Abstract
Peroxisomes are essential organelles responsible for many metabolic reactions, such as the oxidation of very long chain and branched fatty acids, D-amino acids and polyamines, as well as the production and turnover of hydrogen peroxide. They comprise a class of organelles called microbodies, including glycosomes, glyoxysomes and Woronin bodies. Dysfunction of human peroxisomes causes severe and often fatal peroxisome biogenesis disorders (PBDs). Peroxisomal matrix protein import is mediated by receptors that shuttle between the cytosol and peroxisomal matrix using ubiquitination/deubiquitination reactions and ATP hydrolysis for receptor recycling. We focus on the machinery involved in the peroxisomal matrix protein import cycle, highlighting recent advances in peroxisomal matrix protein import, cargo release and receptor recycling/degradation.
Introduction
As ubiquitous single membrane-enclosed organelles of eukaryotic cells, peroxisomes play significant roles in the metabolism of fatty acids, cholesterol, D-amino acids, polyamines and reactive oxygen species. Peroxisomes are a class of structurally and functionally related organelles called microbodies, comprising glyoxysomes of plants and fungi, glycosomes of parasites and Woronin bodies from filamentous fungi [1-4]. Dysfunction of human peroxisomes causes PBDs [5-7]. At least 34 proteins named peroxins (encoded by PEX genes) control peroxisome assembly, division and inheritance.
Peroxisomal matrix protein import pathways
Peroxisomal matrix proteins generally contain one (or rarely both) of two peroxisomal targeting signals (PTS): a C-terminal PTS1 and/or an N-terminal PTS2. Following their synthesis in the cytosol, the PTSs in cargoes are recognized by receptors, Pex5 or Pex7 (and/or co-receptors Pex5L, Pex18/Pex21 or Pex20), that bind and translocate cargoes into the matrix through interactions of the receptor-cargo complexes with a docking subcomplex (Pex13, Pex14 and Pex17), followed by receptor-cargo dissociation. The cargo-free receptors move to the peroxisomal membrane sites, where they are modified by ubiquitination by an E2 ubiquitin-conjugation enzyme (Pex4 or other E2 enzymes) and the peroxisome membrane-associated, RING subcomplex E3 ligases (Pex2, Pex10 and Pex12) [8••,9]. Mono-ubiquitinated receptors/co-receptors are recycled to the cytosol by the AAA (ATPase associated with various cellular activities) peroxins (Pex1 and Pex6) [10] and/or associated proteins [11•] for another round of import (Figure 1). Alternatively, when the receptor recycling is impaired, the receptors/co-receptors are poly-ubiquitinated and degraded by the proteasome via the receptor accumulation and degradation in the absence of recycling (RADAR) pathway [12,13]. Pex8 (in yeast) and Pex3 connect the docking and RING subcomplexes [14,15] (Table 1).
Figure 1.
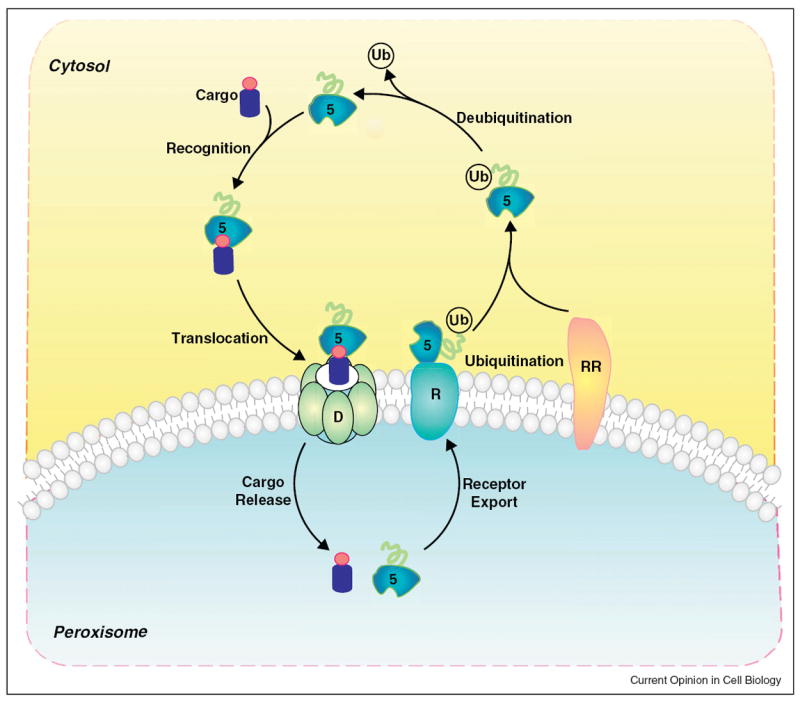
PTS receptor dynamics during peroxisomal matrix protein import. The shuttling of PTS receptors and co-receptors between the cytosol and the peroxisomal matrix can be divided into distinct steps: cargo-receptor recognition, receptor-cargo complex translocation across peroxisomal membrane, dissociation of receptor-cargo complex, receptor export to the cytosolic side of peroxisomal membrane, receptor ubiquitination and release to the cytosol, and deubiquitination for next round import. D: docking subcomplex; R: RING subcomplex; RR: receptor recycling machinery. The circle associated with the cargo denotes the PTS.
