Abstract
As the power of studying mouse genetics and behavior advances, research tools to examine systems level connectivity in the mouse are critically needed. In this study, we compared statistical mapping of the olfactory system in adult mice using manganese-enhanced MRI (MEMRI) and diffusion tensor imaging (DTI) with probabilistic tractography. The primary goal was to determine whether these complementary techniques can determine mouse olfactory bulb connectivity consistent with known anatomical connections. For MEMRI, 3D T1 weighted images were acquired before and after bilateral nasal administration of MnCl2 solution. Concomitantly, high resolution diffusion-tensor images were obtained ex vivo from a second group of mice and processed with a probabilistic tractography algorithm originating in the olfactory bulb. Incidence maps were created by co-registering and overlaying data from the two scan modalities. The resulting maps clearly show pathways between the olfactory bulb and amygdala, piriform cortex, caudate putamen, and olfactory cortex in both the DTI and MEMRI techniques that are consistent with the known anatomical connections. These data demonstrate that MEMRI and DTI are complementary, high-resolution neuroimaging tools that can be applied to mouse genetic models of olfactory and limbic system connectivity.
Keywords: Statistical Mapping, Diffusion Tensor Imaging (DTI), Manganese Enhanced MRI (MEMRI), Olfactory System, Mouse
Introduction
As the power of mouse genetics and behavior push the limits of neuroscience, neuroimaging approaches which can optimally and efficiently utilize these same mouse models are critically needed to determine the strength and pattern of connectivity of different brain systems. The application and development of anterograde and retrograde tracing techniques have been instrumental in understanding structural connectivity of the brain (Callaway 2008; Deller et al., 2000; McDonald, 1992; Raju and Smith, 2006; Sparks et al., 2000). However, classical histological techniques, while a gold standard, present a number of difficulties. In particular, such tracing can take weeks to produce results, are limited by animal and surgical resources, allow for tracing from only one or at most a few areas (depending on the number of different tracers used), require sacrificing the animal, and, especially in the case of the mouse, require high accuracy injections into brain areas that are relatively minute and closely spaced (leading to mice with “missed” or diffuse injections). Subsequently, in a partial redress to these limitations, a number of MRI based techniques have been developed to allow computation of anatomic/structural connections between regions of the brain. Among these are methodologies based on diffusion-tensor imaging (DTI) that rely on the diffusion of water along the lengths of axons in the brain, as well as techniques that use exogenous contrast agents (such as manganese enhanced MRI, MEMRI) that are preferentially taken up by neurons in proximity to where the contrast agent is applied. However, these tools have yet to be sufficiently validated within the mouse brain.
DTI has been extensively used in both human (Douaud et al. 2011; Gutman et al. 2009; Wedeen et al. 2008) and primate studies (Li et al. 2010; Rilling et al. 2008; Wedeen et al. 2008) to compare and contrast the structural connectivity of brain regions. Additional studies, particularly in the porcine brain, have directly compared the results of diffusion tensor tractography to the gold standard tracer-injection studies and have generally shown a high concordance between the histologically defined pathways and those defined based on MRI tractography results (Dyrby et al. 2011; Dyrby et al. 2007). However, at this juncture the use of DTI in mice is limited, and validation of the technique is still underway. For example, Moldrich and colleagues (2010) have recently published an analysis using different DTI processing algorithms to assess general patterns of direct thalamocortical connections.
A complementary technique, manganese-enhanced MRI (MEMRI), has been the subject of increasing interest among the MRI community during the last few years because of the unique dual function of manganese (Mn2+) as a powerful T1 shortening agent and a biological calcium analogue (Silva et al. 2004). Manganese is taken up by depolarized neurons through voltage-gated calcium channels and transported by a microtubule transport system through axons to the synaptic cleft, where it is released along with neurotransmitters and taken up post-synaptically. Therefore, manganese acts as an anterograde tracer that can map an entire neuronal pathway by traveling trans-synaptically (Pautler et al. 1998). By using manganese, contrast can be observed in T1 weighted images as a result of activity dependent uptake and transport of Mn2+ (Lin and Koretsky 1997). These properties have made MEMRI a valuable tool for in vivo neuronal activity dependent tract tracing (Pautler 2004).
Pautler and colleagues were first able to detect enhancement in the olfactory tract after injecting MnCl2 into the nares of mice (Pautler et al. 1998). A similar study demonstrated enhancement of the optical tract after manganese was injected into the vitreous body of the eye of a rat (Watanabe et al. 2001). These two neural tracts have been the focus of several MEMRI studies (Chuang and Koretsky 2009; Cross et al. 2004; Lin et al. 2001; Lin et al. 2003 Olsen et al., 2008; Thuen et al. 2005; Thuen et al. 2009, Watanabe et al. 2004). Other studies have also demonstrated tract-tracing after intracerebral injection (Canals et al. 2008; Dyrby et al. 2007; Leergaard et al. 2003; Pautler et al. 2003;Saleem et al., 2002; Watanabe et al. 2004; Van der Linden et al., 2002). However, intracerebral approaches are limited in ways analogous to the classical tracing studies including manganese diffusion outside of the injection site.
While the results of previous studies are promising, most of the work is not in mice. Additionally, DTI has not been assessed in mice for complex and long tracts in the mouse brain. Furthermore, there has been little work focusing on whether MEMRI and DTI provide corroborating evidence for complex anatomical pathways in the mouse (the closest work is in rats by Lin et al 2001, 2003 and Thuen et al 2009). Since the mouse model is the standard mammalian tool for genetic manipulation, there is the potential for the combination of these methods with genetically targeted approaches as well as following behavioral and developmental manipulations. Therefore, it is imperative that these techniques be validated in a known neural pathway in the mouse brain, which will then allow future studies combining genetic manipulation with neuroimaging approaches for quantitatively and qualitatively comparing connectivity patterns and strength.
