The 2,2,7-trimethylguanosine caps of eukaryal snRNAs and snoRNA are formed by the enzyme Tgs1, which catalyzes sequential guanine-N2 methylations of m7G caps. Atypically, in the divergent unicellular eukaryote Trichomonas vaginalis, spliceosomal snRNAs lack a guanosine cap and the recombinant T. vaginalis trimethylguanosine synthase (TvTgs) produces only m2,7G in vitro. Here, the authors show by direct metabolic labeling that endogenous T. vaginalis RNAs contain m7G, m2,7G, and m2,2,7G caps.
Keywords: Trichomonas vaginalis, snoRNA, capping, trimethylguanosine synthase, pseudouridine
Abstract
The 2,2,7-trimethylguanosine caps of eukaryal snRNAs and snoRNA are formed by the enzyme Tgs1, which catalyzes sequential guanine-N2 methylations of m7G caps. Atypically, in the divergent unicellular eukaryote Trichomonas vaginalis, spliceosomal snRNAs lack a guanosine cap and the recombinant T. vaginalis trimethylguanosine synthase (TvTgs) produces only m2,7G in vitro. Here, we show by direct metabolic labeling that endogenous T. vaginalis RNAs contain m7G, m2,7G, and m2,2,7G caps. Immunodepletion of TvTgs from cell extracts and TvTgs add-back experiments demonstrate that TvTgs produces m2,7G and m2,2,7G caps. Expression of TvTgs in yeast tgs1Δ cells leads to the formation of m2,7G and m2,2,7G caps and complementation of the lethality of a tgs1Δ mud2Δ strain. Whereas TvTgs is present in the nucleus and cytosol of T. vaginalis cells, TMG-containing RNAs are localized primarily in the nucleolus. Molecular cloning of anti-TMG affinity-purified T. vaginalis RNAs identified 16 box H/ACA snoRNAs, which are implicated in guiding RNA pseudouridylation. The ensemble of new T. vaginalis H/ACA snoRNAs allowed us to predict and partially validate an extensive map of pseudouridines in T. vaginalis rRNA.
INTRODUCTION
Eukaryal RNAs synthesized by RNA polymerase II (RNA Pol II) are modified cotranscriptionally by the addition of a 7-methylguanosine (m7G) 5′ cap. The m7G cap is a signature feature of eukaryal mRNA that protects against exonucleolytic decay and promotes translation initiation. The m7G caps of many noncoding RNA pol II transcripts, such as small nuclear (sn) RNAs and small nucleolar (sno) RNAs, are converted post-transcriptionally to 2,2,7-trimethylguanosine (TMG) caps by the enzyme Tgs1 (Mouaikel et al. 2002; Hausmann and Shuman 2005b). Notwithstanding the essentiality of the spliceosomal U1, U2, U4, and U5 snRNAs that receive TMG caps, the TMG modification is apparently dispensable for viability of yeast, insofar as a tgs1Δ mutant of fission yeast grows normally (Hausmann et al. 2007). The tgs1Δ mutation of budding yeast causes a growth defect at cold temperatures, though tgs1Δ cells grow as well as TGS1 cells at 34°–37°C (Mouaikel et al. 2002; Hausmann et al. 2008). The tgs1Δ mutants of budding and fission yeast lack any detectable TMG caps on their U1, U2, U4, and U5 snRNAs (Mouaikel et al. 2002; Hausmann et al. 2007), signifying that there is no Tgs1-independent route to generate TMG caps.
Atypically, but consistent with the apparent inessentiality of TMG caps in fungi, we recently observed that the spliceosomal snRNAs of the divergent unicellular eukaryote Trichomonas vaginalis lack a guanosine cap structure, though these snRNAs are transcribed by RNA Pol II, and T. vaginalis encodes both the enzymatic machinery for m7G capping and a homolog (called TvTgs) of yeast and human Tgs1 (Simoes-Barbosa et al. 2008a,b, 2010). This scenario raises interesting questions about the existence and location of hypermethylated caps in Trichomonas as well as the function of TvTgs1.
In yeast and mammals, biogenesis of TMG cap differs spatially (Cougot et al. 2004). In yeast, Tgs1 resides in the nucleolus, a compartment through which snRNAs and snoRNAs traffic (Mouaikel et al. 2002). Because Tgs1 does not contain known RNA-binding domains, it may rely on protein–protein interactions to target TMG caps to specific cellular transcripts. For instance, yeast Tgs1 interacts with SmB, a core protein component of the U1, U2, U4, and U5 snRNPs, and with Nop58 and Cbf5, which are protein components of box C/D and H/ACA snoRNPs (Mouaikel et al. 2002).
In metazoans, Tgs1 is present in the nucleus and cytoplasm (Cougot et al. 2004). Evidence favors that m7G-capped snRNAs are transported to the cytoplasm, where they are assembled with Sm proteins and the m7G caps are hypermethylated. The TMG cap-binding protein snurportin 1 then promotes transport of the mature RNP particle back to the nucleus (Huber et al. 2002). In contrast, with some exceptions, snoRNAs are retained in the nucleus (Narayanan et al. 2003; Cougot et al. 2004). They are assembled and their caps are hypermethylated in the Cajal bodies, multiple nuclear subcompartments found in animals and plants that orchestrate the assembly and/or modification of the nuclear-transcription and RNA-processing machinery. After RNP assembly and cap hypermethylation, small Cajal body-specific RNAs (a special type of snoRNAs with localization signal) remain in this compartment, whereas other TMG-capped RNAs traffic to the nucleolus (Darzacq et al. 2002; Narayanan et al. 2003; Cougot et al. 2004). The TMG caps of metazoan snRNAs and snoRNAs might promote their nuclear retention and/or return them to the nucleus following mitosis (Cougot et al. 2004).