Table 1.
Proteins involved in the import of peroxisomal matrix proteins.
| Proteins | Functions and properties | Localization |
|---|---|---|
| Receptors | ||
| Pex5 | PTS1 receptor, contains C-terminal TPR domains | Cytosol/peroxisome |
| Pex7 | PTS2 receptor, contains WD domains | Cytosol/peroxisome |
| Pex20/Pex18/Pex21/Pex5L | PTS2 co-receptor, contains Pex7 binding box | Cytosol/peroxisome |
| Docking subcomplex | ||
| Pex14 | Component of the peroxisomal translocon | Integral PMP |
| Pex13 | Contains SH3 motif | Integral PMP |
| Pex17 | Associates with Pex14, no homolog in higher eukaryotes | Integral PMP |
| Pex33 | Contains an N-terminal domain homology to Pex14 | Integral PMP |
| Pex14/17 | Contains N-/C-terminal domain homology to Pex14 and Pex17 | Integral PMP |
| RING subcomplex | ||
| Pex2 | E3 ligase, receptor ubiquitination | Integral PMP |
| Pex10 | E3 ligase, receptor ubiquitination | Integral PMP |
| Pex12 | E3 ligase, receptor ubiquitination | Integral PMP |
| Receptor recycling machineries | ||
| Pex4/Ubc5H | Ubiquitin-conjugating enzyme, E2, for receptor mono-ubiquitination | Peripheral PMP |
| Pex22 | Peroxisomal anchor for Pex4 | Integral PMP |
| Pex1 | AAA ATPase for receptor recycling | Cytosol/PM |
| Pex6 | AAA ATPase for receptor recycling | Cytosol/PM |
| Pex15/Pex26 | Peroxisomal anchor for Pex6 | Integral PMP |
| Ubc4/5 | E2 for receptor poly-ubiquitination | Cytosol |
| Ubp15/USP9X | Deubiquitinase | Cytosol/peroxisome |
| AWP1 | Binds to Pex6 and mono-ubiquitinated Pex5 | Cytosol/peroxisome |
| Linker of the docking and RING subcomplexes | ||
| Pex8 | Contains a PTS1 and a PTS2, no homolog in higher eukaryotes | Peroxisome lumen |
| Pex3 | PMP receptor | Integral PMP |
PM: peroxisomal membrane; PMP: peroxisomal membrane protein.
In most organisms, the PTS1 import pathway functions independently of PTS2 pathway components, but in Arabidopsis thaliana, Pex7 is also required for PTS1 import because it enhances the stability of Pex5 [16]; and in Trypanosomes brucei, pex7 RNA interference lines reduced Pex5 levels and mislocalized both PTS1 and PTS2 cargoes [17]. To perform its role in PTS2 import, Pex7 needs auxiliary proteins (termed co-receptors), such as Pex5L (long isoform of Pex5) in plants and mammals [18,19], Pex18/Pex21 in Saccharomyces cerevisiae [20,21] or Pex20 in other fungi [12,22-24]. All peroxisomal matrix protein import can be mediated only by Pex5 in Phaeodactylum tricornutum [25] and Caenorhabditis elegans [26], or only by Pex20 in Podospora anserina for specific peroxisomal matrix protein import during meiocyte differentiation [27].