Specifically, further understanding the pathways involved in olfactory coding and connectivity in the mouse are particularly important and exciting given the enormous increase in understanding of the molecular mechanisms underlying olfactory processing in recent years (Buck & Axel 1991; Ressler et al. 1993; Ngai et al. 1993; Dulac & Axel 1995). The discovery of the odorant receptor family of genes has lead to breakthroughs in understanding the connectivity between neurons in the nose with the olfactory bulb as well as other olfactory areas (Ressler et al. 1994; Vassar et al. 1994; Mombaerts et al. 1996). A much more detailed understanding of the specific mapping of olfactory information (Sosulski et al., 2011), as well as changes that occur with olfactory learning (Jones et al., 2008), are on the horizon; however progress has been hampered by limited neuroimaging methodology in the mouse.
The aims of this study were to further validate and also directly compare the MEMRI and DTI approaches within the olfactory system of the mouse. The olfactory tract of the mouse was selected for analysis due to its ease of access through the naris (MEMRI), the more complicated geometry and known pattern of functional connections (relative to the aforementioned optic pathway and thalamocortical pathways), as well as for the burgeoning progress in the molecular organization of this system. For MEMRI, manganese was administered to the nasal epithelium and the corresponding enhancement patterns were compared to DTI tracts calculated using a probabilistic tractography algorithm in the FSL analysis suite (Oxford, www.fmrib.ox.ac.uk/fsl). In order to efficiently utilize MRI scanner time, DTI data was acquired using a high throughput ex vivo method (scanning 6 perfused mouse brains) to maximize the spatial resolution and minimize partial volume effects. The results indicate that manganese delivered via the nasal epithelium enhances a large subset of the olfactory network (of known anatomical connections) that was also detected using DTI probabilistic tractography, indicating that MEMRI and DTI have complimentary capabilities for the delineation of known complex white matter pathways. Our data demonstrate the feasibility of probabilistic tractography and MEMRI in small animal imaging, which establishes a foundation for exploring novel networks within the brain with the combined power of mouse genetics tools.
Methods
Animal preparation and imaging
All animals used in this study were treated humanely following a procedure approved by the Institutional Animal Care and Use Committee at Emory University. A total of 6 adult CFW (Carworth Farms White) Swiss Webster mice were used in the experiments for MEMRI and 10 C57BL6 (Jackson Labs) for the DTI probabilistic tractography. Two different strains of mice were used, in part, to demonstrate concordance of anatomical data across strains that have been shown to be behaviorally different in some studies (Van Boegaert et al., 2006).
Group 1 (MEMRI)
All imaging experiments were performed on a Bruker 9.4T horizontal scanner using a 35 mm volume coil (Bruker, Billerica, MA). The 6 mice in Group 1 were imaged before (baseline control) and 42 hours after Mn2+ administration (experimental group) using the following protocol. MnCl2 pellets were weighed and then dissolved into saline via a vortex mixer. No health or behavioral effects were observed after Mn2+ administration. Each mouse was anesthetized with 5% isoflurane initially and maintained on 1.5% isoflurane (30% oxygen) during the imaging procedures. The mice were restrained in a plastic cradle (Bruker, Billerica, MA) with ear and bite bars. Care was taken to align each mouse into the cradle and scanner in a consistent manner. 3D T1 weighted images were obtained using a 3D FLASH sequence with the following parameters: Flip angle = 30 degrees, field of view = 1.92 ×1.92 × 1.92 cm3, acquisition matrix size = 96 × 96 × 96, reconstruction matrix size = 128 × 128 × 128, TR = 70 ms, TE = 4 ms, NEX = 2, imaging time = 21 minutes.
After the pre-contrast image was obtained, the mice were anesthetized with 5% isoflurane, laid in a supine position, and injected with 5μl of 1M MnCl2 solution into each nostril using a 50μl Hamilton syringe inserted at a depth of ~3 mm into the nostril. The MnCl2 mixture was administered in 1–2 μl portions to ensure that it did not overflow from the nostril. After manganese injection the animals were placed back into the anesthesia chamber under 2% isoflurane for an additional 5 minutes, allowing the manganese to be absorbed (when the mice wake up immediately after injection they typically sneeze, expelling the manganese solution). The animals were then returned to their home cage and the post-contrast image was acquired approximately 42 hours after Mn2+ exposure. Following recovery, the animals appeared healthy with no obvious indication of Mn2+ toxicity.
The 42 hour delay was decided empirically based on preliminary studies by our group, as well as by data presented in Paulter et al., 1998. We imaged several mice repeatedly at time points ranging from 24 to 48 hours, and found that the 42 hour time mark provided a nice compromise between extent of enhancement (e.g. the Mn2+ traveling further over time) and signal washout (loss of Mn2+ over time). The goal of our optimization was to image the greatest extent of the olfactory pathway that we could.
Group 2 (DTI)
The mice were euthanized using pentobarbital, and brains were perfused with 4% paraformaldehyde, the brains were extracted from the skull and were rinsed in PBS and then stored at 4 C until embedding in an agarose matrix. Two sets, one of four and another of six mouse brains were embedded in a matrix of 2% agarose (Sigma) doped with an insoluble mixture of 1 mM gadolinium (III) oxide (Fisher Scientific), all contained within a plastic 25 mL tube. Doping with gadolinium (III) oxide serves to suppress the signal (shortens the T1 relaxation time) from the agarose/water solution, providing better separation of the brains from the background.
