Translational regulation is of crucial importance to regulate gene expression in all types of cells. In this paper the authors combine biochemical techniques and electron microscopy to characterize the role of the DEAD-box helicase Rck/p54 in translational repression of mRNAs. Rck/p54 is recruited by sequence-specific translational repressors and multimerizes on mRNA, and this keeps target mRNAs repressed and competent to assemble into P-bodies.
Keywords: DDX6, mRNA storage, mRNA decay, DEAD-box, masking
Abstract
Translational repression is achieved by protein complexes that typically bind 3′ UTR mRNA motifs and interfere with the formation of the cap-dependent initiation complex, resulting in mRNPs with a closed-loop conformation. We demonstrate here that the human DEAD-box protein Rck/p54, which is a component of such complexes and central to P-body assembly, is in considerable molecular excess with respect to cellular mRNAs and enriched to a concentration of 0.5 mM in P-bodies, where it is organized in clusters. Accordingly, multiple binding of p54 proteins along mRNA molecules was detected in vivo. Consistently, the purified protein bound RNA with no sequence specificity and high nanomolar affinity. Moreover, bound RNA molecules had a relaxed conformation. While RNA binding was ATP independent, relaxing of bound RNA was dependent on ATP, though not on its hydrolysis. We propose that Rck/p54 recruitment by sequence-specific translational repressors leads to further binding of Rck/p54 along mRNA molecules, resulting in their masking, unwinding, and ultimately recruitment to P-bodies. Rck/p54 proteins located at the 5′ extremity of mRNA can then recruit the decapping complex, thus coupling translational repression and mRNA degradation.
INTRODUCTION
Translational regulation is a major mechanism of control of gene expression. Some cells rely only on this level of regulation because their transcription is shut down. For instance, mature oocytes and fertilized eggs rely on post-transcriptional control of maternal mRNA until the start of zygotic transcription. Various translational regulators have been identified in this context, such as CPEB, Bruno, Bicoid, Pumilio, and Musashi (Jackson et al. 2010; MacNicol and MacNicol 2010). Their study has led to a generic model where each repressor is recruited to a specific 3′ UTR mRNA sequence and one of its partners interacts with the translation initiation factor eIF4E at the 5′ cap extremity blocking access of the translation initiation machinery. In this model, the closed-loop conformation of the mRNA is maintained from active translation to repression, except that the 5′–3′ bridge made by PABP-eIF4G-eIF4E in polysomes is replaced by one made by the blocking partner and eIF4E in the repressed state. In Xenopus early oocytes, 4E-T bridges CPEB to eIF4E1b, the germ-line homolog of eIF4E. In Drosophila, Cup bridges Bruno to eIF4E. As a variant of this model, Bicoid interacts directly with both the mRNA and eIF4E2 (4EHP) (Jackson et al. 2010). Transcriptionally active cells also regulate expression at the translational level. Local translation in neurons involves some of the factors identified in early development, including members of the CPEB family and somatic factors such as FMRP that interacts with eIF4E via CYFP2 (Napoli et al. 2008). Repressors of the Puf family, which play a role in cell division, differentiation, and development, act by a mechanism that is at least in part similar to the one described above, except that they directly block the cap rather than its ligand eIF4E (Quenault et al. 2011). In vertebrates, the RNAi pathway, at least for a part, also follows a similar scheme, with the Ago2 protein of the RISC complex binding to the cap (Wu and Belasco 2008).
Following translational repression, mRNAs are either stored or degraded. Whether this occurs diffusely throughout the cytoplasm or in specific subcompartments is debated. The recent discovery of ribonucleoprotein granules such as P-bodies, stress granules, germinal and neuronal granules, which are enriched in mRNA degradation factors, mRNA repression factors, or a combination of both, raised the possibility that mRNA decay and storage takes place in these granules. However, various attempts to suppress these granules so far failed to demonstrate their requirement for mRNA decay or translational repression (Decker et al. 2007; Eulalio et al. 2007; Sweet et al. 2007; Stalder and Muhlemann 2009), leaving the question open as to their function.
The Rck/p54 protein is central to P-body assembly in mammals, as its depletion leads to their disappearance and prevents their assembly de novo in response to triggers such as arsenite (Serman et al. 2007). Rck/p54 is a highly conserved member of the DEAD-box RNA helicase family, which are characterized by a core made of two RecA-like domains arranged in tandem. The characteristic DEAD-box motifs present in these domains are required for P-body assembly (Minshall et al. 2009). In addition, the protein contains an N-terminal extension of 61 aa, particularly rich in Q and N residues, reminiscent of a “prion-like” domain. In mammals, Rck/p54 interacts with the RISC complex that mediates translational silencing by miRNAs (Chu and Rana 2006), while its Xenopus Xp54 homolog is known as a component of the repressor CPEB complex in oocytes (Ladomery et al. 1997; Minshall et al. 2001). The protein is therefore at the crossroad between translational repression and P-body formation. Furthermore, its C. elegans CGH1 homolog promotes both mRNA stability in oocytes and mRNA decay in somatic cells (Boag et al. 2008; Noble et al. 2008). Moreover, the yeast Dhh1 homolog interacts with the decapping complex to enhance decapping, which precedes mRNA degradation (Coller et al. 2001). Rck/p54 is therefore also connected to mRNA decay.
We have previously studied P-body ultrastructure in HeLa cells using Rck/p54 as a marker in immunoelectron microscopy (Souquere et al. 2009). The striking abundance of the protein in P-bodies suggested a role more complex than envisioned so far. We therefore investigated the properties of the protein and of its binding to RNA in vitro and in vivo using a combination of microscopy and biochemical techniques. Our results lead us to propose a model where the Rck/p54 protein acts downstream from translational repressors to maintain the repressed state of the mRNA, trigger its localization in P-bodies, and coordinate its repression and ultimate decay.
RESULTS
Quantitative analysis of the Rck/p54 protein in mammalian cells and in P-bodies
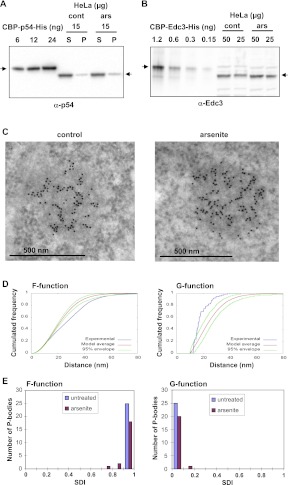
The Rck/p54 protein is prominent in P-bodies in mammalian cells, particularly after a brief arsenite treatment (Souquere et al. 2009). We designed experiments to quantify its abundance in HeLa cells, before and after arsenite treatment. HeLa cells were counted before lysis, and soluble and insoluble proteins were separated by brief centrifugation. As a reference, we produced human Rck/p54 protein tagged with CBP (calmodulin-binding protein) and His (6xHis tag) in Escherichia coli. The recombinant protein was quantified after double purification. Soluble and insoluble HeLa proteins were separated by centrifugation (16,000g) and analyzed by Western blotting with an anti-p54 antibody, along with increasing amounts of the recombinant protein (Fig. 1A). Quantitation of the chemiluminescent signal indicated that 85% of the Rck/p54 protein was in the supernatant. Fifteen micrograms of total soluble proteins, corresponding to 53,000 cells, contained 8.6 ng of Rck/p54 protein, while the cell equivalent amount of insoluble protein contained 1.2 ng. The larger size of the tagged protein (61.2 vs. 54 kDa) was taken into account in the calculation. Thus, each cell contains 0.18 pg of Rck/p54 protein, which corresponds to 2.0×106 Rck/p54 molecules. After arsenite treatment, the global Rck/p54 abundance was not significantly changed, with 2.3×106 molecules per cell.
FIGURE 1.
