Abstract
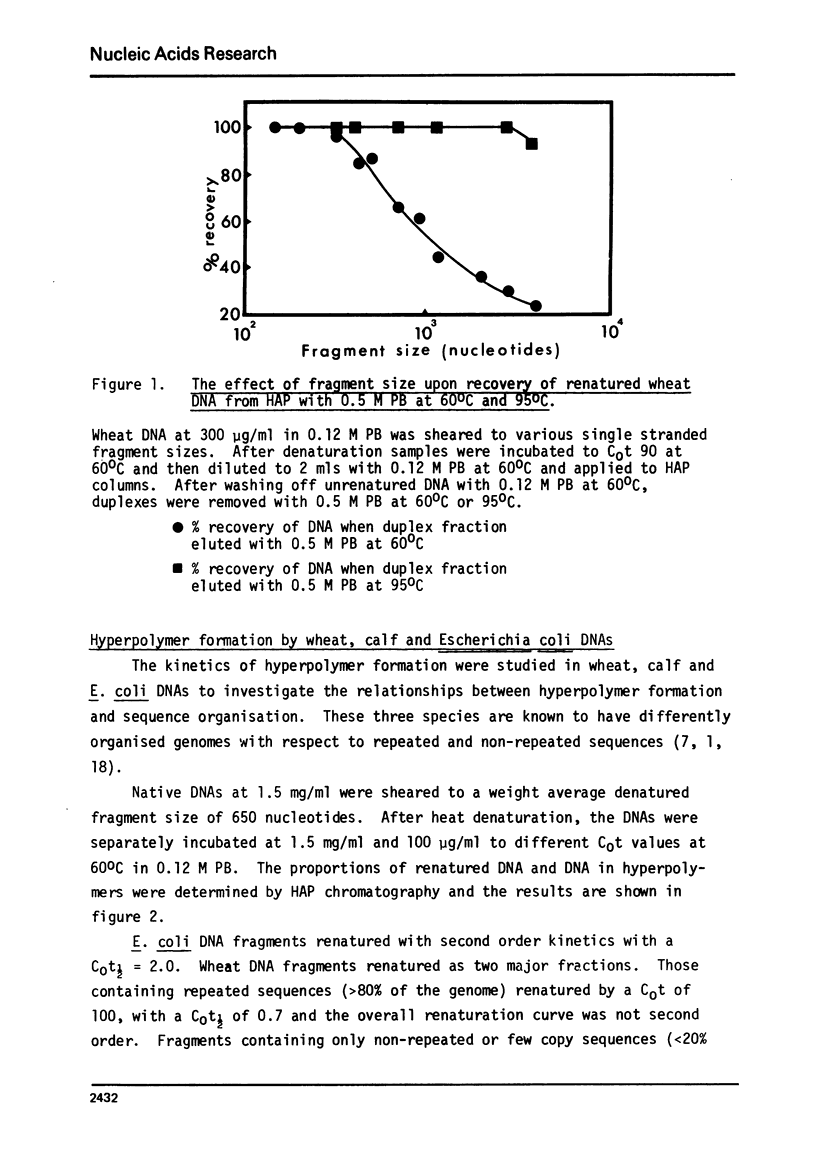
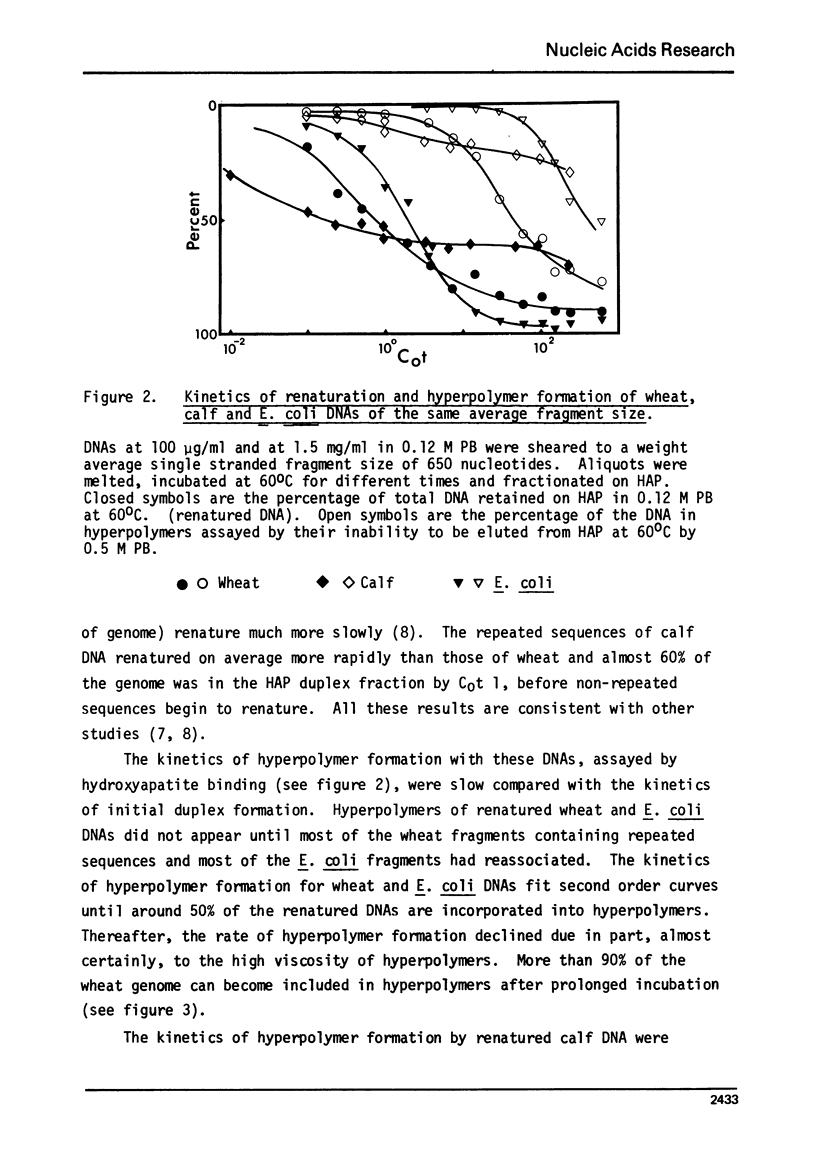
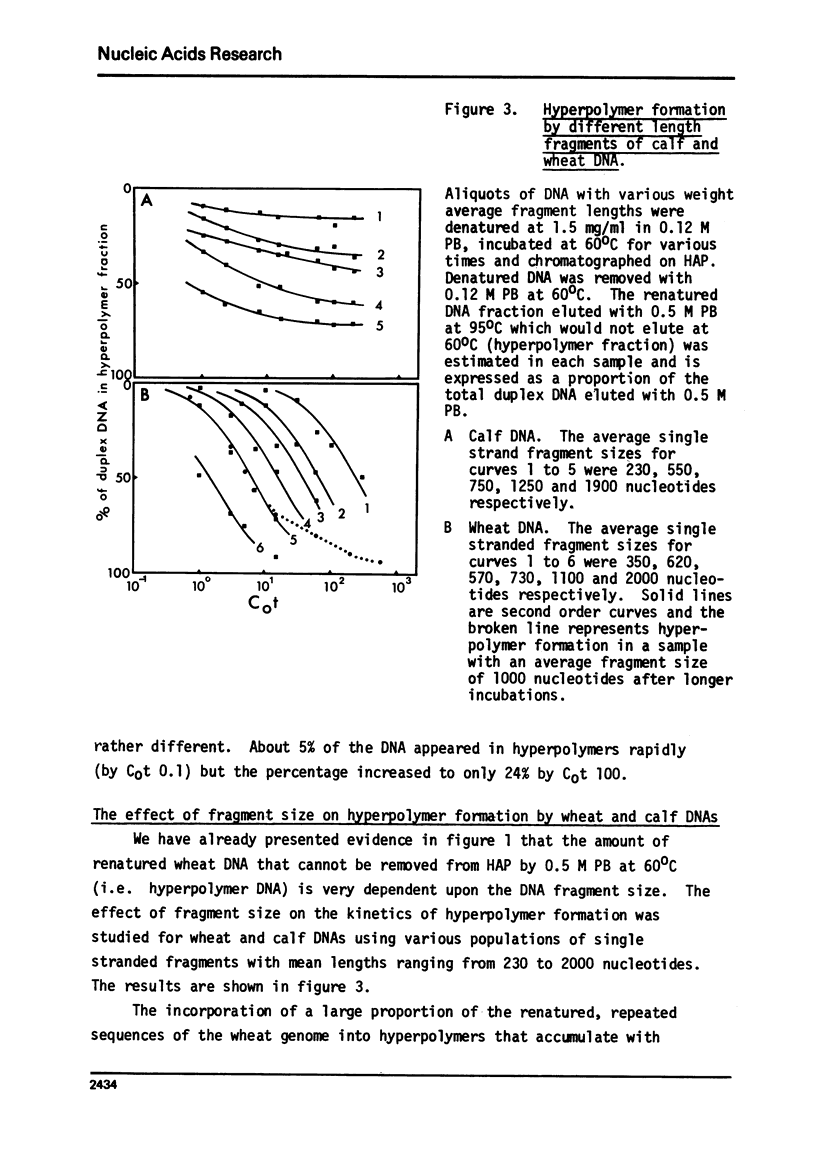
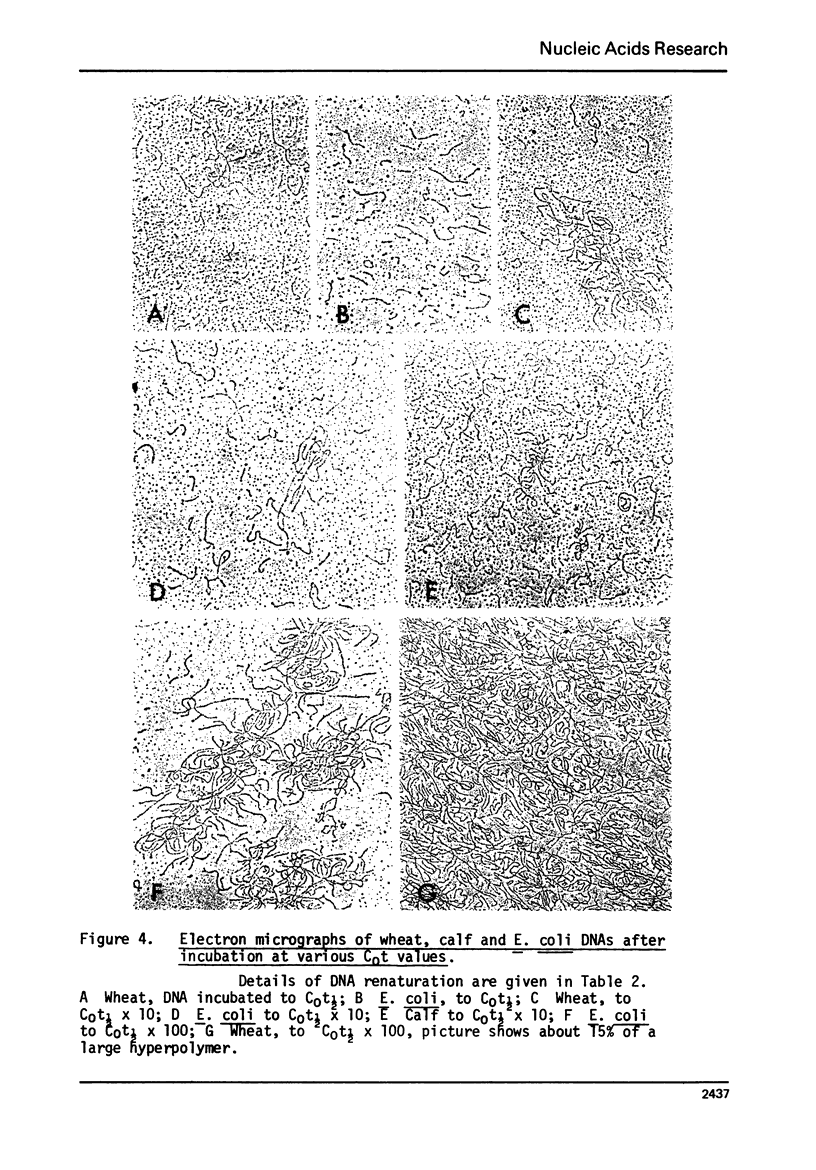
Hyperpolymer formation during the renaturation of DNAs from wheat, calf and E. coli was studied using hydroxyapatite chromatography, electron microscopy and S1 nuclease. Large hyperpolymers could not be eluted from hydroxyapatite with 0.5 M phosphate buffer at 60 degrees C. Large proportions of wheat and E. coli DNAs were incorporated into hyperpolymers when fragments 650 nucleotides long were renatured. A much smaller proportion of calf DNA was incorporated under equivalent conditions. Greater proportions of calf DNA accumulated in hyperpolymers only when longer fragments were incubated. Electron microscopy indicated no obvious differences in the basic structures of hyperpolymers formed by the three DNAs and confirmed the quantitative differences in hyperpolymer formation found by hydroxyapatite chromatography. It is concluded that the proportions and