For each tube of brains, T2-weighted images were first acquired at 100 micron isotropic resolution (TE = 26ms, matrix 256 × 512, 20 averages, scan time ~ 16 hours). Diffusion-weighted images were then acquired using a 2-D spin-echo based sequence with 161 micron isotropic resolution. (TE = 26.9 ms, TR=10000 ms, matrix size of 256 × 128, 60 axial slices collected/tube, 60 gradient directions with a diffusion weighting b=2000, and 3 b=0 images). We used the Massachusetts General Hospital (MGH) 60 direction DTI gradient sequence, and obtained the BVECS from the built-in gradient tables in trackvis.org (Wedeen et al. 2008). The ability to image multiple brains in a single session is the crucial feature that makes this experiment practical. Since the readout direction of the MRI scanner can be increased with no increase in imaging time, brains can be aligned along this direction, allowing 6 brains to be acquired in the same length of time as 2 brains (60 hours in the present study).
Image Processing
MEMRI
One control image was chosen to serve as the standard space for registration. All of the other images were registered to the standard image using automated intensity-based registration, aimed at finding the optimal 9-parameter affine transformation. Normalized cross correlation was used as the objective function.
The standard image was cropped and all the images were normalized to the cropped standard image using the method described by Venot et al. (1983). Individual images were scaled to maximize the number of zero crossings in the image obtained by subtracting the scaled image from a reference image (without any enhancement). This method assumes that most of the brain is not enhanced, which is expected to be true for the methods/parameters we used. This method has been used previously by Cross et al (2004) in Mn2+ enhanced images.
Two tailed, unpaired t-tests were performed between control and contrast-enhanced images on a voxel by voxel basis. Percentage enhancement (PE) relative to the control group was calculated for each significantly enhanced voxel (p < 0.01). The enhanced voxels were clustered based upon their connectivity with the olfactory bulb, and the enhancement maps were overlaid on the average image of all the control images.
DTI
Prior to further processing, the diffusion images from each scan (with one image containing multiple brains) were manually segmented into individual files. For both the T2 weighted scan and the DTI-scans, the brains were isolated from the background using Brain Extraction Toolkit (BET) (FSL, www.fmrib.ox.ac.uk/fsl). A study-specific high-resolution mouse template was generated by nonlinearly registering each mouse brain to a single reference image, and then creating a composite image (FNIRT, www.fmrib.ox.ac.uk/fsl). A transformation matrix for each mouse DTI dataset to this mouse-standard space was subsequently generated (FLIRT, www.fmrib.ox.ac.uk/fsl) using a 12-DOF affine warp. A conservative olfactory bulb (OB) ROI was defined, with anatomical landmarks from the Allen Mouse Brain atlas (http://mouse.brain-map.org/), on this reference image and tractography results were generated for each individual mouse brain (i.e. the ROI was placed in the anterior section of the OB, and did not span the entire cross-section or depth of the olfactory bulb, e.g. the red square on Figure 4G).
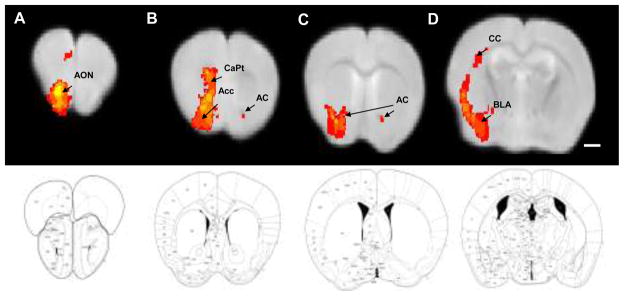
Figure 4. DTI Probabilistic Tractography with Unilateral Olfactory Bulb Seed.
A composite image showing voxels that were common to at least 7/10 mice imaged with DTI probabilistic tractography. DTI images are shown above respective diagrams of mouse coronal sections (Paxinos and Watson, 2001). Connections to the anterior olfactory nucleus and a frontal association area (A), olfactory tubercle, accumbens, and septum (B), pallidum and bilateral connectivity via the anterior commissure (C), as well as connections to the BLA and along the corpus callosum (D) were noted. AON: Anterior Olfactory Nucleus; AC: Anterior Commissure. CC: Corpus Callosum; BLA: Basolateral Amygdala; Acc: accumbens; CaPt: Caudate/Putamen; Scale Bar = 1mm.
The DTI probabilistic tractography was performed using the FSL imaging suite on a Linux workstation. As part of the standard FSL pipeline, the standard OB ROI is projected onto each individual mouse’s DTI scan, tractography is computed for each voxel within the seed mask (using n=25000 streamline fibers/voxel and curvature threshold of 0.2) and then back-transformed into the mouse-standard space. For all tractography, a multi-fiber reconstruction algorithm was implemented in the FSL bedpostx algorithm which allows reconstruction of more geometrically complex pathways, including regions of crossing fibers (Behrens et al. 2007).
Following tractography, each individual’s raw tract map was subsequently smoothed (s=0.167 mm gaussian kernel), thresholded at 1% of the robust mean intensity value, and finally binarized. A composite image was then generated by combining the results from each binarized individual tract map. The value of the composite map at a given voxel corresponds to the number of individual mice with a probabilistic tract passing through that voxel (i.e. the ratio of mice with a tract through that voxel to the total number of mice).
