Abstract
Although alterations in DNA methylation patterns have been associated with specific diseases and environmental exposures, the mediators and signaling pathways that direct these changes remain understudied. The bioactive lipid mediator prostaglandin E2 (PGE2) has been shown to exert a myriad of effects on cell survival, proliferation, and differentiation. Here, we report that PGE2 also signals to increase global DNA methylation and DNA methylation machinery in fibroblasts. HumanMethylation27 BeadChip array analysis of primary fetal (IMR-90) and adult lung fibroblasts identified multiple genes that were hypermethylated in response to PGE2. PGE2, compared with nontreated controls, increased expression and activity (EC50∼107 M) of one specific isoform of DNA methyltransferase, DNMT3a. Silencing of DNMT3a negated the ability of PGE2 to increase DNMT activity. The increase in DNMT3a expression was mediated by PGE2 signaling via its E prostanoid 2 receptor and the second messenger cAMP. PGE2, compared with the untreated control, increased the expression and activity of Sp1 and Sp3 (EC50∼3×107 M), transcription factors known to increase DNMT3a expression, and inhibition of these transcription factors abrogated the PGE2 increase of DNMT3a expression. These findings were specific to fibroblasts, as PGE2 decreased DNMT1 and DNMT3a expression in RAW macrophages. Taken together, these findings establish that DNA methylation is regulated by a ubiquitous bioactive endogenous mediator. Given that PGE2 biosynthesis is modulated by environmental toxins, various disease states, and commonly used pharmacological agents, these findings uncover a novel mechanism by which alterations in DNA methylation patterns may arise in association with disease and certain environmental exposures.—Huang, S. K., Scruggs, A. M., Donaghy, J., McEachin, R. C., Fisher, A. S., Richardson, B. C., Peters-Golden, M. Prostaglandin E2 increases fibroblast gene-specific and global DNA methylation via increased DNA methyltransferase expression.
Keywords: epigenetics, eicosanoids, macrophage
The DNA methylation profile of cells is highly plastic and susceptible to alteration by endogenous mediators and exogenous environmental factors (1, 2); however, the mechanisms by which such factors direct specific changes in the methylation pattern are, in many instances, poorly understood. Both normal cellular differentiation (3) and pathological processes such as tumorigenesis (4, 5), inflammation (6, 7), and fibrosis (8, 9) are associated with changes in the DNA methylation profile. Locally produced lipid mediators are important endogenous participants in numerous homeostatic functions and in disease pathogenesis, but it is not known whether they can alter the DNA methylation pattern in cells.
Prostaglandin E2 (PGE2) is one of the most ubiquitous and important endogenous lipid mediators, and it exerts pleiotropic actions that can vary depending on the context, the tissue environment, and the cell type (10). It is produced constitutively in nearly all tissues, but its synthesis is highly regulated, being increased with the induction of cyclooxygenase (COX) and PGE synthase enzymes in response to microbes, cytokines, hormones, and growth factors and decreased in response to glucocorticoids. Although it has long been recognized during states of inflammation to cause fever, pain, and local tissue edema, PGE2 can also act on epithelial, mesenchymal, and immune cells to influence cellular proliferation (11), differentiation (12), and function (11, 13, 14) and on cancer cells to promote tumorigenesis (15). PGE2 achieves this diversity of function by its ability to ligate 4 different G-protein-coupled receptors, E prostanoid (EP) 1–4, which are coupled to distinct G proteins and signal via alterations in specific second messengers (e.g., cAMP or Ca2+) (16).
In view of the ability of PGE2 to regulate a myriad of fundamental cellular functions in which DNA methylation has been implicated, we sought to determine whether PGE2 is able to directly influence the DNA methylation pattern in cells. In lung fibroblasts, we and others have previously shown that PGE2 inhibits fibroblast to myofibroblast differentiation (12), fibroblast proliferation, and fibroblast production of matrix proteins (11, 17). In this study, we show that PGE2 induces global and gene-specific increases in DNA methylation in lung fibroblasts. This increase in DNA methylation was due to increased expression and activity of the specific DNA methyltransferase (DNMT) 3a. Although the expression of 15-lipoxygenase (18), COX-2 (19), and E prostanoid receptors (9) is known to be regulated by DNA methylation, these findings demonstrate the converse relationship, namely, that PGE2 is capable of increasing global and gene-specific DNA methylation through increased expression and activation of a specific de novo DNMT.
MATERIALS AND METHODS
Cell culture
Primary fetal (IMR-90) and adult (CCL-210, CCL-204, and CCL-209) lung fibroblasts were purchased from American Type Culture Collection (Manassas, VA, USA) and were cultured in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) and 1% penicillin/streptomycin. Cells from the murine monocyte-macrophage leukemic cell line, RAW 264.7, were cultured in Roswell Park Memorial Institute medium. All experiments were performed on cells at passages 5–9. Cells were plated unless otherwise noted at 5 × 105 cells/well in 6-well plates, serum starved overnight, and treated with the following reagents: PGE2 (10−9 to 10−6 M), the prostacyclin [prostaglandin I2 (PGI2)] analogs iloprost (1 μM) and treprostinil (1 μM), PGD2 (1 μM), the thromboxane A2 analog U-4419 (1 μM), and the EP2 agonist butaprost free acid (500 nM) (all from Cayman Chemical, Ann Arbor, MI, USA); aspirin (200 μM), the DNA-damaging agent temozolomide (100 and 250 μM), and actinomycin D (2.5 μg/ml) (all from Sigma-Aldrich, St. Louis, MO, USA); IL-1β (10 ng/ml; BD Biosciences, Sparks, MD, USA); the EP3 agonist ONO-AE3-248 (100 nM), the EP4 agonist ONO-AE1-329 (100 nM), and the EP4 antagonist ONO-AE3-208 (100 nM) (kind gifts from Ono Pharmaceuticals, Osaka, Japan); the adenyl cyclase activator forskolin (100 μM; EMD Chemicals, San Diego, CA, USA); the EP2 antagonist AH6809 (10 μM; Enzo Life Sciences, Farmingdale, NY, USA); and the Sp1/Sp3 inhibitor mithramycin (25–50 nM; Tocris Bioscience, Bristol, UK). For small interfering RNA (siRNA) experiments, cells were cultured to 30–50% confluence and transfected with siRNAs against DNMT1, DNMT 3a, and DNMT 3b (Qiagen, Valencia, CA, USA) using Lipofectamine LTX (Invitrogen) for 48 h in OptiMEM medium (Invitrogen) before being treated with or without PGE2. Specificity of the siRNAs was confirmed by real-time RT-PCR and has been reported previously (9).
Global and gene-specific DNA methylation
Genomic DNA was isolated from 1 × 106 cells using DNeasy (Qiagen). Levels of global DNA methylation were assayed using the MethylFlash Methylated DNA Quantification Kit from Epigentek (Farmingdale, NY, USA) according to the manufacturer's protocol.