Quantitation of the Rck/p54 protein in the cytoplasm and in P-bodies. HeLa cells were treated or not with arsenite to increase the number of P-bodies. (A) Quantitation of Rck/p54 in HeLa cells. Fifteen micrograms of soluble (S) and insoluble (P) proteins were analyzed along with 6–24 ng of recombinant CBP-p54-His protein by Western blotting with an anti-p54 antibody. The arrows indicate the position of the recombinant and human Rck/p54 proteins. (B) Quantitation of Edc3 in HeLa cells. A total of 50 and 25 μg of proteins were analyzed along with 0.15–1.2 ng of recombinant CBP-Edc3-His protein by Western blotting with an anti-Edc3 antibody. The arrows indicate the position of the recombinant and human Edc3 proteins. (C) Concentration of Rck/p54 in P-bodies. HeLa cells fixed with paraformaldehyde were embedded in Lowicryl and immunostained with anti-p54 antibodies coupled to 10-nm gold particles. The labeling is concentrated over the electron-dense P-body. (D) Statistical analysis of Rck/p54 spatial distribution in P-bodies. Immunoelectron microscopy images were analyzed with distance functions F (left) and G (right). The curves obtained for the P-body (C, left) (in blue) were compared with the average curve (in red) and its 95% confidence envelope (in green) obtained with random distributions of the same number of gold-particles in the same contour (hard-core binomial model). (E) Population distribution of SDIs calculated for functions F (left) and G (right), for 46 P-bodies from untreated and arsenite-treated cells, as indicated.
As the amount of mRNA is estimated to be 300,000 molecules per cell (Boulton et. al. 1990; and our own calculation for a mean length of 3 kb), this analysis revealed a surprisingly large molar excess (sevenfold) of the Rck/p54 protein with respect to its potential target mRNAs. For comparison, the same quantification was performed for another P-body component, Edc3 (Fig. 1B). Twenty-five micrograms of total protein contained 0.58 ng of Edc3 protein, which indicated a number of 62,000 Edc3 molecules per cell. Thus, mRNA is in molar excess (fivefold) with respect to Edc3, as expected for a protein involved in mRNA decay. In addition, there are 32-fold fewer Edc3 than Rck/p54 molecules, consistent with Edc3 participating in a subclass of Rck/p54 complexes.
Next, we assessed Rck/p54 enrichment in P-bodies. We have previously reported the highly specific detection of Rck/p54 by immunoelectron microscopy, a technique appropriate to address this issue (Souquere et al. 2009). HeLa cells were treated or not with arsenite, fixed in glutaraldehyde and embedded in Lowicryl K4M. Ultra-thin sections were then incubated with an anti-p54 antibody and a secondary antibody coupled to 10-nm gold particles, and observed by electron microscopy (an example is given in Fig. 1C). In untreated cells, Rck/p54 labeling was scarce in the cytoplasm (3.1 gold particles/μm2, total of 180 μm2) and highly enriched in P-bodies (514 ± 207 gold particles/μm2, 33 P-bodies), indicating that Rck/p54 is on average 170-fold more concentrated in P-bodies than in the surrounding cytoplasm. After arsenite treatment, P-bodies reached a larger size (up to 0.29 μm2 compared with 0.20 μm2 in control cells), but the enrichment was similar (3.9 gold particles/μm2 in the cytosol, total of 212 μm2; 663 ± 169 gold particles/μm2 in the P-bodies, 36 P-bodies). Intriguingly, alignments of gold particles were visible in some P-bodies (Fig. 1C). As thin sections are not oriented, this spatial arrangement has to be frequent.
To test whether this pattern could be produced by random localization of the Rck/p54 protein within P-bodies, each P-body pattern was compared with 999 random patterns obtained with the same number of particles in the same contour, using distance F- and G-functions, as previously described (Andrey et al. 2010). Briefly, F-function is the cumulated distribution of the distance between an arbitrary position in the P-body and its nearest gold particle in the pattern, while G-function is the cumulated distribution of the distance between a gold particle and its nearest neighbor. For most of the 46 P-bodies studied (25 from untreated and 21 from arsenite-treated cells), F and G-functions were outside of the 95% confidence envelope of the random curves (data shown for one P-body in Fig. 1D), indicating nonrandom distribution of the gold particles. Each experimental curve was positioned within the range of variation of its 999 corresponding random curves to compute a spatial distribution index (SDI). The SDI is the probability, under the random model, of exceeding the relative distance between the experimental curve and the average random one, and represents a normalized measure of model fit (Andrey et al. 2010). P-bodies with random spatial repartitions of gold particles would have SDIs uniformly distributed from 0 to 1, while a P-body whose curve fits the random model average curve would have a SDI of 0.5. The highly nonuniform distribution of measured SDIs toward 1 and 0 for F and G functions, respectively, revealed larger empty spaces (F function) and smaller distance to the nearest neighbor (G function) than expected under spatial randomness, thus demonstrating strong particle aggregation in P-bodies (Fig. 1E). These data were consistent with the Rck/p54 protein being densely distributed on fibers, composed of proteins, ribonucleic acids, or a combination of both.
Evidence for Rck/p54 oligomers but no fibrillar autoaggregation in vitro
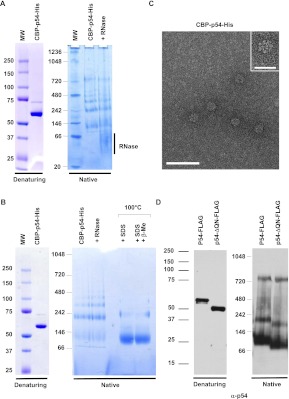
As the Rck/p54 protein contains a QN-rich region similar to the ones that are involved in prion protein autoaggregation, we hypothesized initially that the fibers could result from such a process, and therefore investigated whether the Rck/p54 protein autoaggregates in vitro. The double-purified CBP-p54-His protein described above was analyzed by gel electrophoresis and detected by Coomassie staining. The protein migrated as one major discrete band at the expected size of 61.2 kDa in denaturing Nu-PAGE (Fig. 2A, left). In contrast, in a Native-PAGE gel, the same preparation distributed in several discrete bands between ∼110 and 800 kDa (Fig. 2A, right). For unknown reasons, the band around 800 kDa was not observed in all preparations, despite a similar purity of the protein as judged by migration in Nu-PAGE (Fig. 2B). Overall, the absence of material higher in the gel suggested that the Rck/p54 protein oligomerizes in a limited number of forms, rather than autoaggregates.
FIGURE 2.
RNA-independent oligomerization of recombinant Rck/p54 in vitro. (A) Oligomerization of purified CBP-p54-His. A total of 2 μg of purified recombinant CBP-p54-His was separated on denaturing (left) and native (right) gels, and stained by Coomassie. Where indicated, the protein was treated with RNase A prior to migration. (B) Aberrant migration of Rck/p54 in native gels. A total of 2 μg of purified recombinant CBP-p54-His was separated on denaturing (left) and native (right) gels, stained with Coomassie. Where indicated, the protein was treated with RNase prior to purification, or heated in the presence of SDS with or without β-mercaptoethanol prior to migration. (C) Visualization of the purified protein in electron microscopy. The protein preparation analyzed in A was spread on grids and stained with uranyl acetate prior to observation by electron microscopy in filtered zero loss mode. (Main panel) Magnification ×151,000; bar, 50 nm. (Inset) Magnification ×241,500; bar, 20 nm. (D) Oligomerization is independent of the QN-rich extension of the protein. A total of 50 ng of purified recombinant FLAG-tagged wild-type (p54-FLAG) and truncated (p54-ΔQN-FLAG) Rck/p54 proteins were separated on denaturing (left) and native (right) gels, and analyzed by Western blotting with anti-p54 antibodies.
Importantly, oligomerization was not mediated by any contaminant bacterial RNA that could have been copurified with the protein. Indeed, the incubation of the protein with RNase A prior to migration (Fig. 2A) or within the bacterial lysate prior to affinity purification (Fig. 2B) did not change the migration pattern. It was also not caused by excessive protein concentration, as the same pattern was observed with solutions at 300–0.5 ng/μL (data not shown). In the native gel, the lower form migrated slower than expected for a monomer of 61.2 kDa (∼110 kDa). In order to determine whether this was due to the abnormal migration of the protein or to the formation of a stable dimer, the purified protein was subjected to severe denaturation conditions before migration and treated so as to disrupt potential disulfide bridges. Heating at 100°C in the presence of SDS alone or in the presence of SDS and β-mercaptoethanol, led to a single major band at the same 110-kDa position. Therefore, the smaller Rck/p54 band is a monomer that migrates aberrantly in this type of native gel. The other bands are consistent with the formation of a dimer (∼220 kDa), a trimer (∼330 kDa), a tetramer (∼440 kDa), and in some preparations a heptamer (∼770 kDa). The protein preparations were also deposited on grids and examined by electron microscopy (Fig. 2C, main panel). While the mono-, di-, and trimers are too small to be visualized at this resolution, protein fibers were not seen in any of the preparations. Rather, the detection of the larger oligomer on gels correlated with the presence of a wheel-like structure of 13-nm diameter (Fig. 2C, inset), whose size was consistent with an oligomer made of six to eight CBP-p54-His proteins.