Registration and Comparison of in vivo and ex vivo templates
To allow direct comparison of the MEMRI and tractography results, a 12 DOF affine registration was generated between the reference brains used for the MEMRI and DTI probabilistic tractography. The results from the MEMRI experiments were subsequently registered to the reference image used for the tractography results using nearest-neighbor interpolation.
In order to quantify the overlap of the MEMRI and DTI probabilistic tractography results, a percentage overlap method was selected. The calculations were made for increasing statistical thresholds of the MEMRI analysis (p<0.05, p<0.01, p<0.005, p<0.001, p<0.0001) and increasing concordance of DTI tractography across subjects (from 6 of 10 to 10 of 10 animals showing a particular tract through a voxel).
The DTI Tractography was used as the reference image with the percentage of MEMRI voxels matching those contained in the DTI tractography. Unilateral seeding of the DTI tractography vs. a bilateral intranares manganese injection for the MEMRI allowed for both an ipsilateral and contralateral comparison.
Results
MEMRI
T1-weighted images were obtained from all mice pre- and post-manganese injection. The post-contrast images exhibited a strong increase in signal in the olfactory bulb along with enhancement throughout the olfactory tract. All images were registered to the standard image (normalized correlation coefficient > 0.75). Figure 1 shows representative slices from the average pre- and post-contrast images obtained after registration.
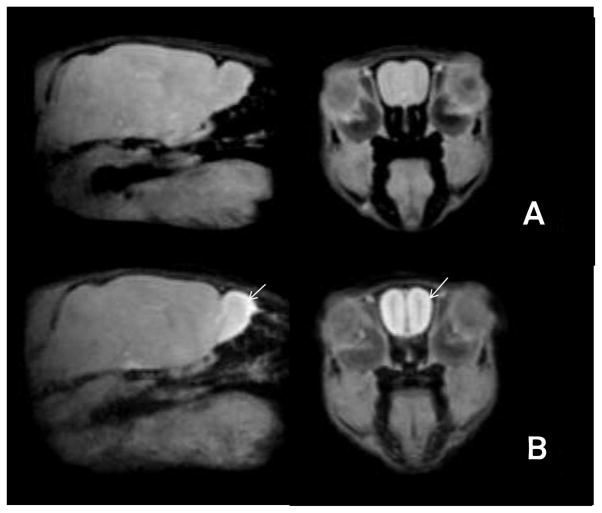
Figure 1. Manganese enhanced imaging (MEMRI) of olfactory system following Mn2+ infusion bilaterally into the nose.
Representative sagittal (left) or coronal (right) slices from average MEMRI images obtained A) pre- and B) post-contrast. Each image is the average of 6 individual images. The edges of the images are crisp and there is no blurring, which suggests accuracy of the registration process. The olfactory bulb and tract are enhanced compared to the rest of the brain in the post-contrast images (arrows).
Maps of voxels in the post-contrast images with statistically significant enhancement compared to pre-contrast images were created and are shown in Figure 2, thresholded at a p-value of 0.01. Maximum enhancement was observed in the olfactory bulb, with a gradual decrease in the intensity of enhancement in the rostral-caudal direction. Statistically significant enhancements, based on comparison to the Allen Brain Atlas, were observed in olfactory bulb (OB), anterior olfactory nucleus (AON), piriform cortex (Pir), olfactory tubercle (OT), anterior commissure (AC), nucleus accumbens (Acc) and basolateral amygdala (BLA). Enhancement was more intense in the outer layers of the OB, as compared with the inner part, comprising axons. The average percentage enhancement for these anatomical regions was also measured and is shown in Figure 3.
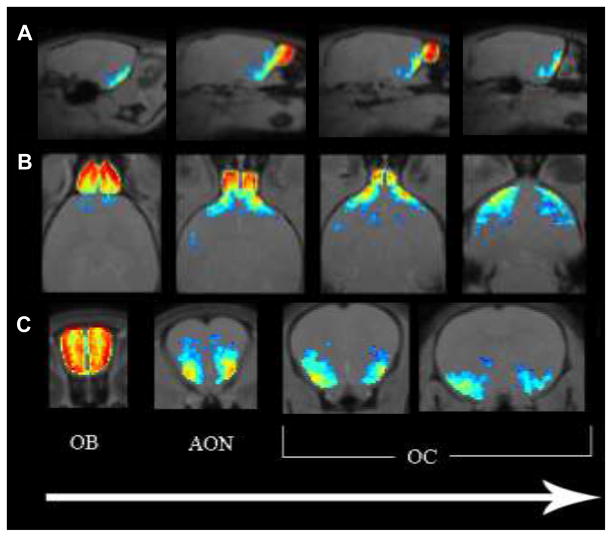
Figure 2. Imaging of Mn2+ enhanced signal throughout olfactory system.
A) sagittal, B) horizontal, and C) coronal slice maps of statistically significant enhancement in the MEMRI data, thresholded at 0.01. The olfactory bulb (OB), anterior olfactory nucleus (AON), and olfactory cortex (OC) are labeled. The direction of the arrow corresponds to anterior to posterior progression through coronal slices, dorsal to ventral progression through horizontal slices, and left to right progression through the sagittal slices.