For gene- and site-specific analysis, 1 μg of genomic DNA was subject to bisulfite conversion using the EZ DNA Methylation Kit from Zymo Research (Irvine, CA, USA). Bisulfite-converted DNA was analyzed for methylation at 27,578 CpG sites using the HumanMethylation27 BeadChip array (Illumina; San Diego, CA, USA) according to the manufacturer's protocol. Signal intensity from methylated and unmethylated probes for all sites was scanned on the Illumina BeadArray Reader and preprocessed using Illumina GenomeStudio software. The methylation status of individual CpG sites was verified by pyrosequencing. Bisulfite-modified DNA was amplified by PCR using biotin-labeled primers specific for the IGFBP2 and MGMT promoter. The amplified product was then mixed with sequence-specific primers and analyzed for methylation at individual CpG sites using a Pyrosequencer (Qiagen). IGFBP2 and MGMT amplification and sequencing-specific primers were obtained from EpigenDx (Worcester, MA, USA). Long interspersed element (LINE)-1 amplification and sequencing primers were obtained from Qiagen.
DNMT activity assay
Cells (3×106) were plated in 100-mm culture dishes and treated with PGE2 for 48 h. Cells were scraped, and nuclear fractions were collected using the Panomics Nuclear Extraction Kit (Affymetrix, Santa Clara, CA, USA). DNMT activity of nuclear extracts was assayed according to the manufacturer's protocol using the DNMT Activity Assay (ActiveMotif, Carlsbad, CA, USA).
Real-time RT-PCR
RNA was isolated from cells using TRIzol (Invitrogen), and quantitative mRNA levels were assayed by real-time RT-PCR using the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Primers for human and murine DNMT1, DNMT3a, and DNMT3b and human Sp1 and Sp3 were obtained from Applied Biosystems. Primer and probes for human and murine β-actin were used as reported previously (11).
Immunoblotting
Cell lysates were collected in lysis buffer (PBS containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM orthovanadate, and protease cocktail inhibitor), resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted using antibodies to the following: DNMT1 (2 μg/ml; Imgenex, San Diego, CA, USA), DNMT3a (2 μg/ml; Abcam, Cambridge, MA), DNMT3b (4 μg/ml; Abcam), O6-methylguanine DNA methyltransferase (MGMT; 1:1000; Cell Signaling Technology, Beverly, MA, USA), poly(ADP-ribose)polymerase (PARP; 1:1000; Cell Signaling Technology), and α-tubulin (1:1000; Sigma-Aldrich). Bound primary antibodies were visualized with appropriate secondary antibody conjugated to horseradish peroxidase and developed with enhanced chemiluminescence reagent (GE Healthcare, Piscataway, NJ, USA). Densitometric analysis was performed on the visualized bands using Scion Image software (U.S. National Institutes of Health, Bethesda, MD, USA).
COMET assay
The COMET assay (Cell Biolabs, San Diego, CA, USA) was used to measure DNA damage, as described previously (20). Cells were pretreated with PGE2 (500 nM) for 48 h. Medium was then removed, and cells were treated for 24 h with the DNA-damaging agent temozolomide (100–250 μM). Cells were then collected, centrifuged, and resuspended in a 1:10 PBS-agarose mixture and placed on glass slides. The cells were washed, permeabilized, and subjected to an electric gradient of 1 V/ml of chamber length for 12 min as per the manufacturer's instructions. Nuclear material was stained and visualized by fluorescent microscopy. The percentage of cells that exhibited a comet tail and the length of the comet tail were measured to quantitate the degree of DNA damage.
Sp1 activity assay
Nuclear extracts were isolated from cells as described above and assayed for Sp1 activity according to the manufacturer's protocol using the Sp1 Transcription Factor Activity Assay Kit (Affymetrix).
Data analysis
Data were analyzed on GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA) using ANOVA or Student's t test, as appropriate, with P < 0.05 defined as statistically significant. Data are expressed as means ± se. For the HumanMethylation27 array data, signal intensities were corrected for red/green color balance, adjusted for background signal, and normalized across the set of arrays. M values were calculated as the log2 ratio of the intensities of the methylated probe vs. unmethylated probe, as described by Du et al. (21). M values were then compared between groups treated with vs. without PGE2, and statistically significant differences were identified using the limma algorithm in the Bioconductor suite (ref. 22; http://www.bioconductor.org). Significant differences in M values were defined by both P < 0.05 and a fold change of >1.5 or 2 as noted. Enrichment analysis was performed using ConceptGen (http://conceptgen.ncibi.org) with Q < 0.05 defined as statistically significant.
RESULTS
PGE2 increases global and gene-specific DNA methylation in fibroblasts
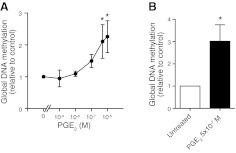
Using the primary fetal lung fibroblast cell line IMR-90, we first determined the effect of PGE2 on global DNA methylation. Cells were treated with PGE2 (10−9 to 10−6 M) for 48 h and assayed for total methylated DNA using an ELISA-based assay. Although PGE2 did not affect levels of global methylation at 24 h (data not shown), it dose dependently increased global DNA methylation by up to 2-fold after 48 h of treatment (Fig. 1A). The increase in global DNA methylation elicited by PGE2 was also observed in primary adult lung fibroblasts (Fig. 1B).
Figure 1.
Effect of PGE2 on global DNA methylation. Primary fetal lung fibroblasts (IMR-90; n=3–5; A) and adult lung fibroblasts (n=5; B) were treated with or without PGE2 at the indicated concentrations for 48 h, and global DNA methylation levels were assayed as described in Materials and Methods. All results are expressed as means ± se. *P < 0.05.
In the mammalian genome, DNA methylation is most abundant along centromeric and interspersing noncoding intergenic regions (23). To determine whether the observed increase in global methylation reflects a change in methylation along these noncoding regions, we performed bisulfite sequencing to assay the methylation status of LINE-1. We did not observe a significant change in LINE-1 methylation in either IMR-90 or adult normal cells treated with PGE2 (Supplemental Fig. S1).
We next sought to determine whether the increase in global DNA methylation elicited by PGE2 reflected an increase in methylation of specific gene promoters. We used the Illumina HumanMethylation27 BeadChip array to survey the methylation status of 27,578 CpG sites (most of which are located within the promoters of genes and many of which are within CpG islands) in IMR-90 and adult lung fibroblasts treated with or without PGE2 for 48 h (n=3 for each cell type). Selecting those CpG sites that exhibited a significant increase or decrease in methylation (P<0.05 and a 2-fold change in M value), we identified 18 genes differentially methylated by PGE2 in IMR-90 cells (Table 1). Of those, more than two-thirds showed an increase rather than a decrease in methylation. Using the same criteria in adult cells, we identified 5 genes that were hypermethylated and no genes that were hypomethylated by PGE2 (Table 2).
TABLE 1.
Genes with >2- or <0.5-fold change in methylation after PGE2 treatment in IMR-90 cells
| Gene ID | Gene symbol | Chromosome | CPG map site | Fold change | P |
|---|---|---|---|---|---|
| 64776 | C11orf1 | 11 | 111255565 | 4.189 | 0.002 |
| 5253 | PHF2 | 9 | 95378980 | 3.527 | 0.033 |
| 1028 | CDKN1C | 11 | 2862072 | 3.150 | 0.040 |
| 985 | CDC2L2 | 1 | 1645098 | 3.041 | 0.005 |
| 55071 | C9orf40 | 9 | 76757181 | 2.719 | 0.043 |
| 4097 | MAFG | 17 | 77478941 | 2.549 | 0.004 |
| 3485 | IGFBP2 | 2 | 217206819 | 2.373 | 0.014 |
| 595 | CCND1 | 11 | 69167000 | 2.226 | 0.032 |
| 8975 | USP13 | 3 | 180853518 | 2.201 | 0.027 |
| 79006 | METRN | 16 | 705435 | 2.123 | 0.034 |
| 126789 | PUSL1 | 1 | 1233923 | 2.072 | 0.047 |
| 29902 | C12orf24 | 12 | 109391110 | 2.066 | 0.021 |
| 26524 | LATS2 | 13 | 20533665 | 2.052 | 0.038 |
| 55693 | JMJD2D | 11 | 94346603 | 0.471 | 0.003 |
| 5649 | RELN | 7 | 103418455 | 0.447 | 0.020 |
| 23518 | R3HDM1 | 2 | 136005195 | 0.444 | 0.019 |
| 51105 | PHF20L1 | 8 | 133857529 | 0.408 | 0.020 |
| 8829 | NRP1 | 10 | 33663793 | 0.336 | 0.039 |
TABLE 2.