Finally, we addressed the question of the role of the QN-rich region in oligomerization. In this experiment, we used FLAG fusion proteins. The single purified human Rck/p54-FLAG protein, deleted or not of its 61 N-terminal aa, was separated by gel electrophoresis and analyzed by Western blotting with anti-p54 antibodies. While the two proteins migrated at their expected size of 56 and 50 kDa on denaturating gels, both migrated as three bands on native gels (Fig. 2D). These bands corresponded to the monomer, dimer, and heptamer observed with the CBP-His tagged protein and were detected in all preparations. It indicated that the oligomerization of the protein was independent of its tag, and that the N-terminal domain was not involved. Thus, unlike prion proteins, the Rck/p54 protein oligomerizes, but does not autoaggregate in vitro. The larger oligomers have a circular and not a fibrillar aspect and form independently of the QN-rich extension of the protein.
The Rck/p54 QN-rich domain is dispensable for P-Body assembly in the cells
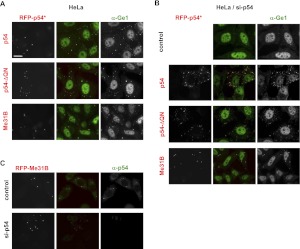
While the QN-rich extension was not involved in the self-association of the purified Rck/p54 protein, it was possible that it mediates association with another cytoplasmic protein, resulting in the formation of fibers. We therefore investigated the role of this region in P-body assembly. We have previously set up a complementation assay to test the capacity of mutant Rck/p54 to assemble P-bodies in mammalian cells (Minshall et al. 2009). Briefly, the endogenous Rck/p54 protein is first depleted by RNAi in HeLa cells, leading to the disappearance of P-bodies. A plasmid encoding RFP-tagged proteins (and insensitive to the Rck/p54 siRNA) is then transfected and the assembly of P-bodies de novo is analyzed. Wild-type RFP-Rck/p54 fully complemented the endogenous proteins, whereas mutations in the characteristic motifs of the DEAD-box helicases abolished this capacity. Here, we tested a RFP-tagged Rck/p54 deleted of its 61 N-terminal aa (p54-ΔQN). When Rck/p54 was present, the truncated protein accumulated like the full-length protein in P-bodies, as labeled with the specific marker Ge1 (Fig. 3A). When Rck/p54 was depleted by RNAi, the truncated protein was sufficient to assemble P-bodies, with the same efficiency as the full-length protein (Fig. 3B).
FIGURE 3.
The Rck/p54 QN-rich extension is dispensable for P-body assembly. (A) Localization in pre-existing P-bodies in the absence of the QN-rich region. HeLa cells were transfected with expression vectors encoding RFP-tagged human wild-type (p54) and truncated (p54-ΔQN) Rck/p54, and Drosophila Me31B. After 40 h, P-bodies were stained with anti-Ge1 antibodies and observed by fluorescence microscopy. Bar, 20 μm. (B,C) De novo assembly of P-bodies in the absence of the QN-rich region. HeLa cells were transfected with si-p54 to deplete Rck/p54 and suppress P-bodies. Twenty-four hours later, cells were transfected as described in A and stained with anti-Ge1 antibodies. (C) P-bodies assembled with Me31B do not contain human Rck/p54. P-bodies were assembled with Me31B as described in B and stained with anti-p54 antibodies.
Interestingly, while the Rck/p54 protein contains a QN-rich region in most species, either at the N (mammals, Xenopus, chicken) or C terminus (yeast), such a region is absent in Drosophila (Me31B), Caenorhabditis (Cgh1), and Trypanosoma (Dhh1). When tested in our assay, Me31B strongly accumulated in P-bodies (Fig. 3A) and efficiently assembled P-bodies after Rck/p54 depletion (Fig. 3B), whose absence was checked using mammalian-specific anti-p54 antibodies (Fig. 3C). In conclusion, the QN-rich domain of Rck/p54 was neither involved in its localization to P-bodies nor in the assembly of new P-bodies. Protein interactions mediated by the globular RecA domains were sufficient for both properties.
Multiple p54 binding along mRNA molecules in vivo
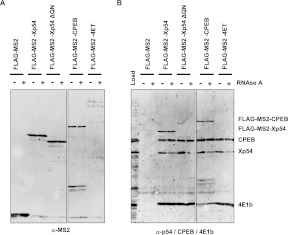
The fact that we found no evidence for prion-like properties of the Rck/p54 protein raised the possibility that the rows of gold particles seen in the P-bodies by immunoelectron microscopy correspond to several Rck/p54 proteins aligning on extended mRNA molecules. As proliferating cells such as HeLa cells are actively translating their pool of mRNA, and as no procedure is available so far to purify P-bodies, we turned to Xenopus oocytes, which are a unique system to study repressed mRNAs. The repression of maternal mRNAs is extensive in these cells and achieved through one well-characterized Xp54-containing complex, the CPEB complex. Previous studies have shown that Xp54 forms both RNA-independent and RNA-dependent dimers or oligomers, in association with other proteins of the CPEB complex, on repressed nonadenylated reporter mRNAs (Minshall and Standart 2004). We first extended these data by repeating the experiment using various proteins of the CPEB complex, including Xp54, CPEB, and 4E-T. Xenopus oocytes were injected with mRNA encoding FLAG-MS2 or FLAG-MS2-fused proteins, lyzed after 16 h, treated or not with RNase A and immunoprecipitated with anti-FLAG antibodies. Western blotting with anti-MS2 antibodies indicated that similar amounts of FLAG-MS2, FLAG-MS2-Xp54, and FLAG-MS2-CPEB were immunoprecipitated, while the FLAG-MS2-4E-T yield was lower, possibly due to its larger size (Fig. 4A). The immunoprecipitates were then analyzed for the presence of CPEB, Xp54, and eIF4E1b, with their respective antibodies (Fig. 4B).
FIGURE 4.
Multiple Rck/p54 binding along mRNA molecules. Xenopus oocytes were injected with an mRNA encoding the indicated proteins. Sixteen hours later, cells were lysed and treated or not with RNAse before immunoprecipitation with anti-FLAG antibody. Immunoprecipitates were analyzed by Western blotting with anti-MS2 (A) or a combination of anti-CPEB, anti-p54, and anti-eIF4E1b antibodies (B). Note that the MS2-Xp54-ΔQN is not detected by the anti-p54 antibody, which recognizes an epitope in the QN region.
FLAG-MS2-Xp54, but not the FLAG-MS2 control protein, coimmunoprecipitated CPEB, eIF4E1b, and Xp54. Some Xp54 was released when lysates were treated with RNase prior to immunoprecipitation, whereas CPEB and eIF4E1b were fully resistant to RNase treatment. The fraction of endogenous Xp54 resistant to RNase indicated the existence of a protein complex containing at least two Xp54 proteins, the tagged and the endogenous ones, consistent with the RNA-independent oligomerization observed with the recombinant human Rck/p54 protein (Fig. 2). FLAG-MS2-CPEB and FLAG-MS2-4E-T coimmunoprecipitated endogenous CPEB, eIF4E1b, and Xp54, and Xp54 was the only partner that was partly RNase sensitive. Therefore, as for Xp54, there are at least two CPEB proteins in the CPEB complex. In addition, the RNase-sensitive fraction of endogenous Xp54, which was pulled-down with the three FLAG-MS2-tagged proteins, indicated that RNAs bound to the CPEB/Xp54/4E-T complex additionally bind other Xp54 proteins. These Xp54 proteins are not contained in a typical CPEB complex, as none of the CPEB and eIF4E1b protein was RNase sensitive. Finally, the QN-rich region of Xp54 did not contribute to these complexes, as the deletion of the 61 N-terminal aa of Xp54 (MS2-Xp54-ΔQN) had no effect on the pulled-down proteins, prior or after RNase treatment. In conclusion, Xp54 is unique among the components of the CPEB complex, as it binds repressed RNA within both the CPEB complex and a distinct complex.