Pex8, found only in yeasts and fungi, has both PTS1 and PTS2, and is required for both PTS1 and PTS2 cargo import. Its import differs from that of normal cargo in that it only requires PTS receptors and Pex14, but not the RING subcomplex proteins or the receptor recycling machinery [28••].
Cargo translocation across the peroxisomal membrane
Pex14, a constituent of the docking subcomplex, is a central component of the minimal translocon for cargo import in Pichia pastoris [28••,29] and S. cerevisiae [30••]. Studies on Leishmania donovani Pex14 (LdPex14) support observations with yeast and mammalian Pex14, that it is a homo-oligomer [31]. LdPex14 undergoes a major conformation change involving reorganization of this complex upon Pex5 binding. A hypothetical model for how Pex5 binding might allow the insertion of a hydrophobic core of Pex14 into the membrane to form a translocation pore remains to be tested [31]. It is still uncertain whether Pex5 or Pex14 oligomers constitute the translocon since both have been proposed to form transient pores and perhaps they may act together [30••,31,32]. It is also unclear if there is a distinct translocon for PTS2 cargo import. This is an important question because if the receptors form, or contribute to, the translocon [30••,32], then there must be two distinct translocons for PTS1 and PTS2 proteins; whereas if Pex14 forms the translocon [31], a single mechanism could operate for matrix protein import. We do not understand how the peroxisomal translocon maintains the impermeability of the peroxisome membrane while importing large receptor-cargo complexes of up to 9 nm in diameter through a protein-conducting channel.
In Arabidopsis, cargo import takes place inefficiently in pex14 alleles lacking detectable Pex14 mRNA and proteins [33], while in the Hansenula polymorpha pex14 deletion strain, Pex5 overexpression restores import of some, but nor all, PTS1 cargo import [34]. It remains to be seen whether some other protein/s can substitute for Pex14 function as a key translocon component in these situations, albeit at a lower efficiency.
There is precedence for Pex14-related proteins, but these do not appear to substitute for Pex14. In Neurospora crassa, a novel Pex14 related protein Pex33, possessing an N-terminal Pex14 homology domain, functions uniquely and non-redundantly with respect to Pex14, as it is essential for the biogenesis of glyoxysomes and Woronin bodies, and it resembles yeast Pex17 despite clear structural differences [35]. In Penicillium chrysogenum, a novel Pex14/17 peroxin, possessing N-terminal and C-terminal domains homologous to those of Pex14 and Pex17, respectively, is required, but not obligatorily, for efficient PTS1 and PTS2 import, but is necessary for conidiospore formation, as well as penicillin production [36]. In Podospora anserina, Pex14/17 is also differentially required for peroxisomal matrix protein import during meiosis and sporulation [27].
The field needs to reconcile what proteins constitute the peroxisomal translocon in these organisms, whether the PTS1 and PTS2 pathways use the same translocon components, and in the event there is more than one translocon, to what extent the blockage of one causes a low level of transport via alternative routes. We must also understand the roles the individual docking subcomplex components play in facilitating and regulating cargo translocation.
Cargo release
The mechanism of cargo release is still a matter of considerable debate with multiple models being proposed. The interactions between the N-terminal region of Pex5 with the docking subcomplex and/or Pex8 could cause a conformational change in the cargo-binding domain of Pex5 and trigger cargo release [37]. The in vitro binding of H. polymorpha Pex5 (HpPex5) to Pex8 resulted in the transition of Pex5 tetramers to monomers, causing a conformational change that may induce some cargo release [38]. The oligomeric state of HpPex5 switches from a cargo-bound tetramer at pH 7.2 (same pH as the cytosol) to a cargo-free monomer at acidic pH 6.0 (pH of the peroxisome lumen). However, the magnitude of Pex8-induced cargo release appears to be insufficient for this to be the sole mechanism of cargo release and its relevance in vivo is unknown. Moreover, our studies do not demonstrate a pronounced effect of pH on cargo release and the measurements of intraperoxisomal pH do not provide any consistent result. In higher eukaryotic cells, Pex5 interacts with monomeric catalase and releases it upon binding the N-terminal domain of Pex14, implicating a role for Pex14 in cargo release [39]. P. pastoris Pex13 interacts more strongly with cargo-free Pex5, subsequent to, or coincident with, cargo release [40]. Pex1 and Pex6 function as drivers of cargo import by coupling ATP-dependent removal of the receptor and cargo translocation into the peroxisomes [41], but they act downstream of cargo translocation into peroxisomes and release in the lumen. How PTS2 cargo is released needs to be addressed in future studies.