Genes with >2- or <0.5-fold change in methylation after PGE2 treatment in adult fibroblasts
| Gene ID | Gene symbol | Chromosome | CpG map site | Fold change | P |
|---|---|---|---|---|---|
| 8726 | EED | 11 | 85633629 | 12.740 | 0.001 |
| 79891 | ZNF671 | 19 | 62930886 | 6.408 | 0.049 |
| 55693 | JMJD2D | 11 | 94346603 | 3.690 | 0.004 |
| 2300 | FOXL1 | 16 | 85169514 | 2.612 | 0.000 |
| 3364 | HUS1 | 7 | 47985966 | 2.196 | 0.011 |
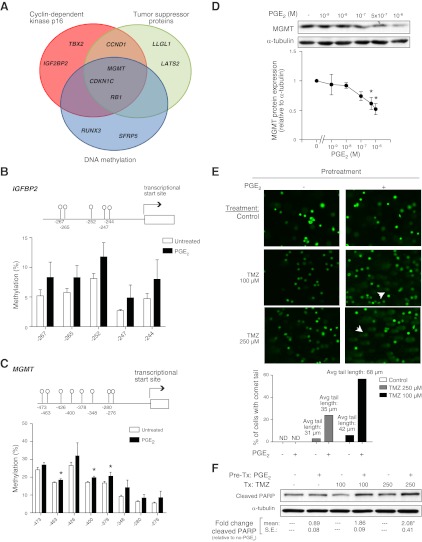
To further explore the observed hypermethylation by PGE2, we broadened our selection criteria from the array to include genes that exhibited only an increase in methylation (P<0.05 but with a fold change >1.5 in M value). This selection yielded 52 genes whose methylation was increased by PGE2 in IMR-90 cells (Supplemental Table S1). Placing these results into biological context, we used ConceptGen (24) to perform enrichment analysis. ConceptGen combines annotations found in gene ontology and gene pathway resources with literature-derived MeSH annotations. ConceptGen identified 3 literature-derived statistically significant (Q<0.05) concept groups—cyclin-dependent kinase inhibitor p16, tumor suppressor proteins, and DNA methylation—with annotations enriched for the hypermethylated genes identified from the array data (Fig. 2A). Interestingly, the function of these enriched groups is congruent with the previously described ability of PGE2 to inhibit fibroblast proliferation (11) and fibroblast survival (25).
Figure 2.
Genes hypermethylated by PGE2. A) Functional cluster analysis was performed on 52 genes identified as exhibiting a 1.5-fold increase in M value methylation (P<0.05) with PGE2 treatment in IMR-90 cells. Three functional clusters were identified (Q<0.05), with the genes associated with each cluster listed. B, C) Methylation status of CpG loci within the IGFBP2 (B) and MGMT (C) promoters, with the location relative to the transcription start site indicated by numbers at bottom, was determined by bisulfite pyrosequencing in IMR-90 cells treated with PGE2 for 48 h. Mean methylation levels from 4 independent experiments at each of the indicated CpG loci is shown. D) MGMT protein expression was assayed by immunoblot and quantitated by densitometric analysis with values normalized to α-tubulin and expressed relative to untreated cells (n=4). Top panel shows a representative immunoblot. IMR-90 cells were pretreated with PGE2 (500 nM) for 48 h, washed, and subsequently treated with or without temozolomide (TMZ) for 24 h. E) DNA damage was assayed by the COMET assay, as described in Materials and Methods, and visualized by fluorescence microscopy. Representative images from 3 independent experiments are shown. Arrowheads indicate representative cells with comet tails, which signify DNA damage. Graph quantifies the percentage of cells that exhibited a comet tail and the mean length of comet tails. F) Apoptosis of cells was assayed by immunoblotting for cleaved PARP. A representative immunoblot from 3 experiments is shown, with the fold increase in densitometry, relative to that with no PGE2 pretreatment, indicated numerically at bottom. *P < 0.05 vs. untreated.
To independently verify the hypermethylation of genes identified from the array, we performed bisulfite sequencing of the IGFBP2 and the MGMT promoters. These genes were chosen based on their fold change in methylation by array analysis and the identification of these genes by the aforementioned functional enrichment analysis. We focused on 5 CpG loci within the IGFBP2 promoter, including one that was identified by the array and 4 adjacent sites. PGE2 treatment resulted in an increase in methylation in all five sites assayed (Fig. 2B), with the average degree of methylation at these sites increasing from 6.7 to 13.1% (P<0.05). We also performed bisulfite sequencing of 22 CpG loci within the MGMT promoter. The mean methylation of all 22 loci increased from 17.3 to 21.5% (P<0.05) with PGE2 treatment, with a more marked increase in methylation observed in 8 particular CpG loci (Fig. 2C). To determine whether the hypermethylation of the MGMT promoter resulted in decreased protein expression, we examined the expression of MGMT by immunoblot analysis and observed that PGE2 dose dependently decreased MGMT expression (Fig. 2D).
MGMT is a DNA repair enzyme up-regulated during DNA damage, and its expression has been shown to increase after injury with temozolomide (26). To determine the functional relevance of hypermethylation of MGMT and diminished MGMT expression by PGE2, we pretreated cells with or without PGE2 for 48 h. Medium was then changed, and cells were subsequently treated with temozolomide. Cells pretreated with PGE2 exhibited increased susceptibility to DNA damage by temozolomide, as shown in COMET assays (Fig. 2E), and to cellular apoptosis, as shown by an increase in cleaved PARP (Fig. 2F). These data are congruent with previous studies that have shown that PGE2 increases fibroblast susceptibility to apoptosis (25) and suggest the hypermethylation of MGMT may be partially responsible for the decrease in cell survival.
PGE2 increases DNMT activity and gene expression
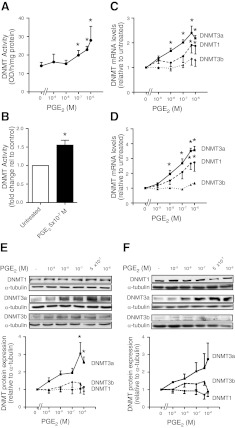
Given the observed increase in global and gene-specific methylation by PGE2, we sought to determine whether the increase in methylation was due to an increase in nuclear activity of DNMTs, the enzymes responsible for the addition of methyl groups to DNA. The nuclear fractions of cells treated for 48 h with or without PGE2 were isolated and assayed for DNMT activity. PGE2 dose dependently increased DNMT activity in IMR-90 (Fig. 3A) and primary adult lung fibroblasts (Fig. 3B).
Figure 3.