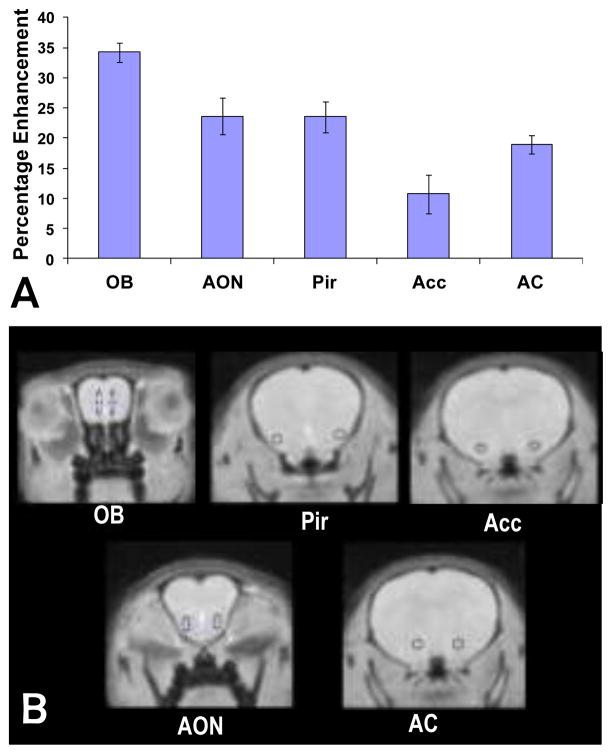
Figure 3. Quantification of Mn2+ enhanced signal across brain regions.
A) Average percent enhancement measured from manually drawn regions of interest in the olfactory bulb (OB), anterior olfactory nucleus (AON), piriform cortex (Pir), anterior commissure (AC), and nucleus accumbens (Acc), based on maps thresholded at p<0.05. Standard deviations across mice are indicated by the error bars. B) Specific regions of interest quantified above are identified on coronal brain images.
In order to estimate the sensitivity of the MEMRI analysis, the minimum percentage difference was measured in brain areas where significant enhancement was observed. For a threshold of p < 0.05, the minimal detectable percent enhancement was 4.1%; for p < 0.01, it was 5.0%.
Probabilistic Tractography
High resolution T2 and DTI scans were obtained ex vivo on all mouse brains. A composite image showing voxels that were found to have a consistent connection in at least 7 of the 10 mice included in this study is shown in Figure 4 (The placement of the seed (in red) and resulting fiber tracts (in blue) are shown below in Figure 5G). Regions shown to be consistently connected to the olfactory bulb seed included the AON, anterior commissure, nucleus accumbens, and BLA, and a projection to the entorhinal cortex.
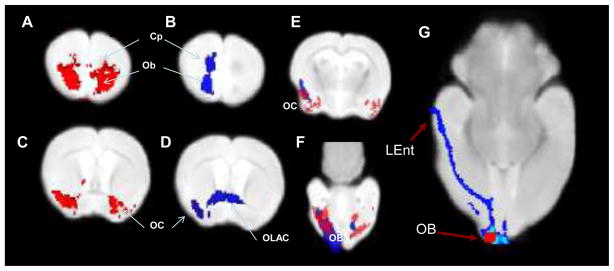
Figure 5. Comparison of results from MEMRI and probabilistic tractography.
Significant enhancement (p>0.05) in the MEMRI data set is shown on two coronal slices in red (A,C) and the DTI probabilistic tractography common to at least 7/10 mice are shown in blue scale with dark blue representing 7 mice and light blue representing 10 mice (B,D). It should be noted that tractography was seeded unilaterally, whereas the Mn solution was injected bilaterally. Panels E and F show both images superimposed on the same map, demonstrating the high degree of overlap between the two methods. An axial view that better depicts the seed region (in red) for the DTI tractography (shown in blue) shown in G. A distal connection to the entorhinal cortex can be seen as well as proximal connections to the anterior olfactory nucleus adjacent to the olfactory bulb seed.
LEnt: Lateral Entorhinal Cortex; OB: Olfactory Bulb seed; OC- Olfactory Cortex; Cp- Caudate-putaman; OLAC- Olfactory limb of the anterior commissure
Overlap of DTI and MEMRI
Figure 5 provides a direct comparison between MEMRI and DTI-based tractography. The results for the two techniques were largely consistent and included areas with known anatomical connections to the olfactory bulb such as the anterior olfactory nucleus, piriform cortex, amygdala, and entorhinal cortex. The most notable differences between the two were that the DTI-tractography also showed a connection through the olfactory limb of the anterior commissure; and the connection to the entorhinal cortex, which is quite caudal from the injection site.
As expected the percentage of MEMRI overlap with DTI varied considerably depending on the statistical threshold of MEMRI and the between subjects concordance of DTI (presented in supplementary table 1). However, use of a high statistical threshold in MEMRI (e.g. P<0.00001) and a lower DTI concordance threshold (6 of 10 mice matching), resulted in an overlap percentage of approximately 75%.
Discussion
This work has demonstrated reproducible and consistent connectivity of the mouse olfactory bulb using two techniques, DTI probabilistic tractography and MEMRI. MEMRI and DTI probabilistic tractography provide complementary information about the network organization of the mouse brain. In MEMRI, contrast is based on both structural integrity and functional synaptic transmission, with the transfer of manganese across synapses dependent upon the existence of activity of the neurons. It is an anterograde tracer, so that enhancement indicates the directionality as well as the functional integrity of the pathway. However, nonspecific enhancement can also occur, either from manganese that is taken up into the blood via the nasal epithelium (Chuang and Koretsky, 2009), or via localized diffusion near synapses. In contrast, DTI probabilistic tractography is sensitive only to the diffusion of water based on the structural properties of white matter, so that tracts can be delineated regardless of their directionality or the level of activity.