Despite the differences in the cell biology of TMG biogenesis in different eukarya, the Tgs1 enzyme is conserved. The tertiary structure, active site, substrate specificity, structure-activity relations, and catalytic mechanism of Tgs1 have been elucidated via biochemical, mutational, and crystallographic studies of Tgs1 enzymes from Schizosaccharomyces pombe (Hausmann and Shuman 2005b; Hausmann et al. 2007), Giardia lamblia (Hausmann and Shuman 2005a; Hausmann et al. 2007), Trichomonas vaginalis (Simoes-Barbosa et al. 2008a), Trypanosoma brucei (Simoes-Barbosa et al. 2008a), Entamoeba histolytica (Simoes-Barbosa et al. 2008a), mimivirus (Benarroch et al. 2009) and humans (Hausmann et al. 2008; Monecke et al. 2009; Benarroch et al. 2010; Chang et al. 2010). It is notable that the enzymes from G. lamblia, T. vaginalis, and mimivirus are limited in vitro to only a single round of methyl transfer to a m7G cap substrate forming a 2,7-dimethylguanosine (DMG) cap product. These studies raised the prospect that Giardia, Trichomonas, and mimivirus might elaborate DMG-capped RNAs (Hausmann and Shuman 2005a; Simoes-Barbosa et al. 2008a; Benarroch et al. 2009).
The finding that Trichomomas spliceosomal snRNAs lack a guanosine cap (Simoes-Barbosa et al. 2008b) and the identification of TvTgs as a DMG synthase in vitro (Simoes-Barbosa et al. 2008a) prompted us to determine whether endogenous Trichomonas RNAs have guanine-N2 methylated caps and, if so, whether they are di- or trimethylated and whether TvTgs is responsible for their formation. Here we report the presence of m7G, DMG, and TMG caps on T. vaginalis endogenous RNAs. Assays of cap methylation using T. vaginalis extracts, and the effects of TvTgs immunodepletion, indicate that TvTgs is capable of converting either an m7G-capped RNA or a DMG-capped RNA to a TMG-capped product. Additionally, we find that TvTgs can genetically complement the synthetic lethal phenotype of a yeast tgs1Δ mud2Δ strain. We present evidence that box H/ACA snoRNAs are the endogenous natural substrates for TvTgs, and we show that TvTgs is present in both nucleus and cytoplasm.
RESULTS AND DISCUSSION
The methylation status of hypermethylated cap RNAs in Trichomonas vaginalis
The observation that Tgs enzymes from Trichomonas, Giardia, and mimivirus (Hausmann and Shuman 2005a; Simoes-Barbosa et al. 2008a; Benarroch et al. 2009) produce only DMG in vitro suggests the possibility that DMG cap structures are present in protozoal or viral RNAs and are potentially biologically relevant. We examined this hypothesis by characterizing the cap structures of endogenous RNAs. T. vaginalis and Saccharomyces cerevisiae RNAs were labeled in vivo by addition of 32P-orthophosphate to the growth medium after a period of growth in phosphate-free medium. The labeled RNAs were immunoprecipitated with an anti-TMG antibody that recognizes TMG caps, but also reacts with m7G caps (Simoes-Barbosa et al. 2008b). Therefore, our assumption was that the anti-TMG antibody would cross-react with DMG caps as well. The immunoprecipitated RNAs were digested with tobacco acid pyrophosphatase (TAP), which releases the guanosine cap nucleotide from the 5′-end of the RNA. The radiolabeled methylated guanylates released by TAP were resolved by two-dimensional thin-layer chromatography (2D-TLC) with specific standards (Simoes-Barbosa et al. 2008a).
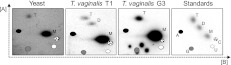
We found that TAP-digestion of anti-TMG precipitated 23P-labeled T. vaginalis RNAs released a mixture of m7GMP, m2,7GMP, and m2,2,7GMP (Fig. 1). It was noteworthy that m2,7GMP was released only from RNAs isolated from the T1 strain of T. vaginalis, which harbors a double-stranded RNA virus (Fig. 1; Supplemental Fig. S1). Digestion of labeled RNAs isolated from the G3 strain of T. vaginalis, which is free of dsRNA virus (Carlton et al. 2007), released m7GMP and m2,2,7GMP, but not m2,7GMP. Total RNA from T. vaginalis G3 contains a low percentage of m2,2,7G (17.8% ± 3.9%), which is comparable to yeast (15.5% ± 5.1%).
FIGURE 1.
The methylation status of guanosine cap nucleotides in Saccharomyces cerevisiae (yeast) and two strains of Trichomonas vaginalis (T1 and G3). In vivo-labeled RNAs were immunoprecipitated by anti-TMG. The guanylate cap was released by TAP and analyzed by two-dimensional thin-layer chromatography (2D-TLC) with solvents [A] and [B], as described in the Materials and Methods. The migration of the four unmodified rNMPs (A, C, G, and U), as detected by UV shading, is plotted on every image for reference (see Materials and Methods). The far right panel (standards, only) denotes the three methylation status of the guanosine cap (M: m7GMP; D: m2,7GMP, and T: m2,2,7GMP) relative to the migration of A, C, G, and U as described in the Materials and Methods.
This finding of DMG caps in the parasite strain T1 harboring the dsRNA virus is in accordance with previous observations that some eukaryal RNA virus transcripts have DMG caps (HsuChen and Dubin 1976; van Duijn et al. 1986). In addition, the same enzymatic limitation of mimivirus Tgs in vitro leads to a speculative scenario in which mimivirus would synthesize DMG-capped viral mRNAs to compete for the host translation machinery. In fact, DMG caps have been shown to be more effective than standard m7G caps in translation systems (Darzynkiewicz et al. 1988; Grudzien et al. 2004) Our data are consistent with the T. vaginalis virus containing a DMG cap; however, the inability to isolate viral particles has precluded definitive analyses.