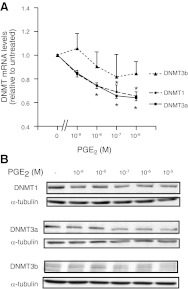
Effect of PGE2 on DNMT activity and expression. A, B) IMR-90 cells (n=4–6; A) and adult lung fibroblasts (n=3; B) were treated for 48 h with the indicated concentrations of PGE2 and assayed for nuclear DNMT activity, as described in Materials and Methods. C, D) DNMT1, DNMT3a, and DNMT3b mRNA levels in IMR-90 fibroblasts (n=6; C) and adult lung fibroblasts (n=4; D) treated for 24 h with the indicated concentrations of PGE2 were assayed by quantitative real-time RT-PCR. DNMT mRNA levels were normalized to β-actin and are expressed as fold change relative to no PGE2-treatment controls. E, F) DNMT1, DNMT3a, and DNMT3b protein expression in IMR-90 cells (n=4; E) and adult lung fibroblasts (n=3; F) treated for 48 h with varying concentrations of PGE2 were assayed by immunoblot analysis. Top panels: representative immunoblots. Bottom panels: fold change in expression based on mean densitometric analysis, normalized to α-tubulin. All results are expressed as means ± se. *P < 0.05.
In mammalian cells, 3 major DNMT isoforms, DNMT1, DNMT3a, and DNMT3b, have been shown to exhibit in vitro and in vivo methyltransferase activity (27). DNMT1, by virtue of its preference for hemimethylated DNA over unmethylated DNA (28), is often considered a maintenance methyltransferase whereas DNMT3a and DNMT3b, by virtue of their lack of preference between unmethylated and hemimethylated DNA, have traditionally been regarded as de novo methyltransferases (27). To determine whether the increase in DNMT activity by PGE2 is associated with a change in expression of specific DNMT isoforms, we performed real-time RT-PCR and observed that 24-h treatment with PGE2 dose dependently increased DNMT1 and especially DNMT3a mRNA levels in both IMR-90 (Fig. 3C) and adult lung fibroblasts (Fig. 3D). The increase by PGE2 in DNMT3a mRNA at 24 h corresponded to an increase in DNMT3a protein expression at 48 h, as assayed by immunoblot, in both IMR-90 (Fig. 3E) and adult fibroblasts (Fig. 3F). There was no change in DNMT1 protein expression despite the increase in its mRNA.
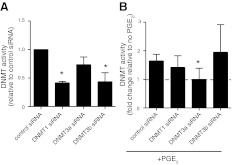
To evaluate the contributions of each DNMT isoform to enzyme activity, we pretreated IMR-90 cells with control and DNMT isoform-specific siRNAs for 48 h followed by PGE2 for an additional 48 h. The selective ability of these isoform-specific siRNAs under the experimental conditions used to knock down expression of their respective target DNMTs was reported previously (9). At baseline, silencing of DNMT1 and DNMT3b, but not of DNMT3a, decreased basal DNMT activity (Fig. 4A). The ability of PGE2 to increase DNMT activity was preserved with control, DNMT1, and DNMT3b siRNA treatment but was selectively abrogated with DNMT3a siRNA treatment (Fig. 4B). These data suggest that although DNMT3a is not a significant contributor to basal DNMT activity, the increase in DNMT activity by PGE2 is predominantly mediated by an increase in DNMT3a expression.
Figure 4.
Effect of silencing DNMT isoforms on DNMT activity. IMR-90 cells were treated with control or isoform-specific DNMT siRNA for 48 h and then were treated with or without PGE2 (500 nM) for an additional 48 h before being assayed for DNMT activity as described in Materials and Methods. A) DNMT activity of cells treated with different target siRNAs at baseline without PGE2 treatment, with relative activity normalized to control siRNA. B) DNMT activity of cells treated with PGE2 (expressed as a fold change relative to no PGE2). All results are expressed as means ± se; n = 3 *P < 0.05 vs. control siRNA.
Endogenous levels of PGE2 regulate expression of DNMT3a
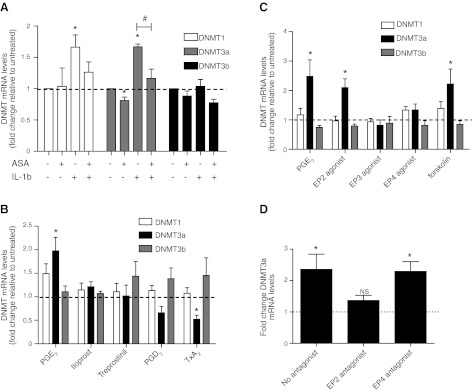
Prostanoids are elaborated constitutively by fibroblasts, and production can increase when expression of key synthetic enzymes such as COX-2 is increased. To determine whether endogenously generated prostanoids also modulate expression of DNMT isoforms, we treated IMR-90 cells for 24 h with the COX inhibitor aspirin and/or the cytokine IL-1β, which potently induces COX-2 expression, and assayed DNMT mRNA levels. IL-1β significantly increased DNMT1 and DNMT3a mRNA levels, and its effect was blocked by concomitant treatment with aspirin (Fig. 5A). Aspirin alone modestly but significantly decreased DNMT3a expression. These findings suggest that increases and decreases in endogenously synthesized prostanoids are sufficient to modulate expression of DNMT3a.
Figure 5.
Role of endogenous prostanoids and specific PGE2 receptors in regulating DNMT expression. A) IMR-90 cells were treated with or without aspirin (ASA, 200 μM) and IL-1β (10 ng/ml) for 24 h and assayed for DNMT1, DNMT3a, and DNMT3b mRNA levels by real-time RT-PCR, with values expressed relative to the no-treatment control. n = 3. B) IMR-90 cells were treated for 24 h with either PGE2 (500 nM), PGI2 analogs iloprost (1 μM) or treprostinil (1 μM), PGD2 (1 μM), or thromboxane A2 (TxA2) analog U-4419 (1 μM) and assayed for DNMT1, DNMT3a, and DNMT3b mRNA levels by real-time RT-PCR, with results expressed as fold change relative to the no-treatment control. n = 7. C) IMR-90 cells were treated with either PGE2 (500 nM), the EP2 agonist butaprost free acid (500 nM), the EP3 agonist ONO-AE3-248 (100 nM), the EP4 agonist ONO-AE1-329 (100 nM), or the adenyl cyclase activator forskolin (100 μM) for 24 h and assayed for DNMT1, DNMT3a, and DNMT3b mRNA levels by real-time RT-PCR. Results are expressed as fold change relative to the untreated control. n = 7. D) IMR-90 cells were pretreated for 30 min in the presence or absence of an EP2 antagonist AH6809 (10 μM) or the EP4 antagonist ONO-AE3-208 (100 nM) before treatment with PGE2 (500 nM) for 24 h. DNMT3a mRNA levels were assayed by real-time RT-PCR, with results expressed as fold change relative to no PGE2 treatment. n = 4. All results are expressed as means ± se. *P < 0.05 vs. untreated. #P < 0.05 for IL-1β-treated with ASA vs. no ASA.
Because prostanoids other than PGE2 can also be produced from COX actions on arachidonic acid, we compared the ability of PGE2 to modulate DNMT expression with that of the PGI2 analogs iloprost and treprostinil, PGD2, and the thromboxane A2 analog U-4419. Among the prostanoids tested, only PGE2 was able to increase DNMT3a mRNA levels (Fig. 5B) and DNMT3a protein expression (as confirmed by immunoblot, data not shown). Because PGE2 is the predominant IL-1β-induced prostanoid synthesized by lung fibroblasts (29), these data suggest that PGE2 is the specific fibroblast-derived prostanoid mediating the effect (Fig. 5A) of IL-1β.