High affinity of Rck/p54 for RNA
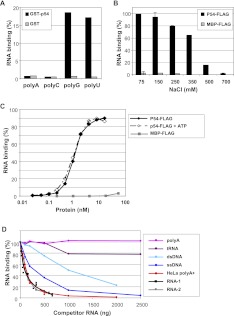
The multiple binding of Rck/p54 along mRNA raised the question of its RNA-binding properties. While the specificity of the CPEB complex is driven by the binding of CPEB to CPE sequences in the 3′ UTR of target mRNAs, direct RNA binding by Rck/p54 has not been characterized in detail. We first investigated the binding of Rck/p54 to RNA homopolymers (∼200–500 nt long) by GST pull-down. GST-fused human Rck/p54 protein bound to glutathione beads was incubated with an excess of radiolabeled synthetic RNA. Only poly(G) and poly(U) were retained by the protein (Fig. 5A), as previously observed with RNA-binding proteins such as CPEB or Pat1b (Marnef et al. 2010). We then performed a quantitative analysis of the binding using the filter retention assay and purified FLAG-tagged Rck/p54, which is closer to the natural Rck/p54 protein (tag of only 8 aa). The maltose-binding protein (MBP) was used as a non-RNA-binding control. A 1350-nt radiolabeled RNA transcribed in vitro was incubated with an excess of Rck/p54 (0.25 ng of RNA and 120 ng of protein, corresponding to a 3500 molar excess of protein) in buffers of increasing salt concentrations (Fig. 5B). Full binding was observed at 75 and 150 mM NaCl. It was largely salt resistant, as it required 500 mM NaCl for severe disruption.
FIGURE 5.
High-affinity and sequence-independent binding of Rck/p54 to RNA. (A) Rck/p54 binding to RNA homopolymers. GST-tagged Rck/p54 and control GST proteins bound to glutathione beads were incubated with radiolabeled RNA homopolymers. The histogram represents the percentage of RNA retained on the beads with the proteins. (B) Sensitivity of RNA binding to salt concentration. Radiolabeled RNA (0.25 ng) was incubated with purified FLAG-tagged Rck/p54 and control MBP proteins (120 ng) in the presence of indicated salt concentrations. RNA binding was assessed by filter-retention assay. The histogram represents the percentage of RNA retained on the filter with the proteins. (C) ATP-independent and high-affinity RNA binding. Radiolabeled RNA (0.25 ng) was incubated with increasing concentrations of the purified FLAG-tagged Rck/p54 and MBP proteins, in the presence or absence of ATP. RNA binding was assessed as in B. (D) Preferential and sequence-independent RNA binding. Radiolabeled RNA (0.25 ng) was incubated with the purified FLAG-tagged Rck/p54 (120 ng) in the presence of various amounts of the indicated RNA and DNA competitors. RNA binding was assessed as in B.
To determine the affinity of Rck/p54 for RNA, the radiolabeled RNA (0.25 ng) was incubated with various concentrations of the Rck/p54 or MBP protein (0.3–160 ng/0.033–17.6 nM). While MBP did not lead to significant RNA binding, maximal binding was reached with 10 nM Rck/p54, with a Kd (corresponding to half of the plateau) of 1 nM (Fig. 5C). The binding curve was sigmoid and could be fitted to the Hill equation with a Hill number of 2.1, indicating that the binding was cooperative. Slight variations were observed between protein preparations, with Kd ranging from 0.8 nM to 2.6 nM. The experiment was repeated in the presence of ATP and Mg++ ions (Fig. 5C). The same plateau and Kd were measured, indicating that ATP had no effect on the affinity of Rck/p54 for RNA in vitro.
We then performed competition experiments to determine Rck/p54 binding preferences. The radiolabeled RNA (0.25 ng) was incubated with Rck/p54 protein (120 ng) in the presence of increasing amounts of cold competitor (Fig. 5D). First, we tested RNAs of various sequence. A distinct in vitro-synthesized 677-nt RNA competed as efficiently as the 1350-nt RNA itself, as did polyadenylated mRNA purified from HeLa cells, demonstrating that the affinity of Rck/p54 measured on the 1350-nt RNA substrate was sequence independent. However, the poly(A) homopolymer did not compete, even at a high concentration, in agreement with the lack of binding observed in the GST pull-down experiment (Fig. 5A). Double-stranded salmon sperm DNA and a synthetic single-stranded 75-nt DNA were 10- and threefold less-efficient competitors, respectively, than RNA, indicating a clear, albeit not complete preference for RNA. Finally, a highly structured RNA such as tRNA was a very inefficient competitor, suggesting that Rck/p54 binds to single-stranded RNA. In conclusion, Rck/p54 binds preferentially RNA of random sequence, probably single-stranded, with a high affinity (Kd of 1 nM), in a manner not requiring ATP binding. These features are consistent with Rck/p54 being able to bind RNA directly in cells, independently of the CPEB complex.
ATP-dependent mRNA relaxing after Rck/p54 binding in vitro
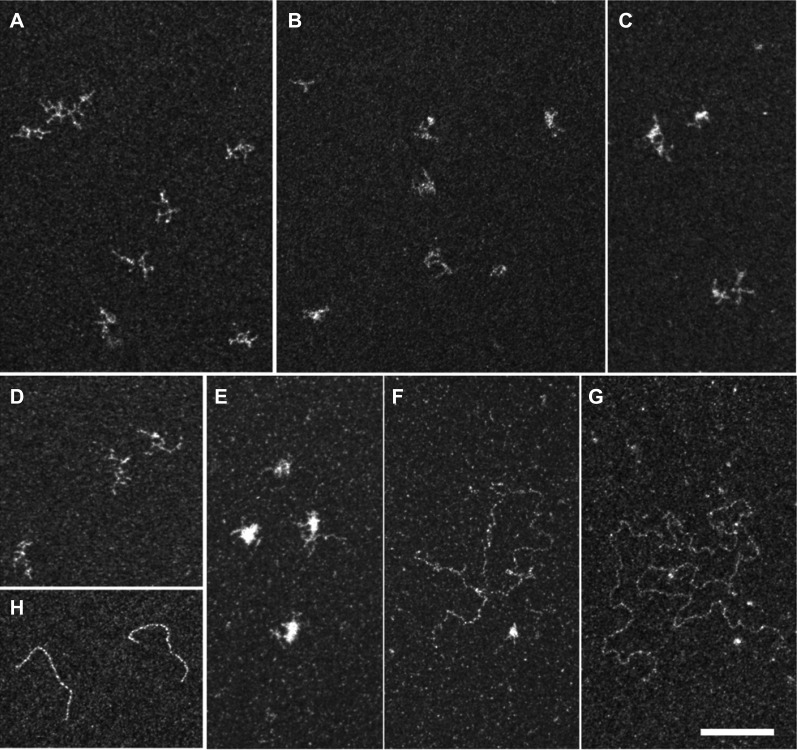
E. coli RecA and the related yeast Rad51 DEAD-box DNA helicases polymerize to form a helical filament on single-stranded DNA. We therefore investigated whether the multiple binding of Rck/p54 to RNA could form a similar nucleofilament. The 1350-nt unlabeled RNA was incubated with or without double-purified CBP-p54-His protein in the presence or absence of ATP, and spread on grids. After staining of the RNA, grids were visualized by electron microscopy (Fig. 6). In the absence of protein and ATP, RNA molecules were highly branched, which reflected their self-organized secondary structures (Fig. 6A). The addition of a 40-fold molar excess of Rck/p54 protein led to slightly more compact molecules, attesting to its binding to RNA (two representative fields in Fig. 6B,C). In the presence of ATP, the morphology of the RNA alone was not changed (Fig. 6D). However, the addition of Rck/p54 led to a mixture of compact (Fig. 6E) and extended (two examples in Fig. 6F,G) mRNP molecules. Relaxed molecules were frequently observed with a 40:1 to 80:1 molar ratio of protein to RNA (∼20% of the molecules) and more rarely with a 4:1 ratio (∼1% of the molecules). Most extended RNAs were longer than expected for a single molecule, as illustrated in Figure 6, F and G, which likely contained three and four RNAs, respectively. Extended RNAs were thinner than a control double-stranded DNA (Fig. 6H). Therefore, while there was no formation of a continuous nucleofilament such as observed with Rad51 (Dupaigne et al. 2008), RNA secondary structures were removed. While the staining and resolution were not sufficient to distinguish Rck/p54 monomers and dimers, the extended RNA molecules were not bound to the higher Rck/p54 oligomers (13 nm diameter) present in the preparation (Fig. 2B). These observations indicated that Rck/p54 binding to RNA in the presence of ATP leads to the removing of secondary structures and the relaxing of the RNA molecule.
