005;15:955–962. doi: 10.1016/j.cub.2005.04.054. [DOI] [PubMed] [Google Scholar]
- 28.Tong X, Zitserman D, Serebriiskii I, Andrake M, Dunbrack R, Roegiers F. Numb independently antagonizes Sanpodo membrane targeting and Notch signaling in Drosophila sensory organ precursor cells. Mol Biol Cell. 2010;21:802–810. doi: 10.1091/mbc.E09-09-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roegiers F, Jan LY, Jan YN. Regulation of membrane localization of Sanpodo by lethal giant larvae and neuralized in asymmetrically dividing cells of Drosophila sensory organs. Mol Biol Cell. 2005;16:3480–3487. doi: 10.1091/mbc.E05-03-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babaoglan AB, O’Connor-Giles KM, Mistry H, Schickedanz A, Wilson BA, Skeath JB. Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions. Development. 2009;136:4089–4098. doi: 10.1242/dev.040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benhra N, Lallet S, Cotton M, Le Bras S, Dussert A, Le Borgne R. AP-1 controls the trafficking of Notch and Sanpodo toward E-cadherin junctions in sensory organ precursors. Curr Biol. 2011;21:87–95. doi: 10.1016/j.cub.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert MM, Robinson BS, Moberg KH. Functional interactions between the erupted/tsg101 growth suppressor gene and the DaPKC and rbf1 genes in Drosophila imaginal disc tumors. PLoS ONE. 2009;4:e7039. doi: 10.1371/journal.pone.0007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Childress JL, Acar M, Tao C, Halder G. Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis. Curr Biol. 2006;16:2228–2233. doi: 10.1016/j.cub.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallagher CM, Knoblich JA. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Developmental Cell. 2006;11:641–653. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 35.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Developmental Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 36.Wilkin M, Tongngok P, Gensch N, Clemence S, Motoki M, Yamada K, Hori K, Taniguchi-Kanai M, Franklin E, Matsuno K, et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Developmental Cell. 2008;15:762–772. doi: 10.1016/j.devcel.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Le Borgne R, Remaud S, Hamel S, Schweisguth F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 2005;3:e96. doi: 10.1371/journal.pbio.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Borgne R, Schweisguth F. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Developmental Cell. 2003;5:139–148. doi: 10.1016/s1534-5807(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 39.Deblandre GA, Lai EC, Kintner C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Developmental Cell. 2001;1:795–806. doi: 10.1016/s1534-5807(01)00091-0. [DOI] [PubMed] [Google Scholar]
- 40.Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Developmental Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- 41.Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- 42.Pitsouli C, Delidakis C. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development. 2005;132:4041–4050. doi: 10.1242/dev.01979. [DOI] [PubMed] [Google Scholar]
- 43.Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. neuralized Encodes a peripheral membrane protein involved in delta signaling and endocytosis. Developmental Cell. 2001;1:807–816. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Struhl G. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development. 2005;132:2883–2894. doi: 10.1242/dev.01860. [DOI] [PubMed] [Google Scholar]
- 45.Glittenberg M, Pitsouli C, Garvey C, Delidakis C, Bray S. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 2006;25:4697–4706. doi: 10.1038/sj.emboj.7601337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fontana JR, Posakony JW. Both inhibition and activation of Notch signaling rely on a conserved Neuralized-binding motif in Bearded proteins and the Notch ligand Delta. Dev Biol. 2009;333:373–385. doi: 10.1016/j.ydbio.2009.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daskalaki A, Shalaby NA, Kux K, Tsoumpekos G, Tsibidis GD, Muskavitch MAT, Delidakis C. Distinct intracellular motifs of Delta mediate its ubiquitylation and activation by Mindbomb1 and Neuralized. The Journal of Cell Biology. 2011;195:1017–1031. doi: 10.1083/jcb.201105166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. By sophisticated mosaic analyses, the authors suggested that Delta is internalized in an Epsin-dependent manner and routed into a recycling pathway. During recycling, Delta is modified into a signaling-competent form that can interact with Notch receptor and lead to the activation of the pathway. [DOI] [PubMed] [Google Scholar]