PGE2 increases DNMT3a expression via EP2-cAMP signaling and increased expression of Sp1 and Sp3
PGE2 treatment resulted in an increase in DNMT3a mRNA at 24 h, and an increase in DNMT3a protein expression, nuclear DNMT activity, and global DNA methylation levels at 48 h. To test whether a prolonged exposure to PGE2 is necessary for the increase in DNMT expression, we treated cells with PGE2 for only 1 h before removing the medium and replacing it with PGE2-free medium (Supplemental Fig. S2). Cells exposed to PGE2 for 1 h exhibited an increase in DNMT3a mRNA levels at 24 h similar to that of cells in which the medium was not removed, suggesting that early PGE2 signaling events are sufficient to increase DNMT3a expression.
PGE2 signals by ligating one of 4 G-protein-coupled EP receptors (16). The EP2 and EP4 receptors signal predominantly through Gαs, leading to activation of adenyl cyclase and increased production of the second messenger cAMP. EP1 and EP3 are predominantly coupled to Gαi, which inhibits adenyl cyclase, or Gαq, which increases intracellular calcium. We and others have previously shown that the inhibitory effects of PGE2 on diverse lung fibroblast functions are primarily mediated via ligation of the EP2 receptor, which is the most abundantly expressed EP receptor in fibroblasts, and increased cAMP (11, 25, 30). To determine the receptors through which PGE2 signals to increase DNMT3a expression, we treated cells with EP receptor-specific ligands and observed that only the EP2 agonist was able to increase DNMT3a mRNA (Fig. 5C). Furthermore, the direct adenyl cyclase activator forskolin also was able to increase DNMT3a expression. Among the two Gαs-associated EP receptors, inhibition of the EP2 receptor, but not of the EP4 receptor, was able to abrogate the effects of PGE2 on DNMT3a expression (Fig. 5D). These findings suggest that PGE2 increases DNMT3a expression via EP2-cAMP signaling. Fittingly, treatment with thromboxane A2, which has been shown to decrease cAMP levels via its Gαi-coupled TP receptor, resulted in a decrease in DNMT3a expression (Fig. 5B).
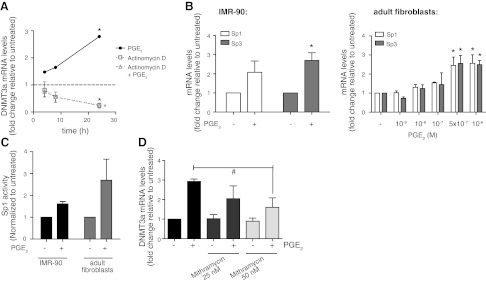
To determine whether the increase in DNMT3a expression by PGE2 is due to increased transcription or decreased degradation of its mRNA, we tested whether the transcription inhibitor actinomycin D interfered with the ability of PGE2 to promote DNMT3a mRNA accumulation over time (Fig. 6A). Data for cells treated with actinomycin D alone indicated that DNMT3a mRNA degraded by ∼80% in 24 h. PGE2 did not change the rate of degradation but was unable to increase DNMT3a mRNA in the presence of actinomycin D treatment. This suggests that the increase in DNMT3a by PGE2 is due to de novo transcription.
Figure 6.
PGE2 regulates DNMT3a expression by enhancing Sp1 and Sp3 transcription factor expression and activity. A) IMR-90 cells were treated alone or in combination with either PGE2 (500 nM) or with the transcription inhibitor actinomycin D (2.5 μg/ml) for the indicated times, and DNMT3a mRNA levels were assayed by real-time RT-PCR. Levels of DNMT3a mRNA are expressed relative to the no-treatment control at each time point. n = 3. B) IMR-90 (left panel, n=3) and adult lung fibroblasts (right panel, n=3) were treated with PGE2 (500 nM unless otherwise indicated) for 24 h, and levels of Sp1 and Sp3 mRNA were assayed by real-time RT-PCR. Levels of Sp1 and Sp3 mRNA are expressed as fold change relative to the no-PGE2 control. C) Sp1 activity was assayed in IMR-90 and adult lung fibroblasts treated with or without PGE2 (500 nM) for 24 h, as described in Materials and Methods. Activity values were normalized to the no-PGE2 control. n = 2 for IMR-90; n = 3 for adult lung fibroblasts. D) IMR-90 cells were treated for 24 h with or without PGE2 and the Sp1/Sp3 inhibitor mithramycin at the indicated concentrations and assayed for DNMT3a mRNA levels by real-time RT-PCR. n = 3. All results are expressed as means ± se. *P < 0.05 vs. untreated. #P < 0.05 for PGE2-treated cells with vs. without 50 mM mithramycin.
Previous studies have shown that DNMT3a expression can be dictated by the transcription factors Sp1 and Sp3 (31). To determine whether the increase in DNMT3a expression by PGE2 is due to up-regulation of Sp1 and/or Sp3, we assayed the expression and activity of these transcription factors. PGE2 increased Sp1 and Sp3 mRNA levels (Fig. 6B) as well as nuclear Sp1 activity (Fig. 6C) in both fetal and adult fibroblasts. The Sp1/Sp3 inhibitor mithramycin attenuated the ability of PGE2 to increase DNMT3a mRNA (Fig. 6D). These data show that the PGE2-mediated increase in DNMT3a is due, at least in part, to increased Sp1/Sp3 transcription factor expression and activity.
PGE2 decreases DNMT expression in RAW 264.7 macrophages
To determine whether the increase in DNMT3a expression by PGE2 is specific to lung fibroblasts, we examined the expression of DNMTs in RAW 264.7 macrophages after treatment with PGE2. Interestingly, PGE2 decreased the expression of DNMT1 and DNMT3a in RAW cells (Fig. 7), suggesting that the PGE2 effects on DNA methylation machinery are cell type-specific.
Figure 7.
Effect of PGE2 on DNMT expression in RAW 264.7 macrophages. RAW 264.7 cells were treated with PGE2 (1 nM to 10 μM) for 24 h and assayed for DNMT mRNA levels by quantitative real-time RT-PCR (n=3; A) or treated for 48 h and assayed for DNMT protein expression by immunoblot (n=2; B). Representative immunoblot is shown. Results are expressed as means ± se. *P < 0.05 vs. untreated.
DISCUSSION
Although patterns of DNA methylation are highly plastic and sensitive to environmental cues (1, 2, 32), the mechanisms by which environmental perturbations direct changes in DNA methylation are not well understood. Synthesized from arachidonic acid via the COX pathway, PGE2 is produced by nearly all cells and exerts a myriad of bioactive actions. PGE2 regulates fever, pain, and local edema during inflammation (10); it inhibits host defense against acute infection (14); it promotes proliferation of epithelial cells (33); it regulates T-cell differentiation (34); and it inhibits fibroblast functions (11, 35). Increased PGE2 promotes carcinogenesis (33), whereas diminished PGE2 promotes fibroproliferation (36), renal dysfunction (37), and gastric cytotoxicity (38). Fundamental processes such as differentiation, proliferation, and tumorigenesis are often associated with distinct changes in DNA methylation patterns. Here we show for the first time that exogenous PGE2 and endogenous PGE2 production can directly increase fibroblast global and gene-specific DNA methylation via increased DNMT3a expression and activity. PGE2 accomplishes this through ligation of the EP2 receptor, resulting in increased cAMP generation and increased expression of Sp1 and Sp3, transcription factors known to increase expression of DNMT3a. The increase in DNMT3a by PGE2 is specific to fibroblasts, as we found PGE2 to decrease DNMT expression in RAW macrophages. These findings provide a mechanism by which PGE2 not only is able to modulate the expression of multiple genes but also is able to maintain these changes over subsequent cell divisions.