FIGURE 6.
Electron microscopy imaging of RNA bound to Rck/p54. RNA was incubated alone (A,D) or with a 40 molar excess of the double-purified CBP-p54-His (B,C,E–G), in the absence (A–C) or presence (D–G) of ATP. The mixture was spread on grids and stained with uranyl acetate prior to observation by electron microscopy in filtered crystallographic dark field mode. RNA molecules alone were highly branched, independently of the presence of ATP (A,D). Upon protein binding in the absence of ATP, only branched RNA molecules were observed, as seen in two representative fields (B,C). In the presence of ATP, a mixture of ∼80% branched (E) and 20% relaxed (F,G) RNA molecules were observed. The characteristic spreading of double-stranded DNA in the same condition is shown for comparison (H). Magnification ×69,100; bar, 100 nm.
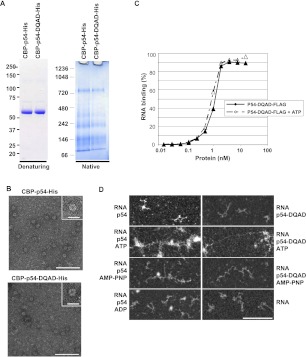
Next, we investigated whether RNA relaxing upon Rck/p54 binding required ATP hydrolysis. We first produced recombinant Rck/p54 protein with mutation in the DEAD motif (DEAD > DQAD), known to suppress ATP hydrolysis, but not ATP binding in yeast Dhh1 (Dutta et al. 2011). The double-purified CBP-p54-DQAD-His was indistinguishable from the wild-type protein in terms of oligomerization pattern in native gels (Fig. 7A) and formation of a wheel-like structure in molecular electron microscopy (Fig. 7B). Like wild-type Rck/p54, the FLAG-tagged DQAD mutant bound RNA with a Kd of 1 nM, independently of the presence of ATP (Fig. 7C). We then analyzed the conformation of RNA bound to CBP-p54-DQAD-His protein in electron microscopy (Fig. 7D). No extended RNA was observed before or after incubation with wild-type or mutant protein alone. In the presence of ATP, while RNA–protein complexes were even more entangled in this experiment, a similar amount of extended molecules was observed with the two proteins. We also replaced ATP by AMP-PNP, which is a nonhydrolyzable ATP analog, or ADP. AMP-PNP enabled RNA relaxing upon wild-type and mutant Rck/p54 binding as did ATP, whereas ADP was fully inactive. Therefore, RNA relaxing upon Rck/p54 binding is dependent on ATP binding but not ATP hydrolysis.
FIGURE 7.
RNA relaxing in the presence of Rck/p54 is independent of ATP hydrolysis. (A) Oligomerization of Rck/p54 DQAD mutant. A total of 2 μg of purified recombinant CBP-p54-His proteins, with or without an E/Q mutation in the DEAD motif, were separated on denaturing (left) and native (right) gels as in Figure 2A. (B) Visualization of Rck/p54 DQAD protein in electron microscopy. Both wild-type and mutant CBP-His-tagged proteins were visualized in electron microscopy as in Figure 2C. (Main panels) Magnification ×69,100; bar, 100 nm. (Insets) Magnification ×151,000; bar, 20 nm. (C) RNA-binding properties of Rck/p54 DQAD mutant. RNA binding was measured in the presence and absence of ATP using FLAG-tagged protein and filter retention assay, as in Figure 5C. (D) Electron microscopy imaging of RNA bound to Rck/p54 in the presence of AMP-PNP, or to Rck/p54 DQAD mutant. RNA was incubated with a 40 molar excess of the double-purified wild-type or mutant CBP-His-tagged proteins in the absence or presence of ATP, AMP-PNP, or ADP, as indicated. The mixture was spread and observed as in Figure 6. RNA alone is also shown. Magnification ×69,100; bar, 100 nm.
DISCUSSION
Rck/p54 abundance in cells and in P-bodies
The abundance of the Rck/p54 protein that we measured in mammalian cells was around 2×106 molecules per cell. This was a surprise in view of the known functions of the protein in repression and decapping complexes. Indeed, this represents a sevenfold molar excess with respect to cellular mRNAs, while one such complex is supposed to repress translation or trigger decapping by binding the mRNA 3′ UTR or 5′ extremity, respectively. The excess is even higher when considering only the mRNAs actually repressed or targeted for degradation. Such excess is not restricted to cultured mammalian cells. Previous estimation in Xenopus has shown that the protein is highly abundant in stage VI oocytes, amounting to ∼60 ng per oocyte (Minshall et al. 2007). This corresponds to a molar excess of 15-fold with respect to its potential targets, ∼2 × 1011 mRNA molecules per oocyte (Smillie and Sommerville 2002). A recent study in Trypanosoma also identified its Dhh1 homolog as a very abundant protein, with an estimated 4×105 Dhh1 molecules per cell, corresponding to an eightfold molar excess over mRNAs (Kramer et al. 2010). Therefore, the large excess of the Rck/p54 is a conserved feature from unicellular organisms to vertebrates, and in various cell types.
We then analyzed the intracytoplasmic distribution of Rck/p54 using immunoelectron microscopy, and measured a 170-fold concentration factor in P-bodies compared with the surrounding cytosol. It is then possible to give an estimate of the Rck/p54 concentration in the P-bodies. A HeLa cell cytoplasm is around 1000 μm3, while a large P-body (500 nm diameter) occupies a volume of 0.065 μm3. If a cell contains 2.0×106 Rck/p54 molecules, which are enriched 170-fold in a single P-body, then this large P-body contains 22,000 Rck/p54 molecules, at a local concentration of 0.56 mM. This estimate is quite robust, as it depends on the enrichment factor in the P-body, which was determined experimentally, but not on its exact size. A cell with 10 large P-bodies will change the calculation by only 10%.
High-affinity binding of Rck/p54 to RNA
Direct binding of Rck/p54 to RNA was maximal at physiological salt concentration. Most of the DEAD-box proteins tested do not show any specificity for a particular RNA (Cordin et al. 2006). The filter retention assay used in our study measures the highest affinity site of the RNA substrate, as RNA retention occurs when the first protein binds. While choosing a long (1350 nt) RNA substrate increased the chance that it could contain a sequence of particularly high affinity, the fact that the binding was competed as efficiently by cellular poly(A)+ RNAs as by itself indicated that it was representative of any mRNA. The measured Kd of 1 nM should therefore apply to cellular mRNA in general. Nevertheless, we showed that Rck/p54 hardly binds poly(A) homopolymer and highly structured RNA such as tRNA. Moreover, our data do not preclude the existence of specific binding sites of higher affinity in some mRNAs. Importantly, the Rck/p54 concentration in the P-bodies (0.5 mM as estimated above) is much higher than the Kd, so that the protein should cover any free mRNA region, except for the poly(A) tail and highly structured regions.
