Abstract
The manipulation of nutrient-signaling pathways in yeast has uncovered the impact of environmental growth conditions in longevity. Studies using calorie restriction show that reducing glucose concentration of the culture media is sufficient to increase replicative and chronological lifespan (CLS). Other components of the culture media and factors such as the products of fermentation have also been implicated in the regulation of CLS. Acidification of the culture media mainly due to acetic acid and other organic acids production negatively impacts CLS. Ethanol is another fermentative metabolite capable of inducing CLS reduction in aged cells by yet unknown mechanisms. Recently, ammonium was reported to induce cell death associated with shortening of CLS. This effect is correlated to the concentration of NH4 + added to the culture medium and is particularly evident in cells starved for auxotrophy-complementing amino acids. Studies on the nutrient-signaling pathways regulating yeast aging had a significant impact on aging-related research, providing key insights into mechanisms that modulate aging and establishing the yeast as a powerful system to extend knowledge on longevity regulation in multicellular organisms.
1. Cellular Pathways Modulating Aging
Aging is common to most living organisms ranging from bacteria, a unicellular prokaryotic organism, to multicellular eukaryotic organisms like humans. It is a complex biological process that involves accumulation of damage at diverse components of the organism leading ultimately to the loss of function and demise [1–3]. In the aging process, cellular activities compromised are modulated by a network of nutrient and energy sensing signaling pathways that are highly conserved among organisms. These pathways include the insulin/insulin-like growth factor 1 (Ins/IGF-1), the protein kinase/target of rapamycin (TOR), and adenylate cyclase/protein kinase A (AC/PKA) pathways [4]. Pioneering studies using mutations in key genes of these pathways have shown an increase by threefold or more in the lifespan of model organisms like yeast [5, 6], fruit flies [7], worms [8, 9], and mice [10, 11]. Many of these mutations which extend lifespan decrease the activity of the nutrient-signaling pathways mimicking a starvation state during which oxidative stress responses are induced, reducing the levels of reactive oxygen species (ROS) and oxidative damage to macromolecules [12]. Accordingly, it has been shown in different aging models (yeast, flies, worms, fish, rodents, and rhesus monkeys) that reducing growth factors/nutrients intake has profound positive effects in extension of lifespan and also improves overall health by delaying or reducing age-related diseases in mammals including diabetes, cancer and cardiovascular diseases (reviewed in [4]). One of the first models to implicate growth- /nutrient-sensing signaling with longevity was the nematode worm Caenorhabditis elegans. In this model it was shown that the recessive mutation in age-I coding for phosphatidylinositol 3-Kinase (PI3K) extends lifespan significantly [8]. Also in C. elegans, the insulin/IGF-1 pathway was linked to longevity by the discovery that mutating the gene coding for an insulin/IGF-1 receptor ortholog, DAF-2, doubled its lifespan [9]. This lifespan extension was dependent on the reduction of activity of Daf-2 and consequently of its downstream effector PI3K (encoded by age-1), and the subsequent activation of Daf-16, a Forkhead FoxO family transcription factor (FOXO), which regulates several genes involved in stress response, antimicrobial activity, and detoxification of xenobiotics and free radicals [4, 9, 13]. Another pathway involved in longevity regulation in C. elegans is the conserved TOR-S6K (Ribosomal S6 protein Kinase) pathway. This pathway interacts with the insulin/IGF-1 pathway converging on the worm ortholog of regulatory associated protein of mTOR, Daf-15, to regulate larval development, metabolism, and longevity [14] and so downregulation of its activity results in extended lifespan [4].
The fruit fly Drosophila melanogaster is a more complex model, allowing studies based on sex differences. As in C. elegans, reducing the activity of the insulin/IGF-1 pathway mediates cellular protection mechanisms and the extension of lifespan in this organism. Mutations in the insulin-like receptor favour the extension of lifespan yielding dwarf sterile flies with females showing up to 85% extension of adult longevity [15]. Downregulation of the TOR pathway in flies, similarly to C. elegans, was shown to increase lifespan when inactivated pharmacologically with rapamycin or with overexpression of dominant-negative forms of S6K or TSC1 or TSC2, which encode negative regulators of TOR [16, 17].