DNMT3a is generally considered a de novo DNMT, targeting previously unmethylated CpG sites for methylation (39). It is therefore not surprising that we observed no increase in methylation of LINE-1 or other repeat elements, which are often considered to be under the control of maintenance methyltransferases such as DNMT1. Instead, we identified a specific group of genes whose promoters exhibited increased methylation with PGE2 treatment. The identification of select targets susceptible to hypermethylation by PGE2 is congruent with the notion that DNMT3a exhibits gene specificity in its methylation targets. Using an unbiased enrichment analysis of genes identified to be hypermethylated by PGE2 treatment, we were able to identify three functional clusters into which these genes can be categorized. Two of these clusters include genes involved in cell cycle and tumor suppression, in keeping with the known ability of PGE2 to inhibit fibroblast proliferation (11) and promote tumorigenesis (15). We independently performed bisulfite sequencing to validate hypermethylation in two of the genes identified from these clusters, IGFBP2 and MGMT. The hypermethylation of MGMT was associated with a decrease in protein expression as well as increased susceptibility to temozolamide, a DNA-damaging agent. Another functional cluster, DNA methylation, was also identified by enrichment analysis, suggesting that PGE2 may regulate, by hypermethylation, other genes involved in DNA methylation machinery. Finally, it has also been suggested that DNMT3a selectively methylates imprinted genes (40), one of which, CDKN1C, was identified in the CpG array we performed. Identifying genes that are hypermethylated by PGE2 may in fact aid in the discovery of previously unrecognized imprinted genes.
Many of the genes that we identified to be hypermethylated by PGE2 in IMR-90 cells differed from those in adult normal fibroblasts, suggesting that these targets are also cell-specific. It has been noted that cellular expression of DNMT3a increases during embryonic development, only to decrease to lower but still measurable levels in adult cell populations (27, 39). These differences in fetal vs. adult fibroblasts suggest that DNMT3a may target different genes for methylation in cells of different ages or in organs at different stages of development. Increases in DNMT3a by PGE2 were also noted to be specific to fibroblasts, because RAW macrophages exhibited a decrease in expression of DNMT1 and DNMT3a with PGE2 treatment. Indeed, a recent article by Xia et al. (41) reported that PGE2 increased expression of a different set of DNMTs, DNMT1 and DNMT3b, in colon cancer cells. Interestingly, MGMT was one of the genes identified to be hypermethylated by PGE2 via DNMT3b in that study (41), suggesting that different DNMT isoforms may also up-regulate similar genes but in different cellular contexts.
To our knowledge, the only transcription factors identified to regulate DNMT3a expression are Sp1 and Sp3 (31). Here, we were able to show that PGE2, via ligation of EP2- and Gαs-dependent increases in cAMP increased Sp1 and Sp3 expression and activity, leading to increased DNMT3a expression. Others have shown that PGE2, via increased cAMP production, increases Sp1 expression and activity in other cellular contexts (42, 43). It is likely that the time required for the transcriptional and translational up-regulation of these genes accounts for why the increase in global DNA methylation by PGE2 was not observed until 48 h. Similar increases in DNMT3a expression were observed in cells whose PGE2 was removed after 1 h of exposure, indicating that early signaling events were sufficient to trigger the downstream changes in DNA methylation. It is not surprising that the increase in DNMT3a expression was mediated by EP2 ligation and signaling, because this is the most abundant EP receptor in lung fibroblasts (11) and has been shown to modulate most of the other inhibitory actions by PGE2 in these cells (11, 12, 25). The half-maximal effective concentration of PGE2 to increase DNMT3a expression was similar to that necessary for the inhibition of other fibroblast functions. The ability of endogenous PGE2, via COX-2 regulation, to elicit these same effects supports the physiological relevance of these findings. The effect of PGE2 on DNMT3a expression may differ in other cell types with different EP receptor profiles. Treatment with the prostanoid thromboxane A2, which signals through the Gαi-coupled TP receptor and decreases cAMP, resulted in decreased DNMT3a expression. The finding that cAMP signaling leads to increased DNMT3a expression suggests that other endogenous or exogenous ligands that activate the cAMP pathway, such as adenosine and β-adrenergic agonists, may also up-regulate DNMT3a expression and global DNA methylation. In contrast and as suggested by our observations with the thromboxane agonist, mediators acting via Gαi-coupled receptors, including, for example, chemokines, may have the opposite effect.
Although mammalian DNMT3a was first cloned and discovered in 1998 (27), many aspects of its structure and function remain poorly understood. Somatic point mutations that are independently associated with a poor outcome in acute myeloid leukemia have been identified in DNMT3A (44). Although DNMT3a is ubiquitously expressed in most tissues, the alternatively spliced variant DNMT3a2 is found predominantly in embryonic tissue (45), and it is not known how its expression is regulated. In our experiments, DNMT3a mRNA levels decrease by 75% by 24 h when transcription is inhibited. There are few studies that address the stability of DNMT3a protein, the regulation of its subcellular localization, the mechanisms by which it selectively targets CpG sites for methylation, or the post-translational modifications of the enzyme. Further studies will be required to ascertain whether PGE2 influences any of these other aspects of enzyme regulation or function.
In our array analysis, setting a modest threshold of a 1.5-fold increase in methylation identified 52 genes in fetal IMR-90 cells and 48 genes in adult cells that were hypermethylated by PGE2. The Illumina HumanMethylation27 array assays, on average, 1 to 2 CpG sites/gene, leaving many CpG sites unevaluated. Given that ELISA-based assays of global methylation showed a 2- to 3-fold increase in methylation by PGE2, we suspect that there may be other unidentified genes or chromosomal regions that are also hypermethylated by this prostanoid. Newer generation (e.g., Illumina HumanMethylation450) bead chip arrays or other methods of genome-wide bisulfite sequencing are required to identify other hypermethylated regions within gene promoters or in pericentromeric heterochromatin. In addition, we were unable to detect mRNA expression of DNMT3L (data not shown), a catalytic enhancer of DNMT3a (46). Enhancement of DNMT3L expression in the presence of PGE2 up-regulation of DNMT3a expression would substantially amplify hypermethylation.