Interestingly, the RNA binding was ATP independent. The same affinity (Kd of 1 nM) was measured in the presence and absence of ATP, using both wild-type and a DEAD motif mutant protein. Similarly, a recent study reported an ATP-independent affinity (Kd of 2 nM) of yeast Dhh1 for a 20-mer oligo(U) substrate (Dutta et al. 2011). More generally, the binding of the DEAD-box proteins Mss116p, CYT-19, Ded1p, SrmB, and DbpA to intron RNA substrates was also found to be ATP independent (Del Campo et al. 2009). We have previously shown that the Rck/p54 ATPase activity is required for P-body assembly in mammalian cells (Minshall et al. 2009). We proposed a model where ATP plays a role in remodeling the interactions between the Rck/p54 protein and its protein partners. The present data confirm that ATP does not play any role in the direct binding of RNA.
Upon Rck/p54 binding, RNA molecules lost secondary structures, as observed in molecular electron microscopy. ATP was required for this, but not ATP hydrolysis. Indeed, an ATPase-defective Rck/p54 mutant also triggered RNA relaxing in the presence of ATP, and ATP could be replaced by a nonhydrolyzable ATP analog. Therefore, RNA unwinding requires a particular Rck/p54 conformation induced by ATP binding (ADP was inactive), but no ATPase-dependent energy transfer. This is similar to the behavior of the DEAD-box protein Ded1p, which promotes RNA strand separation in the presence of nonhydrolyzable ATP analog (Liu et al. 2008). Such DEAD-box proteins behave like RNA chaperones that stabilize single-stranded RNA, rather than true helicases (Jarmoskaite and Russell 2011).
No role of Rck/p54 QN-rich domain in P-body assembly
While the mechanism of P-body assembly is not fully understood, models have been proposed whereby the QN-rich regions of various RNA-binding proteins act as glue that enables the local aggregation of mRNPs. Indeed, in prion proteins, such QN-rich regions have been shown to favor protein autoaggregation in vitro and in vivo. Rck/p54 is central to P-body assembly in mammalian cells (Serman et al. 2007; Minshall et al. 2009). Its QN-rich domain is conserved from yeast to mammals, with the notable exception of the Drosophila, Nematode, and Trypanosoma. Nevertheless, we have not observed any fibrilar aggregation of the human Rck/p54 protein in vitro, even at high concentration. A genomic study intending to detect prion proteins throughout evolution considered as serious candidates QN-rich regions of 80 aa with at least 30 Q/N (Michelitsch and Weissman 2000). In fact, while the yeast sup35 protein contains a prion domain of 123 aa including 52% Q/N, the QN-rich region of Rck/p54 is only 49-aa long and poorer in Q/N (35%). Experimentally, there is no evidence that prion properties can be attributed to such short QN-rich domains.
Even if it does not promote prion-like aggregation, the QN-rich domain could mediate protein–protein interactions. We found that the purified Rck/p54 protein formed oligomers of up to seven protomers, which were RNA and ATP-independent. Mutation of the DEAD motif did not affect the oligomerization pattern. Interestingly, recombinant yeast Dhh1 protein also formed heptamers (data not shown), indicating that oligomerization is a conserved feature of the protein. Dimerization was previously observed for the DEAD box helicase Hera from Thermus thermophilus (Klostermeier and Rudolph 2009). However, in contrast to Hera, the self-interaction motif of Rck/p54 is located within the RecA-like domains, as the N-terminal truncated protein has the same oligomerization pattern as the full-length protein. Together with the fact that none of the known partners of Xp54 in Xenopus oocytes bind the QN-rich domain of the protein (Minshall et al. 2009), there is currently no evidence for a role of this extension in protein–protein interactions.
In yeast, Dhh1 is not required, but favors P-body assembly (Coller and Parker 2005), and deletion of its QN-rich domain decreases P-body localization (Reijns et al. 2008). In contrast, in mammals, Rck/p54 is required for P-body assembly (Serman et al. 2007; Minshall et al. 2009), with no obvious role of its QN-rich domain. As an alternative, we propose that the dimerization capacity of the protein combined to its multiple binding to RNA is the basis of mRNP aggregation within the P-body. Identifying an Rck/p54 mutant unable to multimerize on RNA will help addressing this question. Nevertheless, other Rck/p54 properties are also required, as the DEAD mutant, while oligomerizing and binding RNA like the wild-type protein in vitro, is unable to assemble P-bodies de novo in the cells (Minshall et al. 2009).
Alternative model of translational repression
In this study, we showed that (1) the Rck/p54 protein is in large excess with respect to its possible target mRNA in mammalian cells; (2) in cells it binds repressed mRNA at multiple positions, taking part in previously described repressor complexes like the CPEB complex in Xenopus oocytes, as well as in a distinct complex; (3) in vitro the protein directly binds RNA with high affinity and independently of ATP; (4) in the presence of ATP this binding leads to the relaxing of the RNA molecule, without requiring ATP hydrolysis; (5) the fine localization of Rck/p54 in the cells is consistent with such relaxed organization of Rck/p54 mRNPs in P-bodies. These data lead us to propose an alternative model of translational repression (Fig. 8). First, the binding of a repressor complex, such as CPEB, Bruno, Bicoid, or RISC blocks translation initiation, so that the mRNA is released from polysomes. At this stage, the mRNP is probably looped so that the complex bound in 3′ UTR interferes with the recruitment of initiation factors at the 5′ cap. This repressor complex, which contains Rck/p54 associated through protein–protein interactions, then triggers further binding of Rck/p54 along the length of the mRNA. This recruitment is favored by the abundance of the protein and its high affinity for RNA. The bound mRNA then relaxes in an ATP-dependent manner. As a consequence, RNA-binding proteins with lower affinity for RNA than Rck/p54, and those that bind double-stranded RNA, are chased out or prevented from binding the masked mRNA. The initial repressor complex might become dispensable at this stage. These relaxed molecules aggregate in P-bodies. They would be stored there, unless Rck/p54 proteins were released, in which case they return to translation. When mRNA is maintained in P-bodies, the 5′ cap proximal Rck/p54 protein could recruit the decapping complex leading to RNA degradation. Thus, mRNA decay would be naturally coupled to translational repression, without requiring the remodeling of a translational repressor complex into a degradation complex.
FIGURE 8.
Proposed model for the participation of Rck/p54 to translational repression and mRNA decay. Translational repression is initiated by the specific binding of protein X and its partners, including Rck/p54 (the exact stoichiometry of the repressor complex is not represented here). This triggers the sequence-independent binding of Rck/p54 along the mRNA. RNA relaxing is then favored by the presence of ATP. These extended mRNP complexes are then recruited to P-bodies, giving rise to the Rck/p54-rich fibers observed in electron microscopy. They can either return to translation if Rck/p54 is released, or the Rck/p54 located close to the 5′ cap can recruit the decapping complex, leading to mRNA degradation.
Our model aids in understanding why the depletion of many P-body proteins leads to P-body disappearance, without altering the capacity of forming new P-bodies in response to inducers like arsenite (Serman et al. 2007). Indeed, the number of P-bodies per cell is highly sensitive to the balance between mRNA translation and mRNA storage. Depletion of proteins involved in initial repressor complexes, such as GW182 in the RISC complex, or CPEB in the CPEB complex, would shift this balance toward translation, leading to the disappearance of P-bodies. The extent and the timing of P-body loss would depend on the importance of that repression pathway among others in the cell. Arresting general translation with arsenite would then shift the balance back to mRNA storage, leading to P-body assembly, provided that the underlying mechanism does not involve that particular protein. Rck/p54 is an exception, as its depletion leads to P-body disappearance and prevents their induction following arsenite treatment (Serman et al. 2007). This is consistent with Rck/p54 playing a general role downstream from the initial translational repression and upstream of P-body assembly.