Trichomonas vaginalis Tgs is the enzyme that produces TMG
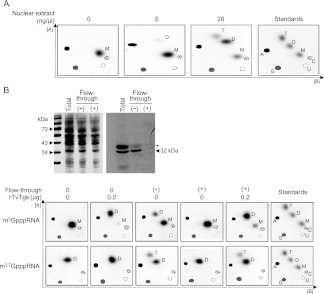
The previous observation that TvTgs produces only DMG in vitro (Simoes-Barbosa et al. 2008a) is at odds with the presence of TMG caps on Trichomonas RNAs shown here. Because there is no additional Tgs paralog in T. vaginalis, it is either the case that TvTgs is able to perform a second guanine-N2 methylation step in vivo (e.g., aided by an activating cofactor) or there is an unrecognizable methyltransferase enzyme that accomplishes this step. To address this, we queried the ability of a T. vaginalis nuclear extract to methylate a m7G-capped T7 RNA polymerase transcript that had been cap-labeled in vitro with [32P]GTP and AdoMet by the vaccinia virus capping enzyme (Simoes-Barbosa et al. 2010). After incubation with the extract and AdoMet, the cap-labeled RNA was recovered and the cap methylation status was analyzed by TAP digestion and 2D-TLC (Fig. 2A).
FIGURE 2.
Trimethylguanosine synthase activity from cell extracts of Trichomonas vaginalis. (A) T. vaginalis nuclear extracts alone can drive formation of DMG (D) and TMG (T) from a m7G (M) capped RNA substrate. The concentration of nuclear extract is indicated at the top of each panel. For resolution of nucleotides, 2D-TLC was used here as described in Figure 1. The migration of the four unmodified ribonucleotides (A, C, G, and U), as detected by UV shading, is plotted on every image for reference (see Materials and Methods). The far right panel for the standards, as in Figure 1, is included here. (B) Immunodepletion of endogenous TvTgs and add-back of recombinant TvTgs demonstrate that TvTgs is the m2,2,7G synthase in T. vaginalis. (Top) SDS-PAGE (left) and Western blot (right) analyses of TvTgs immunodepletion. Molecular weight of protein standards is illustrated on the left side. The total represents an aliquot of nuclear extract prior incubation with anti-TvTgs. After incubation in the absence (−) or presence (+) of anti-TvTgs, the extract is passed through a protein-A column and the flow-through is collected for detection of the remaining endogenous TvTgs. Western blot indicates that virtually all endogenous TvTgs are trapped with the antibody (+) onto the column, as no signal for the 32-kDa TvTgs is left on the flow-through as compared with when the incubation is done in the absence of the antibody (−). A cross-reactive ∼36-kDa species is indicated by an asterisk. (Bottom) Remaining TvTgs activity from immunodepleted nuclear extracts. The flow-through from (−) and (+) immunodepletion were used for Tgs activity in a protein concentration of 100 ng/μL. This was either combined with 0 μg or 0.2 μg of recombinant TvTgs, as denoted at the top of the panels. As indicated left of the figure, m7G- and m2,7GpppRNA substrates were used here and substrate purity is shown when in the absence (0) of rTvTgs and extract (first column). As previously shown (Simoes-Barbosa et al. 2008a), rTvTgs alone can produce DMG (D) from m7G (M), but it is unable to produce TMG (T) from DMG (second column). When there is still endogenous TvTgs in the extract, conversion to DMG and to TMG is observed (third column). Once endogenous TvTgs is depleted, the extract no longer has Tgs activity (fourth column). However, when rTvTgs is added back to the TvTgs-depleted extract, conversion to DMG and TMG is restored (fifth column). For resolution of nucleotides, 2D-TLC was used here as described in Figure 1. As in A, the internal standards (A, C, G, and U) were plotted in each panel, and the far right panel is for standards only.
We found that T. vaginalis nuclear extract converted the input m7G cap to DMG and TMG products (Fig. 2A). DMG caps comprised ∼1% of the 5′ ends when the reactions contained 5 ng/μL of extract and there was no apparent TMG cap. Increasing the extract concentration to 20 ng/μL resulted in an increase in the extent of guanine-N2 methylation and the appearance of TMG caps. Using extract levels that consume all of the input m7G cap substrate (50 ng/μL, data not shown), DMG remained the predominant product. This pattern suggests that the endogenous guanine-N2 methyltransferase acts distributively. Nuclear and cytoplasmic extracts of T. vaginalis displayed similar activity in this assay (Fig. 2A; data not shown).
To examine whether TvTgs was responsible for conversion of m7G to DMG and DMG to TMG, we performed immunodepletion and rTvTgs add-back experiments (Fig. 2B). Rabbit polyclonal antibody was raised against recombinant TvTgs and affinity purified. This antibody was able to detect a polypeptide corresponding to the endogenous 32-kDa TvTgs and a cross-reactive ∼36-kDa species on Western blots of nuclear extracts (Fig. 2B). Passage of the extract through a column of protein-A-anti-TvTgs resin depleted TvTgs from the flow-through fraction (Fig. 2B, +). A control protein-A column without the antibody did not deplete TvTgs (Fig. 2B, –).
We tested the flow-through fractions for guanine-N2 methyltransferase activity with cap-labeled m7GpppRNA and m2,7GpppRNA substrates (Fig. 2B, bottom). The mock-depleted extract (–) converted the input m7G cap to DMG cap and the input DMG cap to a TMG product. In contrast, the immunodepleted extract (+) was unreactive with either substrate. Supplementation of the immunodepleted extract with 0.2 μg of purified recombinant TvTgs restored both methylation reactions: conversion of the m7G to DMG and DMG to TMG (Fig. 2B, bottom). The same amount of recombinant enzyme in the absence of extract efficiently synthesized DMG caps, but could not convert DMG to TMG (Fig. 2B, bottom). This experiment indicates that TvTgs is the enzyme responsible for sequential conversion of a m7GpppRNA to m2,7GpppRNA and m2,2,7GpppRNA and that the ability of TvTgs to execute the second methylation step is dependent on the cellular environment or a specific component of the T. vaginalis extract. It is likely that a cofactor is required for TvTgs full activity.