PTS receptor recycling and the RADAR pathway
The recycling and degradation of PTS receptors/co-receptors (Pex5, Pex18 and Pex20) depends on mono-ubiquitination and poly-ubiquitination pathways, respectively (Figure 2). Pex7 translocation follows an ‘extended shuttle’ mechanism [42], as does Pex5; however, no evidence exists to suggest that recycling and degradation of Pex7 are dependent on ubiquitination.
Figure 2.
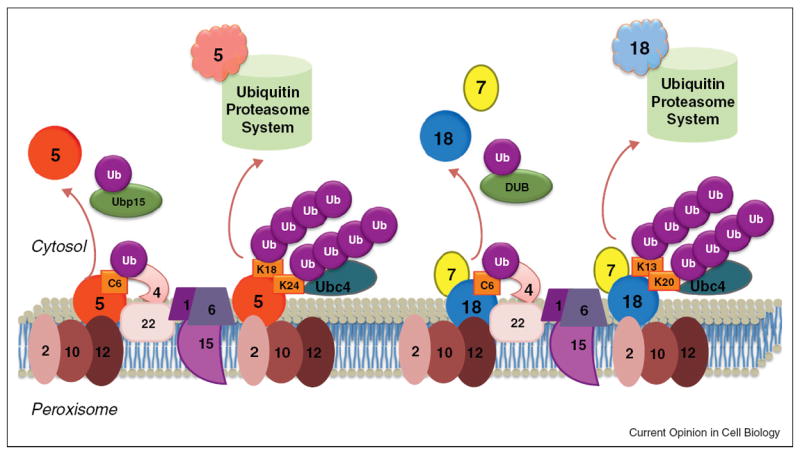
Ubiquitination/deubiquitination of PTS1 receptor (Pex5) and PTS2 co-receptor (Pex18) during receptor recycling and degradation in S. cerevisiae. Three ubiquitin ligases Pex2, Pex10 and Pex12 form the RING subcomplex and together with ubiquitin-conjugating enzymes, Pex4/Ubc4, are responsible for receptor ubiquitination. Pex10 brings the E2 ubiquitin-conjugating enzyme, Pex4, which is anchored to the peroxisomal membrane via Pex22, into association with the RING subcomplex. Pex5 is mono-ubiquitinated on the conserved N-terminal cysteine C6 by Pex4 and Pex12. The AAA peroxins, Pex1 and Pex6, which are anchored at the peroxisomal membrane by binding to Pex15, recognize mono-ubiquitinated Pex5 directly or indirectly and dislocate it from the membrane back to the cytosol in an ATP-dependent manner. The mono-ubiquitinated Pex5 is deubiquitinated by Ubp15, which binds to Pex6, and is recycled for the next round of import. When the receptor recycling is impaired, Pex5 is poly-ubiquitinated on the conserved N-terminal lysines, K18 and K24, by Ubc4 (E2) and Pex2 (E3 ligase) and then degraded by the proteasome via the RADAR pathway. Pex18 is mono-ubiquitinated on the conserved N-terminal cysteine C6 (for receptor recycling), or is poly-ubiquitinated on the conserved N-terminal lysines, K13 and K20, for degradation by the RADAR pathway.
An N-terminal conserved cysteine of Pex5 (C11 in mammals and C6 in S. cerevisiae) is required for its mono-ubiquitination and recycling [43,44]. An N-terminal conserved cysteine of S. cerevisiae Pex18 (C6) is required for its mono-ubiquitination and recycling, as well as for Pex7 import [20]. The N-terminal conserved cysteine of P. pastoris Pex20 (C8) is essential for its recycling [45]; however, whether it is required for mono-ubiquitination has not yet been elucidated. Conserved lysines near the N-termini of Pex5 (K18/24 in S. cerevisiae and K21 in H. polymorpha), Pex18 (K13/20 in S. cerevisiae) and Pex20 (K19 in P. pastoris) are required for their poly-ubiquitination and degradation [12,13,20,44].