One of the primary challenges of DTI tractography is following tracts through a complex pathway. It is often difficult to resolve crossing fibers, although recent advances in crossing fiber algorithms (Behrens et al. 2007) have somewhat ameliorated this problem. In addition, issues related to partial volume effects, selection of thresholds for the tractography results, scan quality, and distance from the seed for a given region, can also significantly influence the results (Moldrich et al. 2010). Despite these limitations, the results from both MEMRI and DTI-based probabilistic tractography are largely consistent with expected neuroanatomical connectivity. Using classic orthograde and retrograde tracing methodology in mice, Shipley and Adamek (1984) described connectivity from the main olfactory bulb to ipsilateral anterior olfactory nucleus, the anterior aspects of the hippocampus, olfactory tubercle, and parts of the medial part of the entorhinal cortex. Of note, contralateral connectivity was noted only to the anterior olfactory nucleus, as well as a connection to the posterolateral and medial amygdala. More recently, Miyamachi et al.(2011) used a trans-synaptic tracing method and showed similar connectivity from the olfactory bulb to the anterior olfactory nucleus, piriform cortex, and cortical amygdala. These classic tracing methods support the data presented herein. Importantly, now that the two techniques are validated for the olfactory system they can be used to provide a quantifiable change in strength and patterns of connectivity with experimental manipulations in genetically modified mice (e.g. Jones et al. 2008).
The results in this study are very similar between MEMRI and DTI probabalistic tractography (up to 75% overlap). The most notable difference was in the caudal portions of the brains, particularly along the corpus callosum and the lateral entorhinal cortex, where DTI probabilistic tractography shows connectivity but MEMRI does not. There are a few technical reasons that may account for the observed difference. First, since the signal enhancement produced by manganese is dependent on the transport of MnCl2 injected, more caudal areas will receive proportionally less manganese resulting in a reduced signal. It is expected that either higher initial volume, higher concentration of MnCl2, or allowing for a longer time period of anaesthesia before the animal awakens and expels the solution should increase caudal enhancement signal. Second, sensitivity profile of the coil used to maximize signal for in vivo studies may also impact MEMRI data from caudal portions of the brain; therefore, increasing the number of averages may improve MEMRI’s sensitivity to distal connections. Finally, from an experimental model perspective, two different mouse species (C57BLJ vs Swiss Webster) were used in these two studies, although this unlikely accounts for any major differences since the olfactory system is a highly conserved across mammals (Scalia and Winans, 1975).
With regards to MEMRI, it is worth noting that significantly higher resolution was obtained in this study than in most previous related work in rodents (Cross et al. 2004; Leergaard et al. 2003). Cross et al. (2004) created 3D enhancement maps with a spatial resolution of 0.273 × 0.273 × 0.5 mm3 after interpolation (0.273 × 0.437 × 1 mm3 without interpolation) with an imaging time of 7 minutes (NEX = 4). Leergaard et al. (2003) used a spatial resolution of 0.390 × 0.390 × 0.390 mm3 for 3D stereotaxic registration, each scan lasting 5.5 minutes (NEX = 1). The MEMRI resolution presented here was 0.200 mm isotropic (0.144 mm isotropic after interpolation) within 20 minutes (NEX = 2). Higher spatial resolution is particularly important for applications which require registration since partial volume effects associated with low spatial resolution will result in registration errors and information loss along the edges. Such errors in registration and data loss make it not feasible to have all the images in a standard space for analysis/comparison within group and across different modalities.
One of the key developments that made this study feasible was the method for obtaining high resolution DTI data from multiple brains during a single scan. Both high spatial resolution and high spectral resolution are important for DTI studies in small animals because the high spatial resolution reduces partial volume effects that reduce the signal from small white matter tracts, and the high spectral resolution facilitates the identification of twisting or crossing white matter pathways. However, to obtain the desired resolution, scan times become quite long (e.g. 60 hours). Therefore, the protocol presented here is designed to take advantage of the fact that acquiring more data points in the readout direction is nearly free in terms of acquisition time, so that an entire tube of six brains could be scanned over a single weekend. Dyrby et al. (2011) describe a similar ex vivo setup, but due to the larger size of the porcine brain, only one brain was imaged in each scan which contrasts with our ability to acquire 6 mouse brains within one imaging session, with the potential to acquire more based on the MRI coil setup.
Conclusions
This study demonstrates high resolution 3D statistical tract mapping for the mouse olfactory system using MEMRI and DTI probabilistic tractography. Progress in mouse molecular genetics and behavior make this a particularly important model for advanced neuroimaging approaches. Additionally, advances in understanding its molecular architecture have made the olfactory system particularly amenable to applying high resolution neuroimaging techniques. The mouse olfactory system contains multiple pathways and more complicated fiber tract geometries than the optical or thalamocortical systems, where the majority of previous studies using MEMRI or DTI probabilistic tractography were performed. Furthermore, the two techniques provided accurate information in the smaller mouse brain volume compared to previous studies using mostly rat or porcine models. The two modalities map similar pathways, although probabilistic tractography may be more sensitive for detecting distal connections. The results confirm that MEMRI and probabilistic tractography provide a similar quality of complementary and accurate information about the function and structure of white matter pathways in the complex mouse olfactory sensory system. The results also lay the foundation for use of these two techniques in experimental questions manipulating the genetics and, therefore, connectivity of the mouse olfactory system with high-throughput neuroimaging techniques.