TvTgs complements the growth of a yeast tgs1Δ mud2Δ strain
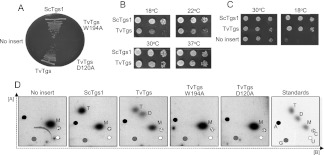
Tgs1 is required for viability of yeast when the gene encoding for the pre-mRNA splicing factor Mud2 is also deleted (Hausmann et al. 2008). Here we tested, by plasmid shuffle, whether expression of TvTgs in yeast could complement the synthetic lethality of tgs1Δ mudΔ (Hausmann et al. 2008; Benarroch et al. 2009). The TvTGS gene was delivered on a 2μ LEU2 plasmid under the transcriptional control of the yeast TPI1 promoter. The empty vector and a plasmid bearing the S. cerevisiae TGS1 gene (ScTGS1) provided negative and positive controls for complementation, respectively. The salient finding was that TvTGS supported growth on FOA-containing medium at 30°C, whereas the vector did not (Fig. 3A). Mutations D120A (in the AdoMet binding site of TvTgs) and W194A (in the m7G binding site) abolished TvTgs complementation activity in vivo (Fig. 3A), We compared growth of the tgs1Δ mud2Δ ScTGS1 and tgs1Δ mud2Δ TvTGS strains on YPD agar at 18°, 22°, 30°, and 37°C (Fig. 3B). The notable finding was that TvTGS cells grew slowly at 18° C and 22°C. Cold-sensitive complementation of tgs1Δ mud2Δ was noted previously for mimivirus Tgs, which, like TvTgs, is exclusively a DMG synthase in vitro (Benarroch et al. 2009). It is not simply the case that TvTgs activity in yeast was intrinsically cold sensitive insofar as the 2μ LEU2 TvTGS plasmid rectified the cold-sensitive growth phenotype of the singly deleted yeast tgs1Δ mutant (Fig. 3C; Mouaikel et al. 2002; Hausmann et al. 2008).
FIGURE 3.
TvTgs is a functional ortholog of S. cerevisiae Tgs1 (ScTgs1). (A) Yeast tgs1Δ mud2Δ cells were transformed with a LEU2 plasmid containing no insert (negative control), ScTgs1 (positive control), TvTgs wild-type, and mutants W194A and D120A. Leu+ transformants were selected at 30°C, streaked to agar medium containing FOA, and photographed after 3 d of incubation at 30°C. (B) Yeast tgs1Δ mud2Δ cells transformed with ScTgs1 and TvTgs were tested for growth on rich medium by spotting serial 10-fold dilutions of liquid cultures (grown in SD-Leu medium) on YPD agar plates, which were then incubated at the temperatures indicated. (C) Yeast tgs1Δ cells were transformed with a LEU2 plasmid containing no insert (negative control), ScTgs1 (positive control), TvTgs. Leu+ transformants were selected at 30°C and tested for growth at 30°C and 18°C by spotting serial 10-fold dilutions of liquid cultures (grown at 30°C in SD-Leu medium) on Leu− agar plates. The plates were photographed after incubation for 3 d at 30°C or 7 d at 18°C. (D) Yeast tgs1Δ cells transformed with LEU2 plasmid containing no insert (negative control), ScTgs1 (positive control), TvTgs wild-type and mutants W194A and D120A were in vivo labeled with phosphorus-32 radionuclide. Total RNA was extracted and immunoprecipitated with anti-TMG. The bound RNA was digested with tobacco acid pyrophosphatase. The released guanosine cap nucleotide in its three possible methylated status (M: m7G, D: DMG, and T: TMG) was resolved by 2D-TLC as in Figure 1. The migration of the four unmodified ribonucleotides (A, C, G, and U), as detected by UV shading, is plotted on every image for reference (see Materials and Methods). The far right panel for the standards only, as in Figure 1, is included here.
m2,2,7G cap nucleotide is produced in yeast tgs1Δ mud2Δ cells transformed with TvTgs
Previous studies of yeast complementation by mimivirus Tgs raised questions about possible differences in vivo between Tgs enzymes that catalyze one versus two cap guanine-N2 methylation steps. It was suggested that yeast cells are uniquely reliant on a TMG synthase for growth at 18°C when the Mud2 splicing factor is absent. Otherwise, a DMG synthase appeared to suffice (Benarroch et al. 2009). The missing link in this supposition was a direct analysis of the cap methylation status of yeast cells complemented with a heterologous cap guanine-N2 methyltransferase.
Here, we performed such an analysis on tgs1Δ cells transformed with TvTGS (wild-type, D120A, or W196A alleles), ScTGS1, or an empty vector control. Yeast cells were labeled with 32P-orthophosphate, RNAs were selected with anti-TMG antibody, and cap status was gauged by TAP digestion and 2D-TLC. Control experiments verified that ScTGS1 cells had TMG and m7G caps (but no DMG caps) and that tgs1Δ cells had no detectable TMG caps (Fig. 3D). The salient finding was that TvTGS cells elaborated a mixture of DMG and TMG caps (Fig. 3D), the production of which was abolished by the active site mutations D120A and W194A.
These results signify that TvTgs is capable of converting DMG to TMG in a heterologous cellular milieu, albeit not with 100% efficiency. The failure of TvTgs to fully convert yeast DMG caps to TMG caps might reflect any of the following: (1) inherent distributivity of the TvTgs enzyme; (2) limiting amount in yeast of some component that promotes the second methylation reaction of TvTgs; or (3) restriction of the second methylation reaction by TvTgs to a particular subset of yeast snRNAs or snoRNAs.
Cellular localization of TvTgs and m2,2,7G-capped RNAs provide insights into putative substrates and cap biogenesis in T. vaginalis
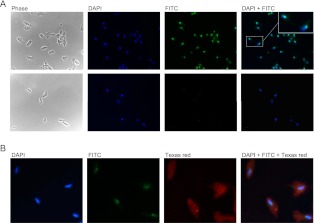
Metazoans and yeast have distinct cellular traffic pathways for maturation of snRNAs and snoRNAs (Cougot et al. 2004). T. vaginalis is distinct from yeast and metazoans insofar as its spliceosomal snRNAs lack a guanosine cap and therefore do not serve as substrates for TvTgs (Simoes-Barbosa et al. 2008a). In order to identify putative substrates for this enzyme and to gain insights into cap biogenesis, immunofluorescence was performed with anti-TMG and anti-TvTgs antibodies.