arrangement of the repeated sequences in the chromosomes of higher organisms determine the extent of rapid hyperpolymer formation during DNA renaturation in vitro.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernardi G. Chromatography of nucleic acids on hydroxyapatite. I. Chromatography of native DNA. Biochim Biophys Acta. 1969 Feb 18;174(2):423–434. doi: 10.1016/0005-2787(69)90273-1. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Galau G. A., Angerer R. C., Britten R. J. Comparative aspects of DNA organization in Metazoa. Chromosoma. 1975 Jul 21;51(3):253–259. doi: 10.1007/BF00284818. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Kuprijanova N. S., Timofeeva M. J. Repeated nucleotide sequences in the loach genome. Eur J Biochem. 1974 May 2;44(1):59–65. doi: 10.1111/j.1432-1033.1974.tb03457.x. [DOI] [PubMed] [Google Scholar]
- Marx K. A., Allen J. R., Hearst J. E. Characterization of the repetitious human DNA families. Biochim Biophys Acta. 1976 Mar 4;425(2):129–147. doi: 10.1016/0005-2787(76)90019-8. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Streeck R. E., Zachau H. G. Investigation of the repetitive sequences in calf DNA by cleavage with restriction nucleases. Eur J Biochem. 1975 Sep 1;57(1):55–68. doi: 10.1111/j.1432-1033.1975.tb02276.x. [DOI] [PubMed] [Google Scholar]
- Rinehart F. P., Wilson V. L., Schmid C. W. Fractionation of the sequence organization of human DNA by preparative HgCl2/Cs2SO4 density gradients. Biochim Biophys Acta. 1977 Jan 3;474(1):69–81. doi: 10.1016/0005-2787(77)90215-5. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Saunders G. F., Shirakawa S., Saunders P. P., Arrighi F. E., Hsu T. C. Populations of repeated DNA sequences in the human genome. J Mol Biol. 1972 Feb 14;63(3):323–334. doi: 10.1016/0022-2836(72)90430-5. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Flavell R. B. Nucleotide sequence organisation in the rye genome. Biochim Biophys Acta. 1977 Jan 3;474(1):82–97. doi: 10.1016/0005-2787(77)90216-7. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Flavell R. B. The relatedness and evolution of repeated nucleotide sequences in the genomes of some Gramineae species. Biochem Genet. 1974 Sep;12(3):243–256. doi: 10.1007/BF00486093. [DOI] [PubMed] [Google Scholar]
- Smith M. J., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: reactivity of single-stranded tails in DNA-DNA renaturation. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4805–4809. doi: 10.1073/pnas.72.12.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton W. D. A crude nuclease preparation suitable for use in DNA reassociation experiments. Biochim Biophys Acta. 1971 Jul 29;240(4):522–531. doi: 10.1016/0005-2787(71)90709-x. [DOI] [PubMed] [Google Scholar]
- Thompson W. F. Aggregate formation from short fragments of plant DNA. Plant Physiol. 1976 Apr;57(4):617–622. doi: 10.1104/pp.57.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wobus U. Molecular characterization of an insect genome: Chironomus thummi. Eur J Biochem. 1975 Nov 1;59(1):287–293. doi: 10.1111/j.1432-1033.1975.tb02454.x. [DOI] [PubMed] [Google Scholar]