Pex4/Pex22 in yeast and plants [46], and UbcH5 in mammals [47] function as the E2 enzymes in mono-ubiquitination of Pex5, while Ubc4 in S. cerevisiae [8••] functions as the E2 enzyme for Pex5 poly-ubiquitination. The roles of the RING subcomplex in Pex5 ubiquitination were determined in S. cerevisiae. Pex12 is required for mono-ubiquitination of Pex5 [8••], while Pex10 [9] or Pex2 [8••] is required for poly-ubiquitination of Pex5.
The release of ubiquitinated receptors from the peroxisomal membrane to the cytosol requires the cooperation of two AAA peroxins, Pex1 and Pex6. Mammalian Pex1 is targeted to peroxisomes dependent on ATP hydrolysis, while Pex6 targeting requires ATP binding. Pex1 and Pex6 are regulated in their peroxisomal localization onto Pex26 (or Pex15 in yeast) via conformational changes during the ATPase cycle [10]. No direct interaction between PTS receptors and Pex6 or Pex1 has been characterized. This raises a question regarding how the ubiquitinated receptors are recognized prior to recycling. This puzzle has been partially solved recently. A novel cofactor of Pex6, AWP1, interacts with Pex6, but not with the Pex1-Pex6 complex. It binds to the cysteine-ubiquitinated form of Pex5 via its A20 zinc-finger domain to catalyze the recycling of Pex5 [11•].
More research is necessary to answer how the E2 and E3 enzymes regulate the choice between the two types of ubiquitination of PTS1 and PTS2 receptors.
Receptor deubiquitination
After receptor export to the cytosol, the ubiquitin must be removed before the next round of import cycle by de-ubiquitinating enzymes (DUBs) – ubiquitin hydrolases or non-enzymatic mechanisms [48]. The ubiquitin hydrolase, Ubp15 from S. cerevisiae (or USP9X from mammalian cells), which binds to the first AAA domain of Pex6, is capable of removing ubiquitin from mono-/poly-ubiquitinated Pex5 [49•,50•]. Although deletion of ubp15 or knock down of USP9X activated the RADAR pathway (as demonstrated by the low level of Pex5), cargo import was not significantly affected, indicating the existence of redundant DUBs acting on Pex5. Which DUB(s) is/are responsible for deubiquitination of PTS2 co-receptors has not been discovered.
Conclusions
The fact that biogenesis of peroxisomal matrix proteins is substantially different from the biogenesis of proteins of other subcellular compartments, such as mitochondria and chloroplasts, makes this an interesting biological problem for investigation. Despite the recent progress described above in defining the steps in the receptor dynamics during the peroxisomal matrix protein import cycle including cargo-recognition, cargo-receptor translocation, cargo release, receptor recycling via ubiquitination and receptor deubiquitination, there are still many unanswered questions ripe for investigation. In addition to the ones mentioned above, further studies must include the molecular and structural characterization of the peroxisomal translocon/s and the role of specific peroxins in many regulatory steps during the import cycle. Defining these molecular events in peroxisome assembly will help us understand the etiology of human PBDs.
Acknowledgments
This work was supported by a National Institutes of Health (NIH) Method to Extend Research in Time (MERIT) award to SS (DK41737).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
-
•
of special interest
-
••
of outstanding interest
- 1.Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011;193:7–16. doi: 10.1083/jcb.201010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rucktäschel R, Girzalsky W, Erdmann R. Protein import machineries of peroxisomes. Biochim Biophys Acta. 2011;1808:892–900. doi: 10.1016/j.bbamem.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Titorenko VI, Terlecky SR. Peroxisome metabolism and cellular aging. Traffic. 2011;12:252–259. doi: 10.1111/j.1600-0854.2010.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goh J, Jeon J, Kim KS, Park J, Park SY, Lee YH. The PEX7-mediated peroxisomal import system is required for fungal development and pathogenicity in Magnaporthe oryzae. PLoS ONE. 2011;6:e28220. doi: 10.1371/journal.pone.0028220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimozawa N. Molecular and clinical findings and diagnostic flowchart of peroxisomal diseases. Brain Dev. 2011;33:770–776. doi: 10.1016/j.braindev.