Anti-TMG antibody stained a single spot in the nucleus of T. vaginalis (Fig. 4A, top) and detection was lost if permeable cells were treated with RNase A prior to incubation with the antibody (Fig. 4A, bottom). The anti-TMG staining colocalizes with the nucleolar marker AC40, a subunit of RNA polymerases I and III (data not shown) (Simoes-Barbosa et al. 2008b). Therefore, TMG-capped RNAs or TvTgs substrates are associated with the nucleolus. Other TMG-capped RNAs are found to localize in the nucleolus as well (Jady et al. 2004).
FIGURE 4.
Immunolocalization of m2,2,7GpppRNAs and TvTgs in Trichomonas vaginalis. (A) Immunofluorescence using mouse monoclonal anti-m2,2,7G (1200× dilution) in T. vaginalis cells at 630× magnification. (Top) m2,2,7GpppRNAs were detected by FITC staining as indicated. An inset shows a strong immunoreactive spot in the nucleus of two cells. (Bottom) The RNA nature of anti-m2,2,7G recognition in cells was demonstrated here, where permeable cells were treated with RNase A prior to incubation with this antibody. DAPI was used to stain nuclei. (B) Immunofluorescence using mouse monoclonal anti-m2,2,7G (2400× dilution) and affinity-purified rabbit polyclonal anti-rTvTgs (1000× dilution) in T. vaginalis cells at magnification of 1000×. m2,2,7GpppRNAs were detected by FITC and endogenous TvTgs was detected by Texas red staining, as indicated.
Simultaneous probing with anti-TMG and anti-TvTgs antibodies showed that TMG caps and TMG synthase do not colocalize; rather, TvTgs is distributed diffusely in the cytoplasm and nucleoplasm (Fig. 4B). Finding of TvTgs in both compartments is in agreement with the fact that similar levels of Tgs activity are observed in cytosolic (data not shown) and nuclear extracts. In addition, endogenous TvTgs was found in both compartments after cell fractionation and Western blotting (data not shown). These data are consistent with the trafficking of TvTgs between the nucleus and the cytosol; alternatively, the protein may be dually localized without dynamic exchange between these compartments.
Trichomonas vaginalis box H/ACA snoRNAs have TMG caps
To identify specific Trichomonas TMG-capped RNAs, we performed preparative immunoprecipitation with anti-TMG antibody and radiolabeled the 3′ ends of the RNAs by using T4 RNA ligase and [32P]pCp. Analysis of labeled RNA content by high-resolution PAGE revealed several distinct small RNAs migrating between 100 and 300 nucleotides (Supplemental Fig. S2). After cloning and sequencing, a total of 18 novel noncoding RNAs were identified as unique sequences with perfect matches in the genome of this organism (Supplemental Fig. S3). Sixteen of these RNAs possess features typical of box H/ACA snoRNAs (hence, we named them TVsnoR1 to R16). The two others were designated as orphan RNAs (TVOrphan1 and TVOrphan2), for which we could not predict a function.
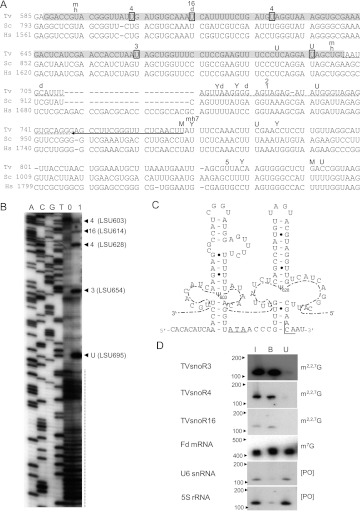
T. vaginalis box H/ACA snoRNAs were predicted to contain a “hairpin-hinge–hairpin-tail” secondary structure with conserved boxes and to guide formation of pseudouridines in 32 positions across T. vaginalis ribosomal RNAs (Supplemental Figs. S3, S4). Most of T. vaginalis snoRNAs identified here display functional homology with others found in yeast and humans, although some of these snoRNAs were found to modify novel positions in the rRNAs (Fig. 5; Supplemental Figs. S3–S6). The majority of these snoRNAs guide pseudouridylation in more than one position of ribosomal RNAs and their guide regions are located either in the 5′ or 3′ hairpin (Supplemental Fig. S3). TVsnoR12 and TVsnoR13 are homologs; they diverge in sequence in their 3′ segments, but are predicted to modify the same uridines in T. vaginalis SSU and LSU rRNAs (Supplemental Fig. S3). TVsnoR1 and TVsnoR2 are partial homologs that may have arisen by gene duplication. They share high similarity on their 3′ halves and most of the uridines they predict to modify.
FIGURE 5.
LSU rRNA pseudouridines, a box H/ACA snoRNA and anti-TMG analysis in Trichomonas vaginalis. (A) Pseudouridines in the LSU rRNA of T. vaginalis (Tv) as compared with S. cerevisiae (Sc) and human (Hs). MUSCLE alignment of sequences is illustrated. The arrow underlines the oligonucleotide sequence used for pseudourinide mapping in T. vaginalis. The dashed line denotes the region of RNA structural inaccessibility for the mapping technique used here. The gray highlighted sequence indicates the extent of pseudouridine mapping with confidence using this oligonucleotide. The boxed “U” in the sequence corresponds to pseudouridines that were validated experimentally in T. vaginalis. The letters M, Y, m, d, and h on the top of the sequences correspond to pseudouridines that are found in metazoans, yeast, mouse, Drosophila, and humans, respectively. The letter U on the top of the sequences corresponds to pseudouridines that are universally conserved in all species. The numbers on the top of the sequences identify the unique box H/ACA snoRNA in T. vaginalis that guides this pseudouridylation as described in Supplemental Figure S3. (B) Mapping of pseudourines by primer extension. This experiment was done with the oligonucleotide shown on A. A, C, G, and T indicate one of the four nucleotides in the sequencing reaction. 0 and 1 are the primer extension reactions with the untreated RNA and CMC-treated RNA, respectively. The dashed vertical line denotes the region of RNA structural inaccessibility as described in A. The arrowheads point to the pseudouridines with positions in T. vaginalis LSU rRNA, indicated between parentheses, found in this mapping experiment. Numbers outside of the parentheses identify the unique box H/ACA snoRNA in T. vaginalis that guides this pseudouridylation, as described in Supplemental Figure S3. (C) The sequence and predicted secondary structure of TVsnoR4. The lateral symmetrical loops of TVsnoR4 hairpins are shown in interaction with T. vaginalis LSU rRNA (dashed line) and pseudouridines (Ψ603 and Ψ628) are indicated. Box H and ACA are underlined. (D) Anti-TMG immunoprecipitation of T. vaginalis RNAs and Northern blot hybridization. I, B, and U correspond to the input RNA (the same RNA amount used in the immunoprecipitation reaction), bound RNA, and unbound RNA, respectively. Blots were hybridized with specific probes against three T. vaginalis box H/ACA snoRNAs (TVsnoR3, TVsnoR4, and TVsnoR16), of which the respective guided-pseudouridines were shown in A and B, ferredoxin (Fd) mRNA, U6 snRNA, and 5S rRNA. For reference, the molecular weight of RNA standards are shown on the left and the 5′-end status is shown on the right.