Part of this model is reminiscent of the maskosome hypothesis proposed earlier in Xenopus oocytes (Sommerville 1999). In these cells, a small set of abundant proteins, including Xp54 and FRGY2a/b, are associated with most, if not all, repressed mRNAs. It was proposed that the Xp54 helicase unwinds the RNA molecule so that the CSD (Cold Shock Domain) containing proteins FRGY2a/b, which have higher affinity for single-stranded RNA, can cover the entirety of the RNA, resulting in its packaging and masking. Based on our data, we propose that translational repression in human somatic cells ends up with Rck/p54 stably bound to the repressed mRNA, and inducing mRNA relaxing rather than packaging. At first glance, a major difference with respect to oocytes is the multiplicity of repression pathways in somatic cells. In fact, Rck/p54 is found in an increasing number of specific repressor complexes. Beside the CPEB complex, which regulates CPE-containing mRNAs (Richter 2007), it is involved in the repression of the r15-lox mRNA in erythroid precursor cells (Naarmann et al. 2010). In this pathway, the hnRNPK complex binds the DICE sequence in the 3′ UTR of the mRNA, which recruits Rck/p54 in an RNA-dependent manner. Both hnRNPK and Rck/p54 are required for the repression. Furthermore, Rck/p54 is associated with RISC and functionally required for regulation by miRNAs, which is likely to concern a large number of mRNAs (Chu and Rana 2006). Finally, in some cell lines, Rck/p54 silencing leads to a major increase in global translation (up to 50%), indicating that it is widely involved in repression (Jangra et al. 2010). Altogether, these observations are in support of Rck/p54 being involved in a large number of repression pathways and eventually playing the same central role in translational repression in somatic cells as in oocytes.
MATERIALS AND METHODS
Cell culture and transfection
Epithelioid carcinoma HeLa cells were maintained in DMEM supplemented with 10% fetal calf serum. Transient transfections were performed with 3 μg of plasmid DNA or siRNA per 60-mm diameter dish using a standard calcium phosphate procedure. The RFP-p54 plasmid contains the human Rck/p54 ORF, while the siRNA targets the Rck/p54 3′ UTR, as previously described (Minshall et al. 2009). The RFP-p54-ΔQN was derived from RFP-p54 by PCR, while the RFP-Me31B plasmid was generated by replacing the human ORF by its Drosophila homolog.
Preparation of recombinant proteins in E. coli
The bacterial expression vectors GST-p54 and p54-FLAG contain the full human Rck/p54 ORF cloned into pGEX-6P-3 (GE Healthcare) and pT7-FLAG-2 (Sigma), respectively. To obtain the bacterial expression vector CBP-p54-His, the full human Rck/p54 ORF was cloned between the N-terminal CBP and the C-terminal His tags, as previously described (Ballut et al. 2005). The DQAD mutation was introduced in the CBP-p54-His and p54-FLAG vectors by mutagenesis, as previously described (Minshall et al. 2009). Both plasmids were verified by sequencing. The MBP-FLAG expression vector contains the MBP ORF synthesized by PCR from pMAL-P2 vector (New England Biolabs) and cloned into pT7-FLAG-2.
The GST fusion proteins were expressed in E. coli TG1, and induced by adding 1 mM isopropyl-b-D-thiogalactopyranoside (IPTG) overnight at 25°C. Crude protein extracts were prepared and purified as described previously (Dreuillet et al. 2008). The GST fusion proteins bound to glutathione beads were used as 50% slurry in the appropriate buffer.
CBP-His and FLAG fusion proteins were expressed in E. coli BL21-CodonPlus (Novagen). CBP-His proteins were induced overnight in Luria broth with 1 mM IPTG at 25°C and FLAG proteins in MagicMedia (Invitrogen) at 37°C. The CBP-p54-His protein was sequentially purified on nickel-nitrilotriacetic acid affinity resin (Qiagen) and calmodulin affinity resin (Stratagene), as previously described (Puig et al. 2001). The double-purified CBP-p54-His was dialyzed against 10 mM HEPES (pH 7.5), 75 mM NaCl, 0.1 mM EDTA, and 10% (w/v) glycerol before storage at –80°C. FLAG fusion proteins were prepared and purified on anti-FLAG M2 affinity gel (Sigma). The purity and amount of the recombinant proteins were determined in Nu-PAGE gel (Invitrogen) stained with Coomassie Brilliant Blue or by Western blotting.
RNA-binding assay
RNA was synthesized with T7 RNA polymerase (Promega) according to the manufacturer, in the presence or absence of 50 μCi of [α-32P]rCTP (Perkin-Elmer). After passage through a G50 spin column (Roche), the RNA was ethanol precipitated and dissolved in H2O. RNA homopolymers (Sigma) were 5′ end-labeled with [γ-32P]ATP after dephosphorylation and separated from unincorporated nucleotides.
The immobilized GST fusion protein was incubated with radiolabeled RNA homopolymer at 4°C for 60 min in a total volume of 500 μL of binding buffer (10 mM HEPES at pH 7.5, 75 mM NaCl, 0.1 mM EDTA, 10% [w/v] glycerol) in the presence of 25 μg/mL of BSA on a rotating wheel. After four washings with binding buffer, the bound radioactive material was quantified in a scintillation counter. For filter-binding assay, the purified proteins were incubated with the radiolabeled RNA in 150 μL of binding buffer containing 25 μg/mL of BSA for 30 min at 20°C and filtered through a wet nitrocellulose filter (Whatman Protan BA85) under gentle suction. The filter was washed four times with 1 mL of binding buffer without glycerol and counted in a scintillation counter.
Protein analysis by Western blotting
HeLa cells were lyzed in a buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, 1 mM EDTA, 1% NP-40 and protease inhibitors (Roche). Soluble and insoluble proteins were separated by centrifugation at 16,000g for 10 min at 4°C. Soluble proteins were quantified by the Coomassie protein assay (Pierce). Immunoprecipitation experiments using lysates prepared from Xenopus oocytes injected with FLAG-MS2-fusion protein mRNAs were performed essentially as described previously (Minshall et al. 2007) with the substitution of anti-FLAG antibody (Sigma-Aldrich) used at a concentration of 2.5 μL per 10 μL of protein G-Sepharose. Xenopus protein samples were separated on 15% SDS–polyacrylamide gels and transferred to PVDF membrane prior to probing with primary antibody, followed by IRDye 680 goat anti-rabbit antibody (Li-Cor) at a dilution of 1:10,000 and detection on an Odyssey Infra-Red Imager (Li-Cor).
Other proteins were separated on Nu-PAGE 4%–12% Bis-Tris gels (Invitrogen) and stained with Coomassie blue, or separated on Native-PAGE 4%–16% Bis-Tris gels (Invitrogen) and stained with Simply blue safe stain (Invitrogen), as indicated. For Western blotting, proteins were then transferred to a PVDF membrane (Perkin Elmer). After blocking in PBS-T (PBS, 0,1% Tween-20) containing 5% (wt:vol) nonfat dry milk for 1 h at room temperature, the membrane was incubated with the primary antibody for 1 h at 37°C, rinsed in PBS-T, and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. After washing in PBS-T, immune complexes were detected using the Supersignal West Pico Chemiluminescent Signal kit (Pierce) and visualized by exposure to CL-XPosure film (Pierce) or using a Fluor-S Max MultiImager with Quantity One software (Bio-Rad).
Antibodies included rabbit polyclonal to Rck/p54 from Bethyl Laboratories, Inc., CPEB (Thom et al. 2003), Xp54 (Minshall and Standart 2004), eIF4E1b (Minshall et al. 2007), MS2 (gift from C.W.J. Smith), and Edc3 (gift from Jens Lykke-Andersen) (Fenger-Gron et al. 2005). Secondary antibodies were purchased from Immunoresearch Laboratories (Immunotech).
Immunofluorescence
Cells grown on glass coverslips were fixed in methanol for 3 min at −20°C. After rehydratation, cells were incubated with the primary antibody for 1 h, rinsed with PBS, incubated with the secondary antibody for 45 min, and rinsed with PBS, all steps being performed at room temperature. Slides were mounted in Citifluor (Citifluor). Rabbit polyclonal anti-p54 and mouse monoclonal anti-Ge1 antibodies were purchased from Bethyl Laboratories, Inc. and Santa Cruz, respectively. Secondary Cy2-conjugated antibody was purchased from Immunoresearch Laboratories (Immunotech).
Microscopy was performed on a Leica DMR microscope (Leica) using a 63X1.32 oil immersion objective. Photographs were taken using a Micromax CCD camera (Princeton Instruments) driven by Metamorph software. Images were processed using Adobe Photoshop software.