Acknowledgments
Funding for this project was provided by two Venture Grants from the Science and Technology Center (The Center for Behavioral Neuroscience of the National Science Foundation under Agreement No. IBN-9876754), NIH grant (DA019624), and the Yerkes National Primate Research Center Base Grant 2P51RR000165-51.
References
- Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–87. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18:617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals S, Beyerlein M, Keller AL, Murayama Y, Logothetis NK. Magnetic resonance imaging of cortical connectivity in vivo. Neuroimage. 2008;40:458–472. doi: 10.1016/j.neuroimage.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Chuang KH, Koretsky AP. Accounting for nonspecific enhancement in neuronal tract tracing using manganese enhanced magnetic resonance imaging. Magn Reson Imaging. 2009;27:594–600. doi: 10.1016/j.mri.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DJ, Minoshima S, Anzai Y, Flexman JA, Keogh BP, Kim Y, Maravilla KR. Statistical mapping of functional olfactory connections of the rat brain in vivo. Neuroimage. 2004;23:1326–1335. doi: 10.1016/j.neuroimage.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Deller T, Naumann T, Frotscher M. Retrograde and anterograde tracing combined with transmitter identification and electron microscopy. J Neurosci Methods. 2000 Nov 15;103(1):117–26. doi: 10.1016/S0165-0270(00)00301-0. [DOI] [PubMed] [Google Scholar]
- Douaud G, Jbabdi S, Behrens TE, Menke RA, Gass A, Monsch AU, Rao A, Whitcher B, Kindlmann G, Matthews PM, Smith S. DTI measures in crossing-fibre areas: increased diffusion anisotropy reveals early white matter alteration in MCI and mild Alzheimer’s disease. Neuroimage. 2011;55:880–890. doi: 10.1016/j.neuroimage.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Dyrby TB, Baare WF, Alexander DC, Jelsing J, Garde E, Sogaard LV. An ex vivo imaging pipeline for producing high-quality and high-resolution diffusion-weighted imaging datasets. Hum Brain Mapp. 2011;32:544–563. doi: 10.1002/hbm.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrby TB, Sogaard LV, Parker GJ, Alexander DC, Lind NM, Baare WF, Hay-Schmidt A, Eriksen N, Pakkenberg B, Paulson OB, Jelsing J. Validation of in vitro probabilistic tractography. Neuroimage. 2007;37:1267–1277. doi: 10.1016/j.neuroimage.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Gutman DA, Holtzheimer PE, Behrens TE, Johansen-Berg H, Mayberg HS. A tractography analysis of two deep brain stimulation white matter targets for depression. Biol Psychiatry. 2009;65:276–282. doi: 10.1016/j.biopsych.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SV, Choi DC, Davis M, Ressler KJ. Learning-dependent structural plasticity in the adult olfactory pathway. J Neurosci. 2008;49:13106–11. doi: 10.1523/jneurosci.4465–08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leergaard TB, Bjaalie JG, Devor A, Wald LL, Dale AM. In vivo tracing of major rat brain pathways using manganese-enhanced magnetic resonance imaging and three-dimensional digital atlasing. Neuroimage. 2003;20:1591–1600. doi: 10.1016/j.neuroimage.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Li L, Preuss TM, Rilling JK, Hopkins WD, Glasser MF, Kumar B, Nana R, Zhang X, Hu X. Chimpanzee (Pan troglodytes) precentral corticospinal system asymmetry and handedness: a diffusion magnetic resonance imaging study. PLoS One. 2010;5:e12886. doi: 10.1371/journal.pone.0012886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Tseng WYI, Cheng HC, Chen JH. Validation of Diffusion Tensor Magnetic Resonance Axonal Fiber Imaging with Registered Manganese-Enhanced Optic Tracts. Neuroimage. 2001;14:1035–1047. doi: 10.1006/nimg.2001.0882. [DOI] [PubMed] [Google Scholar]
- Lin CP, Wedeen VJ, Chen JH, Yao C, Tseng WYI. Validation of diffusion spectrum magnetic resonance imaging with manganese-enhanced rat optic tracts and ex vivo phantoms. Neuroimage. 2003;19:482–495. doi: 10.1016/S1053-8119(03)00154-X. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Koretsky AP. Manganese ion enhances T1-weighted MRI during brain activation: an approach to direct imaging of brain function. Magn Reson Med. 1997;38:378–388. doi: 10.1002/mrm.1910380305. [DOI] [PubMed] [Google Scholar]
- Ngai J, Chess A, Dowling MM, Necles N, Macagno ER, Axel R. Coding of olfactory information: topography of odorant receptor expression in the catfish olfactory epithelium. Cell. 1993;72:667–80. doi: 10.1016/0092-8674(93)90396-8. [DOI] [PubMed] [Google Scholar]
- McDonald AJ. Neuroanatomical labeling with biocytin: a review. Neuroreport. 1992 Oct;3(10):821–7. doi: 10.1097/00001756-199210000-00001. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, Callaway EM, Horowitz MA, Luo L. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldrich RX, Pannek K, Hoch R, Rubenstein JL, Kurniawan ND, Richards LJ. Comparative mouse brain tractography of diffusion magnetic resonance imaging. Neuroimage. 2010;51:1027–1036. doi: 10.1016/j.neuroimage.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–86. doi: 10.1016/S0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Olsen O, Thuen M, Martin Berry, Kovalev V, Petrou P, Goa P, Sandvig A, Haraldseth O, Brekken C. Axon tracing in the adult rat optic nerve and tract after intravitreal injection of MnDPDP using a semiautomatic segmentation technique. J Magn Reson Imaging. 2008;24:34–42. doi: 10.1002/jmri.21234. [DOI] [PubMed] [Google Scholar]