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Liang JS, Lu JF. Peroxisomal disorders with infantile seizures. Brain Dev. 2011;33:777–782. doi: 10.1016/j.braindev.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Fidaleo M. Peroxisomes and peroxisomal disorders: the main facts. Exp Toxicol Pathol. 2010;62:615–625. doi: 10.1016/j.etp.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 8••.Platta HW, El Magraoui F, Bäumer BE, Schlee D, Girzalsky W, Erdmann R. Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009;29:5505–5516. doi: 10.1128/MCB.00388-09. This paper shows that the three components of the RING subcomplex display distinct roles in Pex5 ubiquitination. Pex12 is required for mono-ubiquitination of Pex5, while Pex2, with the assistance of Pex10, is required for poly-ubiquitination of Pex5. Ref. [9] shows that Pex10 is the E3 ligase responsible for the poly-ubiquitination of Pex5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams C, van den Berg M, Geers E, Distel B. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun. 2008;374:620–624. doi: 10.1016/j.bbrc.2008.07.054. [DOI] [PubMed] [Google Scholar]
- 10.Fujiki Y, Nashiro C, Miyata N, Tamura S, Okumoto K. New insights into dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p in shuttling of PTS1-receptor Pex5p during peroxisome biogenesis. Biochim Biophys Acta. 2012;1823:145–149. doi: 10.1016/j.bbamcr.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 11•.Miyata N, Okumoto K, Mukai S, Noguchi M, Fujiki Y. AWP1/ZFAND6 functions in Pex5 export by interacting with cys-monoubiquitinated Pex5 and Pex6 AAA ATPase. Traffic. 2012;13:168–183. doi: 10.1111/j.1600-0854.2011.01298.x. The release of Pex5 from the peroxisomal membrane to the cytosol requires an ubiquitin binding protein, AWP1, which interacts with Pex6. Therefore, it serves as a linker between the PTS1 receptor and receptor recycling machinery. This study solves the puzzle of how mono-ubiquitinated Pex5 is exported to the cytosol by AAA ATPases, Pex1 and Pex6, without a direct interaction. [DOI] [PubMed] [Google Scholar]
- 12.Léon S, Zhang L, McDonald WH, Yates J, Cregg JM, Subramani S. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J Cell Biol. 2006;172:67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol. 2007;177:197–204. doi: 10.1083/jcb.200611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agne B, Meindl NM, Niederhoff K, Einwächter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH. Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell. 2003;11:635–646. doi: 10.1016/s1097-2765(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 15.Hazra PP, Suriapranata I, Snyder WB, Subramani S. Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 2002;3:560–574. doi: 10.1034/j.1600-0854.2002.30806.x. [DOI] [PubMed] [Google Scholar]
- 16.Ramón NM, Bartel B. Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell. 2010;21:1263–1271. doi: 10.1091/mbc.E09-08-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galland N, Demeure F, Hannaert V, Verplaetse E, Vertommen D, Van der Smissen P, Courtoy PJ, Michels PA. Characterization of the role of the receptors PEX5 and PEX7 in the import of proteins into glycosomes of Trypanosoma brucei. Biochim Biophys Acta. 2007;1773:521–535. doi: 10.1016/j.bbamcr.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Honsho M, Hashiguchi Y, Ghaedi K, Fujiki Y. Interaction defect of the medium isoform of PTS1-receptor Pex5p with PTS2-receptor Pex7p abrogates the PTS2 protein import into peroxisomes in mammals. J Biochem. 2011;149:203–210. doi: 10.1093/jb/mvq130. [DOI] [PubMed] [Google Scholar]
- 19.Khan BR, Zolman BK. pex5 mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol. 2010;154:1602–1615. doi: 10.1104/pp.110.162479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hensel A, Beck S, El Magraoui F, Platta HW, Girzalsky W, Erdmann R. Cysteine-dependent ubiquitination of Pex18p is linked to cargo translocation across the peroxisomal membrane. J Biol Chem. 2011 doi: 10.1074/jbc.M111.286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein K, Schell-Steven A, Erdmann R, Rottensteiner H. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol Cell Biol. 2002;22:6056–6069. doi: 10.1128/MCB.22.17.6056-6069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otzen M, Wang DY, Lunenborg MGJ, van der Kiel IJ. Hansenula polymorpha Pex20p is an oligomer that binds the peroxisomal targeting signal 2 (PTS2) J Cell Sci. 2005;118:3409–3418. doi: 10.1242/jcs.02463. [DOI] [PubMed] [Google Scholar]