Following these predictions, we chose four regions of the 28S rRNA of T. vaginalis to validate experimentally the presence of pseudouridines. These regions included pseudouridine positions that are either predicted to be unique to this organism or universally conserved (Fig. 5; Supplemental Figs. S5, S6). We used a primer extension-derived technique where pseudouridines remain with a carbodiimide adduct after the RNA is treated with CMC, or 1-cyclohexyl-3-(do-morpholinoethyl)carbodiimide metho-p-toluenosulfonate, followed by alkali. Chemically modified pseudouridines induce premature stops at the adjacent 3′ nucleotide when reverse transcription is carried by Avian myeloblastosis virus (AMV) reverse transcriptase. This is then resolved in high-resolution sequencing gels (Ofengand et al. 2001a).
Not only were all predictions confirmed, but also 10 additional pseudouridines were detected within a coverage of 546 nt across the four regions analyzed here (Fig. 5; Supplemental Figs. S5, S6). For instance, we confirmed that TVsnoR4 uses both 5′ and 3′ hairpins to guide pseudouridylation of two novel positions on LSU of T. vaginalis, LSU 603 and 628, respectively (Fig. 5A–C; Supplemental S3). In this same region, we confirmed a novel modification (LSU 654) guided by TVsnoR3 and a pseudouridine also present in Drosophila (T. vaginalis LSU 614) guided by TVsnoR16 (Fig. 5A,B). A universally conserved pseudouridine (yeast rRNAs LSU 902 and human LSU 1670) was found in T. vaginalis LSU 695 (Fig. 5A,B) despite no guide snoRNA being found here for this modification. In contrast, another conserved pseudouridine (yeast LSU 896 and human LSU 1664) was absent in T. vaginalis (Fig. 5A,B). A third phylogenetically conserved pseudouridine (yeast LSU 941, human LSU 1741, T. vaginalis LSU 732) was found to reside in a structured region of T. vaginalis LSU that would not allow mapping by this primer extension technique (Fig. 5A,B). This structured region strongly induced premature stops even in the absence of carbodiimide-modified pseudouridines (Fig. 5B). For this reason, this phylogenetic conserved pseudouridine and another predicted pseudouridine in T. vaginalis (LSU 723) guided by both TVsnR1 and TVsnR2 (Fig. 5A) could not be confirmed. In conclusion, our mapping analysis shows a high density of pseudouridines in T. vaginalis rRNA. Additionally, it indicates that other box H/ACA snoRNAs are present in the T. vaginalis genome that would lead to additional modifications.
Next, to demonstrate the methylation status of their guanosine caps, we probed Northern blots of anti-TMG immunoprecipitated RNAs. We confirmed that all box H/ACA snoRNAs described here were found predominantly in the bound fraction of immunoprecipitated RNA (Fig. 5D; Supplemental Fig. S7). As previously reported (Simoes-Barbosa et al. 2008b), spliceosomal snRNAs and 5S rRNA that do not contain a guanosine cap were found in the unbound fraction, whereas mRNAs which contain m7G caps were found to cross-react with this antibody (Fig. 5D; Supplemental Fig. S7). In agreement with the immunofluorescence results, we conclude that small RNAs located in the nucleolus—box H/ACA snoRNAs—are preferred substrates for Tgs in this divergent unicellular eukaryote.
Most of the T. vaginalis snoRNAs identified represent single copy genes. They are likely to represent independent genes and are not derived from other coding gene transcripts or introns (Supplemental Fig. S3). These snoRNA transcripts can potentially acquire m7G caps, which are then converted to TMG. Curiously, a manual investigation of the few introns (Vanacova et al. 2005) predicted 15 intronic snoRNAs (Supplemental Fig. S8), all of which represent box C/D snoRNAs, and not a single box H/ACA snoRNA was found in the introns. If their synthesis is directed by the promoter of the intron-containing gene and they are matured by processing of the excised intron, then it is expected that such snoRNAs would have 5′-monophosphate ends instead. We have confirmed this experimentally (data not shown). Therefore, box H/ACA snoRNAs identified here are the only RNA species known to harbor TMG caps in T. vaginalis. Although we cannot completely rule out that other species of RNAs with TMG caps can be discovered, our data suggest that there might have been a strong selection toward box H/ACA snoRNAs acquiring TMG caps in contrast to spliceosomal snRNAs (Simoes-Barbosa et al. 2008b) and box C/D snoRNAs in T. vaginalis.