- Pautler RG. In vivo, trans-synaptic tract-tracing utilizing manganese-enhanced magnetic resonance imaging (MEMRI) NMR Biomed. 2004;17:595–601. doi: 10.1002/nbm.942. [DOI] [PubMed] [Google Scholar]
- Pautler RG, Mongeau R, Jacobs RE. In vivo trans-synaptic tract tracing from the murine striatum and amygdala utilizing manganese enhanced MRI (MEMRI) Magn Reson Med. 2003;50:33–39. doi: 10.1002/mrm.10498. [DOI] [PubMed] [Google Scholar]
- Pautler RG, Silva AC, Koretsky AP. In vivo neuronal tract tracing using manganese-enhanced magnetic resonance imaging. Magn Reson Med. 1998;40:740–748. doi: 10.1002/mrm.1910400515. [DOI] [PubMed] [Google Scholar]
- Raju DV, Smith Y. Anterograde axonal tract tracing. Curr Protoc Neurosci. 2006 Nov;Chapter 1(Unit 1.14) doi: 10.1002/0471142301.ns0114s37. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-G. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–55. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TE. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat Neurosci. 2008;11:426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Saleem KS, Pauls JM, Augath M, Trinath T, Prause BA, Hashikawa T, Logothetis NK. Magnetic resonance imaging of neuronal connections in the macaque monkey. Neuron. 2002;34:685–700. doi: 10.1016/S0896-6273(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Scalia F, Winans SS. The differential projections of the olfactory bulb and accessory olfactory bulb in mammals. J Comp Neurol. 1975;161:31–55. doi: 10.1002/cne.901610105. [DOI] [PubMed] [Google Scholar]
- Shipley MT, Adamek GD. The connections of the mouse olfactory bulb: a study using orthograde and retrograde transport of wheat germ agglutinin conjugated to horseradish peroxidase. Brain Res Bull. 1984;12:669–688. doi: 10.1016/0361-9230(84)90148-5. [DOI] [PubMed] [Google Scholar]
- Silva AC, Lee JH, Akoi I, Koretsky AP. Manganese-enhanced magnetic resonance imaging (MEMRI): Methodological and Practical Considerations. NMR Biomed. 2004;17:527–531. doi: 10.1002/nbm.945. [DOI] [PubMed] [Google Scholar]
- Sosulski DL, Bloom ML, Cutforth T, Axel R, Datta SR. Distinct representations of olfactory information in different cortical centres. Nature. 2011;472:213–6. doi: 10.1038/nature09868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks DL, Lue LF, Martin TA, Rogers J. Neural tract tracing using Di-I: a review and a new method to make fast Di-I faster in human brain. J Neurosci Methods. 2000 Nov 15;103(1):3–10. doi: 10.1016/S0165-0270(00)00291-0. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB. The chromosomal distribution of mouse odorant receptor genes. Proc Natl Acad Sci USA. 1996;93:884–8. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuen M, Olsen O, Berry M, Pedersen TB, Kristoffersen A, Haraldseth O, Sandvig A, Brekken C. Combination of Mn(2+)-enhanced and diffusion tensor MR imaging gives complementary information about injury and regeneration in the adult rat optic nerve. J Magn Reson Imaging. 2009;29:39–51. doi: 10.1002/jmri.21606. [DOI] [PubMed] [Google Scholar]
- Thuen M, Singstad TE, Pedersen TB, Haraldseth O, Berry M, Sandvig A, Brekken C. Manganese-enhanced MRI of the optic visual pathway and optic nerve injury in adult rats. J Magn Reson Imaging. 2005;22:492–500. doi: 10.1002/jmri.20400. [DOI] [PubMed] [Google Scholar]
- van Bogaert MJ, Groenink L, Oosting RS, Westphal KG, van der Gugten J, Olivier B. Mouse strain differences in autonomic responses to stress. Genes Brain Behav. 2006;5(2):139–49. doi: 10.1111/j.1601-183X.2005.00143.x. [DOI] [PubMed] [Google Scholar]
- Van der Linden A, Verhoye M, Van Meir V, Tindemans I, Eens M, Absil P, Balthazart J. In vivo manganese-enhanced magnetic resonance imaging reveals connections and functional properties of the songbird vocal control system. Neuroscience. 2002;112:467–474. doi: 10.1016/S0306-4522(02)00070-2. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nuñez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–91. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Venot A, Lebruchec JF, Golmard JL, Roucayrol JC. An Automated Method for the Normalization of Scintigraphic Images. The Journal of Nuclear Medicine. 1983;24:529–531. [PubMed] [Google Scholar]
- Watanabe T, Frahm J, Michaelis T. Functional mapping of neural pathways in rodent brain in vivo using manganese-enhanced three-dimensional magnetic resonance imaging. NMR in Biomedicine. 2004;17:554–568. doi: 10.1002/nbm.937. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Michaelis T, Frahm J. Mapping of retinal projections in the living rat using high-resolution 3D gradient-echo MRI with Mn2+-induced contrast. Magn Reson Med. 2001;46:424–429. doi: 10.1002/mrm.1209. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, Pandya DN, Hagmann P, D’Arceuil H, de Crespigny AJ. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41:1267–1277. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]