The ability of the environment to shape the DNA methylome is well recognized, and methylation is felt to represent a major mechanism by which environmental exposures contribute to disease susceptibility (1, 47). Diet and levels of micronutrients, such as folate, have been shown to exert a profound effect on the methylation patterns found in mothers and their offspring (2, 32). In contrast, understanding how the methylome is affected by environmental signals that do not affect methyl donor levels is more challenging. Aging (48), smoking (49), and heavy metal exposure (50) have all been associated with changes in the DNA methylome, and, in some cases, with changes in DNMT expression, but the mechanisms by which these changes occur are largely unknown. Here, we show that PGE2 can alter the methylation landscape through direct up-regulation and increased activity of DNMT3a. These changes have significant clinical relevance, because PGE2 generation is up-regulated in inflammation, infection, and cancer and down-regulated in fibrosis. Polymorphisms in COX-2 have been shown to modulate levels of PGE2 production (51) and diminished EP2 receptor expression, which has been observed in pulmonary fibrosis, has been shown to impair PGE2 responsiveness (52). A recent study showed that bisphenol A decreases PGE2 biosynthesis (53), which may therefore explain the DNA hypomethylating effect of this environmental toxin (32). Finally, nonsteroidal anti-inflammatory drugs, which inhibit the catalytic activity of COX, are over-the-counter medications used virtually indiscriminately for fever and pain. Our findings allow us to speculate that changes in PGE2 biosynthesis or signaling may contribute to alterations in DNA methylation patterns associated with a wide variety of diseases and environmental exposures. Moreover, the ability of cAMP to modulate DNA methylation suggests that our findings with PGE2 may extend to other G-protein-coupled receptor ligands that signal by increasing or decreasing this critical second messenger.
Supplementary Material
Acknowledgments
The authors thank Drs. Robert Lyons and Susan L. Dagenais for their assistance with the HumanMethylation27 CpG array.
This work was funded by a grant from the American Thoracic Society Coalition for Pulmonary Fibrosis Foundation and by grants HL094657 (to S.K.H.), HL094311 (to M.P.-G.), AG013283, and P30 ES017885 from the U.S. National Institutes of Health.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- COX
- cyclooxygenase
- DNMT
- DNA methyltransferase
- EP
- E prostanoid
- LINE
- long interspersed element
- MGMT
- O6-methylguanine DNA methyltransferase
- PARP
- poly(ADP-ribose) polymerase
- PGE2
- prostaglandin E2
- PGI2
- prostaglandin I2 (prostacyclin)
- siRNA
- small interfering RNA
REFERENCES
- 1. Fraga M. F., Ballestar E., Paz M. F., Ropero S., Setien F., Ballestar M. L., Heine-Suner D., Cigudosa J. C., Urioste M., Benitez J., Boix-Chornet M., Sanchez-Aguilera A., Ling C., Carlsson E., Poulsen P., Vaag A., Stephan Z., Spector T. D., Wu Y. Z., Plass C., Esteller M. (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U. S. A. 102, 10604–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waterland R. A., Jirtle R. L. (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meissner A., Mikkelsen T. S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B. E., Nusbaum C., Jaffe D. B., Gnirke A., Jaenisch R., Lander E. S. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Esteller M. (2008) Epigenetics in cancer N. Engl. J. Med. 358, 1148–1159 [DOI] [PubMed] [Google Scholar]
- 5. Feinberg A. P., Cui H., Ohlsson R. (2002) DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin. Cancer Biol. 12, 389–398 [DOI] [PubMed] [Google Scholar]
- 6. Ballestar E., Esteller M., Richardson B. C. (2006) The epigenetic face of systemic lupus erythematosus. J. Immunol. 176, 7143–7147 [DOI] [PubMed] [Google Scholar]
- 7. North M. L., Ellis A. K. (2011) The role of epigenetics in the developmental origins of allergic disease. Ann. Allergy Asthma Immunol. 106, 355–361; quiz 362 [DOI] [PubMed] [Google Scholar]
- 8. Hagood J. S., Prabhakaran P., Kumbla P., Salazar L., MacEwen M. W., Barker T. H., Ortiz L. A., Schoeb T., Siegal G. P., Alexander C. B., Pardo A., Selman M. (2005) Loss of fibroblast Thy-1 expression correlates with lung fibrogenesis. Am. J. Pathol. 167, 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang S. K., Fisher A. S., Scruggs A. M., White E. S., Hogaboam C. M., Richardson B. C., Peters-Golden M. (2010) Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am. J. Pathol. 177, 2245–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Funk C. D. (2001) Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294, 1871–1875 [DOI] [PubMed] [Google Scholar]
- 11. Huang S., Wettlaufer S. H., Hogaboam C., Aronoff D. M., Peters-Golden M. (2007) Prostaglandin E2 inhibits collagen expression and proliferation in patient-derived normal lung fibroblasts via E prostanoid 2 receptor and cAMP signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L405–L413 [DOI] [PubMed] [Google Scholar]
- 12. Kolodsick J. E., Peters-Golden M., Larios J., Toews G. B., Thannickal V. J., Moore B. B. (2003) Prostaglandin E2 inhibits fibroblast to myofibroblast transition via E. prostanoid receptor 2 signaling and cyclic adenosine monophosphate elevation. Am. J. Respir. Cell Mol. Biol. 29, 537–544 [DOI] [PubMed] [Google Scholar]
- 13. White E. S., Atrasz R. G., Dickie E. G., Aronoff D. M., Stambolic V., Mak T. W., Moore B. B., Peters-Golden M. (2005) Prostaglandin E2 inhibits fibroblast migration by E-prostanoid 2 receptor-mediated increase in PTEN activity. Am. J. Respir. Cell Mol. Biol. 32, 135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aronoff D. M., Canetti C., Peters-Golden M. (2004) Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J. Immunol. 173, 559–565 [DOI] [PubMed] [Google Scholar]
- 15. Greenhough A., Smartt H. J., Moore A. E., Roberts H. R., Williams A. C., Paraskeva C., Kaidi A. (2009) The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 30, 377–386 [DOI] [PubMed] [Google Scholar]
- 16. Sugimoto Y., Narumiya S. (2007) Prostaglandin E receptors. J. Biol. Chem. 282, 11613–11617 [DOI] [PubMed] [Google Scholar]
- 17. Fine A., Poliks C. F., Donahue L. P., Smith B. D., Goldstein R. H. (1989) The differential effect of prostaglandin E2 on transforming growth factor-β and insulin-induced collagen formation in lung fibroblasts. J. Biol. Chem. 264, 16988–16991 [PubMed] [Google Scholar]
- 18. Liu C., Xu D., Sjoberg J., Forsell P., Bjorkholm M., Claesson H. E. (2004) Transcriptional regulation of 15-lipoxygenase expression by promoter methylation. Exp. Cell Res. 297, 61–67 [DOI] [PubMed] [Google Scholar]
- 19. Toyota M., Shen L., Ohe-Toyota M., Hamilton S. R., Sinicrope F. A., Issa J. P. (2000) Aberrant methylation of the cyclooxygenase 2 CpG island in colorectal tumors. Cancer Res. 60, 4044–4048 [PubMed] [Google Scholar]
- 20. Ostling O., Johanson K. J. (1984) Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 123, 291–298 [DOI] [PubMed] [Google Scholar]