Because the structure of rRNAs in T. vaginalis is not available, the role of pseudouridines that we were able to predict and validate here is only speculative at the moment. However, the noticeable abundance and distribution of pseudouridines in this organism give support for the function of this “fifth” nucleoside as proposed (Ofengand et al. 2001b). Pseudouridines coordinate water molecules via their free N1-H and, as a summation of forces, they exert a significant “rigidifying” influence on the nearby sugar-phosphate backbone (Ofengand et al. 2001b). Their influence on enhancing base stacking explains why this modification is so often found in structured RNA molecules. In fact, certain genetic mutants lacking specific pseudoridines in tRNA or rRNA can cause protein translation impairment (Ofengand et al. 2001b). Here we have isolated and characterized T. vaginalis box H/ACA snoRNAs by mapping their pseudouridine sites. A recent genome-wide survey predicted the presence box H/ACA snoRNAs in G. lamblia and T. vaginalis (Chen et al. 2011). These studies indicate that RNA pseudouridylation has an important biological role and confers a selective advantage, as it has been maintained in deep-branching eukaryotes.
MATERIALS AND METHODS
Cell culture and 32P-labeling
T. vaginalis was maintained in TYM media supplemented with 10% horse serum and iron (Clark and Diamond 2002), and 32P-labeling was done as described previously (Simoes-Barbosa et al. 2010). Yeast cells were grown in standard YPD and phosphate-depleted YPD was used for 32P-labeling of yeast cultures as described (Grosjean et al. 2007). A total of 50–100 mL of phosphate-depleted cultures were supplemented with 1 mCi of 32P-orthophosphate and incubated for 2 h at 30°C when >90% of incorporation was achieved.
RNA manipulation and anti-m2,2,7G immunoprecipitation
In vivo-labeled and unlabeled RNAs were isolated from T. vaginalis (strain G3) and yeast cultures using TRIzol (Invitrogen). Yeasts were predigested with zymolyase before lysis in TRIzol. Immunoprecipitation of RNA with anti-TMG was done as previously described (Simoes-Barbosa et al. 2008b). The unlabeled RNA immunoprecipitated by anti-TMG was 3′-end labeled with [32P]pCp, resolved in 6% polyacrylamide DNA sequencing gels, and RNA bands were subsequently gel-purified, cloned, and sequenced as described previously (Simoes-Barbosa et al. 2008b). For blotting, unlabeled anti-TMG precipitated RNAs were resolved on TBE-Urea 6% polyacrylamide gels (20 × 20 cm) and transferred to nylon membranes using standard procedures followed by hybridization with 32P-labeled probes using the recommended stringency (GE Healthcare).
Trimethylguanosine synthase activity and analysis of methylated guanosine caps
Cell extracts from T. vaginalis strain G3 were prepared as previously described (Simoes-Barbosa et al. 2008b). Fresh extracts were used for Tgs activity using 32P-radiolabeled RNA substrate in 50–100-μL reactions as reported (Simoes-Barbosa et al. 2008a). After purification of the RNA product, guanylate caps were released by digestion with tobacco acid pyrophosphatase (TAP) and the methylation status of the guanylate cap nucleotide was determined by two-dimensional thin-layer chromatography (2D-TLC) with solvents A/B or A/C, as previously described (Grosjean et al. 2007; Simoes-Barbosa et al. 2008a). For the 2D-TLC analysis, the four ribonucleotides (A, U, C, and G) were used as internal standards and were detected by UV shading. The 32P-labeled guanosine cap standards were prepared with the use of recombinant Tgs enzymes as previously described (Simoes-Barbosa et al. 2008a). The relative abundance of the methylated guanylates was measured by densitometry using ImageJ software and the percentage of guanylate caps, m2,2,7G/(m7G + m2,2,7G), was estimated.
Rabbit polyclonal antibody anti-TvTgs and immunodepletion
Rabbit polyclonal antibody against TvTgs was produced by Pacific Immunology. TvTgs-specific antibody was purified from the last bleed (20 mL) by incubation with a PVDF strip containing 1 mg of rTvTgs (Simoes-Barbosa et al. 2008a). The specific antibody was eluted from the membrane by acid glycine solution and immediately neutralized. The antibody was dialyzed against phosphate-saline buffer (PBS) and concentrated to ∼1.5 mL (100 μg/mL) (Centricon, Millipore). Approximately 0.25 mg of freshly prepared nuclear extract was incubated with the affinity-purified anti-TvTgs in a 1:50 dilution. A final volume of 0.5 mL was adjusted with the nuclear extract dialysis buffer without glycerol. In parallel, another 0.25 mg of the same extract was mock incubated. Next, 0.25 mL of protein-A packed beads (Pierce) was added to both, and the reactions were then reincubated for 40 min each at 4°C with agitation. The flow-through, collected by a gravity column, was analyzed by Western blot and for Tgs activity and compared with the same extract prior to incubation.
Indirect immunofluorescence
Parasites (strain G3) were fixed onto microscopy slides, permeabilized, and incubated with antibodies as previously described (Simoes-Barbosa et al. 2008b). When needed, RNAs were digested with 0.2 mg of RNAse A for 30 min at room temperature. Affinity purified rabbit anti-TvTgs and monoclonal mouse anti-TMG were diluted in PBS by serial dilution varying concentrations from 200× to 2400× and 500× to 4000×, respectively, and tested in the immunofluorescence assay. Texas red-conjugated anti-rabbit IgG and FITC-conjugated anti-mouse IgG (Invitrogen), respectively, were used at a 1:2000 dilution as secondary antibodies for fluorescent detection of anti-rTvTgs and anti-TMG.
Computational prediction of RNA structure and RNA-guided pseudoridylation
A pattern search algorithm was used initially to screen for snoRNAs (Grillo et al. 2003). Secondary structures were predicted manually using standard RNA base-pairing rules, and the position of the H and ACA box motifs. Unpaired nucleotides in the predicted guide region were then used to write Perl scripts (http://www.perl.org) to locate uridines in the rRNA flanked by sequences able to base pair with the guides on both sides of an snoRNA guide pocket. These scripts relied on standard RNA base-pairing rules and secondary structure models described earlier for snoRNAs (Ganot et al. 1997). Predicted modifications were given a confidence level based on the length of complementarity between the rRNA and the snoRNA, and whether pseudouridine has been found at the homologous position in other organisms.