- 49.Overstreet E, Chen X, Wendland B, Fischer JA. Either part of a Drosophila epsin protein, divided after the ENTH domain, functions in endocytosis of delta in the developing eye. Curr Biol. 2003;13:854–860. doi: 10.1016/s0960-9822(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 50.Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- 51.Hawryluk MJ, Keyel PA, Mishra SK, Watkins SC, Heuser JE, Traub LM. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic. 2006;7:262–281. doi: 10.1111/j.1600-0854.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- 52.Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 53.Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 54.Xie X, Cho B, Fischer JA. Drosophila Epsin’s role in Notch ligand cells requires three Epsin protein functions: The lipid binding function of the ENTH domain, a single Ubiquitin interaction motif, and a subset of the C-terminal protein binding modules. Dev Biol. 2012;363:399–412. doi: 10.1016/j.ydbio.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 56.Eun SH, Banks SML, Fischer JA. Auxilin is essential for Delta signaling. Development. 2008;135:1089–1095. doi: 10.1242/dev.009530. [DOI] [PubMed] [Google Scholar]
- 57.Kandachar V, Bai T, Chang HC. The clathrin-binding motif and the J-domain of Drosophila Auxilin are essential for facilitating Notch ligand endocytosis. BMC Dev Biol. 2008;8:50. doi: 10.1186/1471-213X-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. Using live cell imaging, this study showed that the recycling endosome, marked by Rab11, accumulates around the centrosome exclusively in the pIIb cell following mitosis. [DOI] [PubMed] [Google Scholar]
- 59.Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, Knoblich JA, Bellen HJ. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Developmental Cell. 2005;9:351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 60.Giagtzoglou N, Yamamoto S, Zitserman D, Graves HK, Schulze KL, Wang H, Klein H, Roegiers F, Bellen HJ. dEHBP1 controls exocytosis and recycling of Delta during asymmetric divisions. The Journal of Cell Biology. 2012;196:65–83. doi: 10.1083/jcb.201106088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajan A, Tien A-C, Haueter CM, Schulze KL, Bellen HJ. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol. 2009;11:815–824. doi: 10.1038/ncb1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nichols JT, Miyamoto A, Olsen SL, D’Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. The Journal of Cell Biology. 2007;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Banks SML, Cho B, Eun SH, Lee J-H, Windler SL, Xie X, Bilder D, Fischer JA. The functions of auxilin and Rab11 in Drosophila suggest that the fundamental role of ligand endocytosis in notch signaling cells is not recycling. PLoS ONE. 2011;6:e18259. doi: 10.1371/journal.pone.0018259. In this study, by showing recycling of Delta is not required in germline cells, the authors suggest that the mechanism of Notch activation might vary between cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinmaster G, Fischer JA. Notch ligand ubiquitylation: what is it good for? Developmental Cell. 2011;21:134–144. doi: 10.1016/j.devcel.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Windler SL, Bilder D. Endocytic internalization routes required for delta/notch signaling. Curr Biol. 2010;20:538–543. doi: 10.1016/j.cub.2010.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heuss SF, Ndiaye-Lobry D, Six EM, Israël A, Logeat F. The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc Natl Acad Sci US A. 2008;105:11212–11217. doi: 10.1073/pnas.0800695105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Widau RC, Herring BP, Gallagher PJ. Delta-like 1-Lysine613 regulates notch signaling. Biochim Biophys Acta. 2011;1813:2036–2043. doi: 10.1016/j.bbamcr.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68•.Benhra N, Vignaux F, Dussert A, Schweisguth F, Le Borgne R. Neuralized promotes basal to apical transcytosis of delta in epithelial cells. Mol Biol Cell. 2010;21:2078–2086. doi: 10.1091/mbc.E09-11-0926. This study showed localization of Notch and Delta at the apical and basolateral plasma membranes respectively in Drosophila SOPs and polarized MDCK cells. They further showed that Delta is transcytosed to the apical membrane in a Neuralized-dependent manner, suggesting that the relocalization of Delta facilitates its interaction with Notch. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Coumailleau F, Fürthauer M, Knoblich JA, González-Gaitán M. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 2009;458:1051–1055. doi: 10.1038/nature07854. Reports a sub-population of early endosomes, marked by the Sara protein, are asymmetrically segregated to the pIIa cell during mitosis. These endosomes initially contain Sanpodo, Delta, and both the Notch ICD and ECD, at later time points, only the Notch ECD is detected, suggesting that Notch cleavage may occur within these endosomes to promote Notch activation in the pIIa cell. [DOI] [PubMed] [Google Scholar]