Cellular immunoelectron microscopy and statistical spatial analysis
Cell embedding in Lowicryl K4M (Chemische Werke Lowi) and post-embedding Rck/p54 immunolocalization were performed using secondary anti-rabbit antibodies coupled to 10-nm gold-particles, as described previously (Souquere et al. 2009). Ultrathin sections of two untreated and three arsenite-treated cell samples were analyzed with a FEI Tecnai Spirit transmission electron microscope. Digital images were taken with a SIS MegaviewIII CCD camera.
For statistical analysis of gold-particle spatial distribution, P-body contours were manually delineated based on density, and the positions of gold particles were detected using automated image thresholding with ImageJ software (Rasband, W.S., ImageJ, US National Institutes of Health, http://rsb.info.nih.gov/ij/, 1997–2008). Each experimental pattern was compared with a hard-core binomial model, conditioned on the number of observed particles and the P-body contour, with gold-particles modeled as hard disks of 10-nm diameter. Observed and simulated patterns were analyzed with distance functions F and G (Diggle 2003).
Molecular electron microscopy
For protein visualization, samples were processed for negative staining. Five microliters of a 0.37-mg/mL (6 μM) CBP-p54-His solution were deposited onto a 300 mesh copper grid coated with a collodion covered with thin carbon film, activated by glow-discharge. The grids were washed with aqueous 2% (w/vol) uranyl acetate (Merck) and dried with ashless filter paper (VWR). Observations were carried out on a Zeiss 912AB transmission electron microscope in filtered zero loss mode.
For nucleic acid visualization, samples were processed for positive staining as previously described (Le Cam 2000). The in vitro synthesized RNA (1350 nt) was incubated with the CBP-p54-His protein in binding buffer (10 mM Hepes at pH 7.5, 75 mM NaCl, 2.5 mM MgCl2) in the presence or absence of 0.1 mM ATP, AMP-PNP or ADP, as indicated. The RNA concentration was set at 2.5 nM (1.1 μg/mL), while the protein was at 10, 100, or 200 nM (0.6, 6, or 12 μg/mL). After 10 min at 37°C, the solution was diluted 10 times in binding buffer, and 5 μL were deposited onto a 600 mesh copper grid coated with a thin carbon film, activated by glow-discharge in the presence of pentylamin (Merck). After 1 min, grids were washed with aqueous 2% (w/vol) uranyl acetate (Merck) and then dried with ashless filter paper (VWR). Observations were carried out on a Zeiss 912AB transmission electron microscope in filtered crystallographic dark field mode. Electron micrographs were obtained using a ProScan 1024 HSC digital camera and Soft Imaging Software system.
ACKNOWLEDGMENTS
We thank Hervé Le Hir for his kind help in the production of the CBP-His fusion proteins. We also thank C.W.J. Smith for providing the MS2 antibody, and Jens Lykke-Andersen for providing the Edc3 antibody. This work was supported by the Ligue Nationale Contre le Cancer, the Association pour la Recherche sur le Cancer, an international joint grant from the Royal Society/CNRS, the CNRS and the University Pierre et Marie Curie.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034314.112.
REFERENCES
- Andrey P, Kiêu K, Kress C, Lehmann G, Tirichine L, Liu Z, Biot E, Adenot PG, Hue-Beauvais C, Houba-Herin N, et al. 2010. Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLoS Comput Biol 6: e1000853 doi: 10.1371/journal.pcbi.1000853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H 2005. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol 12: 861–869 [DOI] [PubMed] [Google Scholar]
- Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK 2008. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol 182: 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton AA, Baker GB, Campagnoni AT, ed. 1990. Molecular neurobiological techniques. In Neuromethods, Vol 16. Humana Press, Clifton, NJ [Google Scholar]
- Chu CY, Rana TM 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4: e210 doi: 10.1371/journal.pbio.0040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P 2006. The DEAD-box protein family of RNA helicases. Gene 367: 17–37 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM 2009. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol 389: 674–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle PJ, ed. 2003. Statistical analysis of spatial point patterns. Edward Arnold, London, UK [Google Scholar]
- Dreuillet C, Harper M, Tillit J, Kress M, Ernoult-Lange M 2008. Mislocalization of human transcription factor MOK2 in the presence of pathogenic mutations of lamin A/C. Biol Cell 100: 51–61 [DOI] [PubMed] [Google Scholar]
- Dupaigne P, Lavelle C, Justome A, Lafosse S, Mirambeau G, Lipinski M, Pietrement O, Le Cam E 2008. Rad51 polymerization reveals a new chromatin remodeling mechanism. PLoS ONE 3: e3643 doi: 10.1371/journal.pone.0003643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Zheng S, Jain D, Cameron CE, Reese JC 2011. Intermolecular interactions within the abundant DEAD-box protein Dhh1 regulate its activity in vivo. J Biol Chem 286: 27454–27470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM 2010. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J Virol 84: 6810–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R 2011. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA 2: 135–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostermeier D, Rudolph MG 2009. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res 37: 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Queiroz R, Ellis L, Hoheisel JD, Clayton C, Carrington M 2010. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J Cell Sci 123: 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomery M, Wade E, Sommerville J 1997. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res 25: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam E, Delain E, Larquet E, Culard F, Cognet JAH. 2000. DNA-protein complexes analysed by electron microscopy and cryo-microscopy (ed. A Travers, M Buckle), pp. 337–350. Oxford University Press, Oxford, UK [Google Scholar]
- Liu F, Putnam A, Jankowsky E 2008. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci 105: 20209–20214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNicol MC, MacNicol AM 2010. Developmental timing of mRNA translation–integration of distinct regulatory elements. Mol Reprod Dev 77: 662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Maldonado M, Bugaut A, Balasubramanian S, Kress M, Weil D, Standart N 2010. Distinct functions of maternal and somatic Pat1 protein paralogs. RNA 16: 2094–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS 2000. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci 97: 11910–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Standart N 2004. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res 32: 1325–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Thom G, Standart N 2001. A conserved role of a DEAD box helicase in mRNA masking. RNA 7: 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Reiter MH, Weil D, Standart N 2007. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem 282: 37389–37401 [DOI] [PubMed] [Google Scholar]
- Minshall N, Kress M, Weil D, Standart N 2009. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell 20: 2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naarmann IS, Harnisch C, Muller-Newen G, Urlaub H, Ostareck-Lederer A, Ostareck DH 2010. DDX6 recruits translational silenced human reticulocyte 15-lipoxygenase mRNA to RNP granules. RNA 16: 2189–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. 2008. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134: 1042–1054 [DOI] [PubMed] [Google Scholar]
- Noble SL, Allen BL, Goh LK, Nordick K, Evans TC 2008. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J Cell Biol 182: 559–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Quenault T, Lithgow T, Traven A 2011. PUF proteins: Repression, activation and mRNA localization. Trends Cell Biol 21: 104–112 [DOI] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci 121: 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD 2007. CPEB: A life in translation. Trends Biochem Sci 32: 279–285 [DOI] [PubMed] [Google Scholar]
- Serman A, Le Roy F, Aigueperse C, Kress M, Dautry F, Weil D 2007. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res 35: 4715–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie DA, Sommerville J 2002. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J Cell Sci 115: 395–407 [DOI] [PubMed] [Google Scholar]
- Sommerville J 1999. Activities of cold-shock domain proteins in translation control. Bioessays 21: 319–325 [DOI] [PubMed] [Google Scholar]
- Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D 2009. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci 122: 3619–3626 [DOI] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O 2009. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet TJ, Boyer B, Hu W, Baker KE, Coller J 2007. Microtubule disruption stimulates P-body formation. RNA 13: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom G, Minshall N, Git A, Argasinska J, Standart N 2003. Role of cdc2 kinase phosphorylation and conserved N-terminal proteolysis motifs in cytoplasmic polyadenylation-element-binding protein (CPEB) complex dissociation and degradation. Biochem J 370: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Belasco JG 2008. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell 29: 1–7 [DOI] [PubMed] [Google Scholar]