Mapping of pseudouridines in Trichomonas vaginalis ribosomal RNA
Pseudouridines were mapped in four regions of T. vaginalis LSU (nt 587–701; nt 1651–1818; nt 2168–2320; nt 2362–2471) following a standard protocol (Ofengand et al. 2001a). Chemically modified pseudouridines were identified by primer extension in high-resolution 5% polyacrylamide DNA-sequencing gels. The primer extension reactions were run side by side with a DNA sequence ladder that originated from T. vaginalis LSU when primed with the same oligonucleotide. All reactions were run twice with a short and long run for confirmation of the mapped pseudouridines. Oligonucleotides used in the primer extension reactions are described in Figure 5, Supplemental Figure S5, and Supplemental Figure S6 for reference.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI30537 (P.J.) and GM52470 (S.S.). S.S. is an American Cancer Society Research Professor. A.S.-B. was supported by a Faculty Research Development Fund from the University of Auckland.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034249.112.
REFERENCES
- Benarroch D, Qiu ZR, Schwer B, Shuman S 2009. Characterization of a mimivirus RNA cap guanine-N2 methyltransferase. RNA 15: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch D, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Shuman S 2010. Cap analog substrates reveal three clades of cap guanine-N2 methyltransferases with distinct methyl acceptor specificities. RNA 16: 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis.. Science 315: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Schwer B, Shuman S 2010. Mutational analyses of trimethylguanosine synthase (Tgs1) and Mud2: Proteins implicated in pre-mRNA splicing. RNA 16: 1018–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Penny D, Collins LJ 2011. Characterization of RNase MRP RNA and novel snoRNAs from Giardia intestinalis and Trichomonas vaginalis. BMC Genomics 12: 550 doi: 10.1186/1471-2164-12-550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CG, Diamond LS 2002. Methods for cultivation of luminal parasitic protists of clinical importance. Clin Microbiol Rev 15: 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, van Dijk E, Babajko S, Séraphin B 2004. ‘Cap-tabolism'. Trends Biochem Sci 29: 436–444 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T 2002. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 21: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz E, Stepinski J, Ekiel I, Jin Y, Haber D, Sijuwade T, Tahara SM 1988. β-globin mRNAs capped with m7G, m22.7G or m32.2.7G differ in intrinsic translation efficiency. Nucleic Acids Res 16: 8953–8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11: 941–956 [DOI] [PubMed] [Google Scholar]
- Grillo G, Licciulli F, Liuni S, Sbisà E, Pesole G 2003. PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res 31: 3608–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Droogmans L, Roovers M, Keith G 2007. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol 425: 55–101 [DOI] [PubMed] [Google Scholar]
- Grudzien E, Stepinski J, Jankowska-Anyszka M, Stolarski R, Darzynkiewicz E, Rhoads RE 2004. Novel cap analogs for in vitro synthesis of mRNAs with high translational efficiency. RNA 10: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Shuman S 2005a. Giardia lamblia RNA cap guanine-N2 methyltransferase (Tgs2). J Biol Chem 280: 32101–32106 [DOI] [PubMed] [Google Scholar]
- Hausmann S, Shuman S 2005b. Specificity and mechanism of RNA cap guanine-N2 methyltransferase (Tgs1). J Biol Chem 280: 4021–4024 [DOI] [PubMed] [Google Scholar]
- Hausmann S, Ramirez A, Schneider S, Schwer B, Shuman S 2007. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe. Nucleic Acids Res 35: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Zheng S, Costanzo M, Brost RL, Garcin D, Boone C, Shuman S, Schwer B 2008. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: Functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways. J Biol Chem 283: 31706–31718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HsuChen CC, Dubin DT 1976. Di- and trimethylated congeners of 7-methylguanine in Sindbis virus mRNA. Nature 264: 190–191 [DOI] [PubMed] [Google Scholar]
- Huber J, Dickmanns A, Lührmann R 2002. The importin-β binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol 156: 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body–specific localization signal. J Cell Biol 164: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke T, Dickmanns A, Ficner R 2009. Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1. Nucleic Acids Res 37: 3865–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonné R 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 9: 891–901 [DOI] [PubMed] [Google Scholar]
- Narayanan A, Eifert J, Marfatia KA, Macara IG, Corbett AH, Terns RM, Terns MP 2003. Nuclear RanGTP is not required for targeting small nucleolar RNAs to the nucleolus. J Cell Sci 116: 177–186 [DOI] [PubMed] [Google Scholar]
- Ofengand J, Del Campo M, Kaya Y 2001a. Mapping pseudouridines in RNA molecules. Methods 25: 365–373 [DOI] [PubMed] [Google Scholar]
- Ofengand J, Malhotra A, Remme J, Gutgsell NS, Del Campo M, Jean-Charles S, Peil L, Kaya Y 2001b. Pseudouridines and pseudouridine synthases of the ribosome. Cold Spring Harb Symp Quant Biol 66: 147–159 [DOI] [PubMed] [Google Scholar]
- Simoes-Barbosa A, Louly C, Franco OL, Rubio MA, Alfonzo JD, Johnson PJ 2008a. The divergent eukaryote Trichomonas vaginalis has an m7G cap methyltransferase capable of a single N2 methylation. Nucleic Acids Res 36: 6848–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Barbosa A, Meloni D, Wohlschlegel JA, Konarska MM, Johnson PJ 2008b. Spliceosomal snRNAs in the unicellular eukaryote Trichomonas vaginalis are structurally conserved but lack a 5′-cap structure. RNA 14: 1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Barbosa A, Hirt RP, Johnson PJ 2010. A metazoan/plant-like capping enzyme and cap modified nucleotides in the unicellular eukaryote Trichomonas vaginalis. PLoS Pathog 6: e1000999 doi: 10.1371/journal.ppat.1000999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn LP, Kasperaitis M, Ameling C, Voorma HO 1986. Additional methylation at the N(2)-position of the cap of 26S Semliki Forest virus late mRNA and initiation of translation. Virus Res 5: 61–66 [DOI] [PubMed] [Google Scholar]
- Vanacova S, Yan W, Carlton JM, Johnson PJ 2005. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proc Natl Acad Sci 102: 4430–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]