- 21. Du P., Zhang X., Huang C. C., Jafari N., Kibbe W. A., Hou L., Lin S. M. (2010) Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 23. Kochanek S., Renz D., Doerfler W. (1993) DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 12, 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sartor M. A., Mahavisno V., Keshamouni V. G., Cavalcoli J., Wright Z., Karnovsky A., Kuick R., Jagadish H. V., Mirel B., Weymouth T., Athey B., Omenn G. S. (2010) ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics 26, 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang S. K., White E. S., Wettlaufer S. H., Grifka H., Hogaboam C. M., Thannickal V. J., Horowitz J. C., Peters-Golden M. (2009) Prostaglandin E2 induces fibroblast apoptosis by modulating multiple survival pathways. FASEB J. 23, 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Balana C., Ramirez J. L., Taron M., Roussos Y., Ariza A., Ballester R., Sarries C., Mendez P., Sanchez J. J., Rosell R. (2003) O6-methyl-guanine-DNA methyltransferase methylation in serum and tumor DNA predicts response to 1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastoma multiforme. Clin. Cancer Res. 9, 1461–1468 [PubMed] [Google Scholar]
- 27. Okano M., Xie S., Li E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219–220 [DOI] [PubMed] [Google Scholar]
- 28. Pradhan S., Bacolla A., Wells R. D., Roberts R. J. (1999) Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 274, 33002–33010 [DOI] [PubMed] [Google Scholar]
- 29. Wilborn J., Crofford L. J., Burdick M. D., Kunkel S. L., Strieter R. M., Peters-Golden M. (1995) Cultured lung fibroblasts isolated from patients with idiopathic pulmonary fibrosis have a diminished capacity to synthesize prostaglandin E2 and to express cyclooxygenase-2. J. Clin. Invest. 95, 1861–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choung J., Taylor L., Thomas K., Zhou X., Kagan H., Yang X., Polgar P. (1998) Role of EP2 receptors and cAMP in prostaglandin E2 regulated expression of type I collagen α1, lysyl oxidase, and cyclooxygenase-1 genes in human embryo lung fibroblasts. J. Cell. Biochem. 71, 254–263 [PubMed] [Google Scholar]
- 31. Jinawath A., Miyake S., Yanagisawa Y., Akiyama Y., Yuasa Y. (2005) Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem. J. 385, 557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dolinoy D. C., Huang D., Jirtle R. L. (2007) Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. U. S. A. 104, 13056–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gupta R. A., Dubois R. N. (2001) Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat. Rev. Cancer 1, 11–21 [DOI] [PubMed] [Google Scholar]
- 34. Okano M., Sugata Y., Fujiwara T., Matsumoto R., Nishibori M., Shimizu K., Maeda M., Kimura Y., Kariya S., Hattori H., Yokoyama M., Kino K., Nishizaki K. (2006) E prostanoid 2 (EP2)/EP4-mediated suppression of antigen-specific human T-cell responses by prostaglandin E2. Immunology 118, 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang S. K., Wettlaufer S. H., Chung J., Peters-Golden M. (2008) Prostaglandin E2 inhibits specific lung fibroblast functions via selective actions of PKA and Epac-1. Am. J. Respir. Cell Mol. Biol. 39, 482–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borok Z., Gillissen A., Buhl R., Hoyt R. F., Hubbard R. C., Ozaki T., Rennard S. I., Crystal R. G. (1991) Augmentation of functional prostaglandin E levels on the respiratory epithelial surface by aerosol administration of prostaglandin E. Am. Rev. Respir. Dis. 144, 1080–1084 [DOI] [PubMed] [Google Scholar]
- 37. DiBona G. F. (1986) Prostaglandins and nonsteroidal anti-inflammatory drugs. Effects on renal hemodynamics. Am. J. Med. 80, 12–21 [DOI] [PubMed] [Google Scholar]
- 38. Robert A., Nezamis J. E., Lancaster C., Hanchar A. J. (1979) Cytoprotection by prostaglandins in rats. Prevention of gastric necrosis produced by alcohol, HCl, NaOH, hypertonic NaCl, and thermal injury. Gastroenterology 77, 433–443 [PubMed] [Google Scholar]
- 39. Okano M., Bell D. W., Haber D. A., Li E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 40. Hata K., Okano M., Lei H., Li E. (2002) Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 [DOI] [PubMed] [Google Scholar]
- 41. Xia D., Wang D., Kim S. H., Katoh H., DuBois R. N. (2012) Prostaglandin E2 promotes intestinal tumor growth via DNA methylation. Nat. Med. 18, 224–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zheng Y., Ritzenthaler J. D., Sun X., Roman J., Han S. (2009) Prostaglandin E2 stimulates human lung carcinoma cell growth through induction of integrin-linked kinase: the involvement of EP4 and Sp1. Cancer Res. 69, 896–904 [DOI] [PubMed] [Google Scholar]
- 43. Bradbury D., Clarke D., Seedhouse C., Corbett L., Stocks J., Knox A. (2005) Vascular endothelial growth factor induction by prostaglandin E2 in human airway smooth muscle cells is mediated by E prostanoid EP2/EP4 receptors and SP-1 transcription factor binding sites. J. Biol. Chem. 280, 29993–30000 [DOI] [PubMed] [Google Scholar]
- 44. Ley T. J., Ding L., Walter M. J., McLellan M. D., Lamprecht T., Larson D. E., Kandoth C., Payton J. E., Baty J., Welch J., Harris C. C., Lichti C. F., Townsend R. R., Fulton R. S., Dooling D. J., Koboldt D. C., Schmidt H., Zhang Q., Osborne J. R., Lin L., O'Laughlin M., McMichael J. F., Delehaunty K. D., McGrath S. D., Fulton L. A., Magrini V. J., Vickery T. L., Hundal J., Cook L. L., Conyers J. J., Swift G. W., Reed J. P., Alldredge P. A., Wylie T., Walker J., Kalicki J., Watson M. A., Heath S., Shannon W. D., Varghese N., Nagarajan R., Westervelt P., Tomasson M. H., Link D. C., Graubert T. A., DiPersio J. F., Mardis E. R., Wilson R. K. (2010) DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weisenberger D. J., Velicescu M., Preciado-Lopez M. A., Gonzales F. A., Tsai Y. C., Liang G., Jones P. A. (2002) Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells. Gene 298, 91–99 [DOI] [PubMed] [Google Scholar]
- 46. Gowher H., Liebert K., Hermann A., Xu G., Jeltsch A. (2005) Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 280, 13341–13348 [DOI] [PubMed] [Google Scholar]
- 47. Jirtle R. L., Skinner M. K. (2007) Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y., Liu Y., Strickland F. M., Richardson B. (2010) Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity and acute coronary syndromes. Exp. Gerontol. 45, 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu H., Zhou Y., Boggs S. E., Belinsky S. A., Liu J. (2007) Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene 26, 5900–5910 [DOI] [PubMed] [Google Scholar]
- 50. Jiang G., Xu L., Song S., Zhu C., Wu Q., Zhang L., Wu L. (2008) Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology 244, 49–55 [DOI] [PubMed] [Google Scholar]
- 51. Szczeklik W., Sanak M., Szczeklik A. (2004) Functional effects and gender association of COX-2 gene polymorphism G-765C in bronchial asthma. J. Allergy Clin. Immunol. 114, 248–253 [DOI] [PubMed] [Google Scholar]
- 52. Huang S. K., Wettlaufer S. H., Hogaboam C. M., Flaherty K. R., Martinez F. J., Myers J. L., Colby T. V., Travis W. D., Toews G. B., Peters-Golden M. (2008) Variable prostaglandin E2 resistance in fibroblasts from patients with usual interstitial pneumonia. Am. J. Respir. Crit. Care Med. 177, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kristensen D. M., Skalkam M. L., Audouze K., Lesne L., Desdoits-Lethimonier C., Frederiksen H., Brunak S., Skakkebaek N. E., Jegou B., Hansen J. B., Junker S., Leffers H. (2011) Many putative endocrine disruptors inhibit prostaglandin synthesis. Environ. Health Perspect. 119, 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.