- 23.Sichting M, Schell-Steven A, Prokisch H, Erdmann R, Rottensteiner H. Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol Biol Cell. 2003;14:810–821. doi: 10.1091/mbc.E02-08-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Einwächter H, Sowinski S, Kunau WH, Schliebs W. Yarrowia lipolytica Pex20p, Saccharomyces cerevisiae Pex18p/Pex21p and mammalian Pex5pL fulfil a common function in the early steps of the peroxisomal PTS2 import pathway. EMBO Rep. 2001;2:1035–1039. doi: 10.1093/embo-reports/kve228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez NH, Felsner G, Schramm FD, Klingl A, Maier UG, Bolte K. A single peroxisomal targeting signal mediates matrix protein import in diatoms. PLoS ONE. 2011;6:e25316. doi: 10.1371/journal.pone.0025316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motley AM, Hettema EH, Ketting R, Plasterk R, Tabak HF. Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 2000;1:40–46. doi: 10.1093/embo-reports/kvd010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peraza-Reyes L, Arnaise S, Zickler D, Coppin E, Debuchy R, Berteaux-Lecellier V. The importomer peroxins are differentially required for peroxisome assembly and meiotic development in Podospora anserina: insights into a new peroxisome import pathway. Mol Microbiol. 2011;82:365–377. doi: 10.1111/j.1365-2958.2011.07816.x. [DOI] [PubMed] [Google Scholar]
- 28••.Ma C, Schumann U, Rayapuram N, Subramani S. The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol Biol Cell. 2009;20:3680–3689. doi: 10.1091/mbc.E09-01-0037. Defines peroxin/s essential for the import of Pex8, a special cargo containing a PTS1 as well as a PTS2. The authors demonstrated that the minimal peroxisomal translocon is composed of the PTS receptors and Pex14, but not the entire importomer. Although not essential, other components of the importomer are required for efficiently targeting Pex8 to peroxisomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Léon S, Subramani S. Two independent pathways traffic the intraperoxisomal peroxin PpPex8p into peroxisomes: mechanism and evolutionary implications. Mol Biol Cell. 2006;17:690–699. doi: 10.1091/mbc.E05-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Meinecke M, Cizmowski C, Schliebs W, Krüger V, Beck S, Wagner R, Erdmann R. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010;12:273–277. doi: 10.1038/ncb2027. As demonstrated by a planar lipid bilayer assay, the authors show that Pex14 of the docking subcomplex and the membrane-bound PTS1 receptor, Pex5, form a transient peroxisomal translocon upon induction by the Pex5/cargo complex. This inducible protein channel is 9 nm in diameter when induced by a 750 kDa complex, Pex5 with its cargo, Fox1. [DOI] [PubMed] [Google Scholar]
- 31.Cyr N, Madrid KP, Strasser R, Aurousseau M, Finn R, Ausio J, Jardim A. Leishmania donovani peroxin 14 undergoes a marked conformational change following association with peroxin 5. J Biol Chem. 2008;283:31488–31499. doi: 10.1074/jbc.M803529200. [DOI] [PubMed] [Google Scholar]
- 32.Erdmann R, Schliebs W. Peroxisomal matrix protein import: the transient pore model. Nat Rev Mol Cell Biol. 2005;6:738–742. doi: 10.1038/nrm1710. [DOI] [PubMed] [Google Scholar]
- 33.Monroe-Augustus M, Ramón NM, Ratzel SE, Lingard MJ, Christensen SE, Murali C, Bartel B. Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol Biol. 2011;77:1–15. doi: 10.1007/s11103-011-9782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salomons FA, Kiel J, Faber KN, Veenhuis M, van der Klei IJ. Overproduction of Pex5p stimulates import of alcohol oxidase and dihydroxyacetone synthase in a Hansenula polymorpha pex14 null mutant. J Biol Chem. 2000;275:12603–12611. doi: 10.1074/jbc.275.17.12603. [DOI] [PubMed] [Google Scholar]
- 35.Managadze D, Würtz C, Wiese S, Schneider M, Girzalsky W, Meyer HE, Erdmann R, Warscheid B, Rottensteiner H. Identification of PEX33, a novel component of the peroxisomal docking complex in the filamentous fungus Neurospora crassa. Eur J Cell Biol. 2010;89:955–964. doi: 10.1016/j.ejcb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Opaliński L, Kiel JA, Homan TG, Veenhuis M, van der Klei IJ. Penicillium chrysogenum Pex14/17p – a novel component of the peroxisomal membrane that is important for penicillin production. FEBS J. 2010;277:3203–3218. doi: 10.1111/j.1742-4658.2010.07726.x. [DOI] [PubMed] [Google Scholar]
- 37.Stanley WA, Wilmanns M. Dynamic architecture of the peroxisomal import receptor Pex5p. Biochim Biophys Acta. 2006;1763:1592–1598. doi: 10.1016/j.bbamcr.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 38.Wang D, Visser NV, Veenhuis M, van der Klei IJ. Physical interactions of the peroxisomal targeting signal 1 receptor Pex5p, studied by fluorescence correlation spectroscopy. J Biol Chem. 2003;278:43340–43345. doi: 10.1074/jbc.M307789200. [DOI] [PubMed] [Google Scholar]
- 39.Freitas MO, Francisco T, Rodrigues TA, Alencastre IS, Pinto MP, Grou CP, Carvalho AF, Fransen M, Sá-Miranda C, Azevedo JE. PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J Biol Chem. 2011;286:40509–40519. doi: 10.1074/jbc.M111.287201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urquhart AJ, Kennedy D, Gould SJ, Crane DI. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J Biol Chem. 2000;275:4127–4136. doi: 10.1074/jbc.275.6.4127. [DOI] [PubMed] [Google Scholar]
- 41.Grimm I, Saffian D, Platta HW, Erdmann R. The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in. Biochim Biophys Acta. 2012;1823:150–158. doi: 10.1016/j.bbamcr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Nair DM, Purdue PE, Lazarow PB. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol. 2004;167:599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okumoto K, Misono S, Miyata N, Matsumoto Y, Mukai S, Fujiki Y. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic. 2011;12:1067–1083. doi: 10.1111/j.1600-0854.2011.01217.x. [DOI] [PubMed] [Google Scholar]
- 44.Williams C, van den Berg M, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem. 2007;282:22534–22543. doi: 10.1074/jbc.M702038200. [DOI] [PubMed] [Google Scholar]
- 45.Léon S, Subramani S. A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol. J Biol Chem. 2007;282:7424–7430. doi: 10.1074/jbc.M611627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams C, van den Berg M, Panjikar S, Stanley WA, Distel B, Wilmanns M. Insights into ubiquitin-conjugating enzyme/co-activator interactions from the structure of the Pex4p:Pex22p complex. EMBO J. 2011;31:391–402. doi: 10.1038/emboj.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grou CP, Carvalho AF, Pinto MP, Wiese S, Piechura H, Meyer HE, Warscheid B, Sá-Miranda C, Azevedo JE. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J Biol Chem. 2008;283:14190–14197. doi: 10.1074/jbc.M800402200. [DOI] [PubMed] [Google Scholar]
- 48.Grou CP, Carvalho AF, Pinto MP, Huybrechts SJ, Sá-Miranda C, Fransen M, Azevedo JE. Properties of the ubiquitin-Pex5p thiol ester conjugate. J Biol Chem. 2009;284:10504–10513. doi: 10.1074/jbc.M808978200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Debelyy MO, Platta HW, Saffian D, Hensel A, Thoms S, Meyer HE, Warscheid B, Girzalsky W, Erdmann R. Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J Biol Chem. 2011;286:28223–28234. doi: 10.1074/jbc.M111.238600. This paper shows that S. cerevisiae Ubp15, an ubiquitin hydrolase partially associated with the peroxisomal membrane, is capable of removing ubiquitin moieties from mono-, as well as poly-ubiquitinated, Pex5. The authors also show that Ubp15 is a subunit of the Pex6 complex. However, a PTS1 import defect is only observed in the Ubp15 deletion strain under oxidative stress condition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Grou CP, Francisco T, Rodrigues TA, Freitas MO, Pinto MP, Carvalho AF, Domingues P, Wood SA, Rodríguez-Borges JE, Sá-Miranda C, et al. Identification of ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting on the ubiquitin-peroxin 5 (PEX5) thioester conjugate. J Biol Chem. 2012 doi: 10.1074/jbc.M112.340158. This paper shows that USP9X of mammals, a close homolog of ScUbp15, is the most active cytosolic ubiquitin hydrolase acting on ubiquitinated mammalian Pex5. [DOI] [PMC free article] [PubMed] [Google Scholar]