Identical outcomes for genetic or pharmacologic manipulation of insulin/IGF-1 and TOR pathways and for dietary restriction regimes were observed in D. melanogaster and C. elegans, as well as in yeasts, establishing the evolutionary conserved roles of these pathways in determining lifespan and implicating them as mediators of the protective effects of dietary restriction in different species [4, 18].
In mammals, hormones of the endocrine system, the growth hormone, insulin-like growth factor-1 (IGF-1), and insulin pathways are key players in the hormonal control of aging in association with an increase of antioxidant defenses and increased stress resistance (reviewed in [19]). Deficiency in levels of circulating growth hormone has been shown to enhance antioxidant defenses and stress resistance, reduce tumor burden, and to increase insulin sensitivity (reviewed in [20]). Enhanced insulin sensitivity is a common feature of long-living mutant mice and in humans, studies of centenarian populations strongly correlates this increase in insulin sensitivity with longevity [21, 22]. IGF-1 and insulin also modulate TOR activity through Akt kinase which is a downstream effector of the insulin/IGF-1 pathway [23], and inhibition of mTOR pathway by rapamycin [24] or deletion of its downstream effector S6K, increases mice lifespan [25]. The lifespan extension due to the deletion of S6K was accompanied by slower progression of age-related pathologies and in particular slower loss of insulin sensitivity [25].
Another pathway involved in longevity regulation is the AC/PKA pathway that is conserved from yeast to mammals. Downregulation of the Ras/AC/PKA pathway was first shown in yeast to have a major effect on lifespan extension [4, 6, 26]. Only recently, studies correlating AC/PKA pathway with aging and age-related diseases started to emerge in mammals. Deletion of the mouse AC type 5, which mediates PKA activity by modulating cAMP levels, was reported to significantly increase lifespan, as it does in yeast [27], and improve cardiac stress resistance [28]. Likewise as described for yeast [26], deletions of PKA subunits in mice have recently been shown to increase lifespan while protecting against age-related deleterious changes such as weight gain, hypertrophic liver, and cardiac dysfunction [29]. Although the subunits deleted are the regulatory subunits (RIIβ) and not the catalytic subunits like in yeast (TPK), loss of RIIβ in mice causes a concomitant and compensatory decrease in catalytic subunits showing a mechanistic association between loss of these subunits and lifespan extension [29]. This converging result in such divergent models suggests a highly conserved role for PKA in longevity and opens the possibility for new therapeutic targets for aging and obesity.
In mammals the Ras proteins do not directly signal to PKA through AC [30] as it occurs in yeast [31]; however, a recent study reported that homozygous deletion of Ras-GRF1 promotes both median and maximum longevity in mice [32]. Ras-GRF1 is a guanine nucleotide exchange factor (GEF) responsible for activating Ras by favouring its GTP-bound state [33], suggesting that the cause of longevity extension of the Ras-GRF1 deletion could be the reduction of Ras activity [34]. Therefore, the Ras pathway appears as a conserved pathway in the aging process from yeast to mammals [34].
Yeast has emerged as a highly exploited model to study the environmental and genetic factors affecting longevity. In particular, the genetic tools now available make yeast one of the best established experimental model organisms for screening genes involved in the regulation of fundamental cellular process including the pathways controlling lifespan. In the following sections we focus on the particular case of the lifespan in yeast and its modulation triggered by extrinsic culture medium factors. We start with basic aspects of both replicative and chronological lifespan in yeast. The subsequent sections are dedicated to an overview highlighting the impact of culture medium and products of fermentation on the yeast chronological lifespan.
2. Chronological and Replicative Lifespan in Yeast
Two yeast lifespan models have been characterized: replicative lifespan (RLS) and chronological lifespan (CLS) (reviewed in [35]). RLS is defined as the total number of times a single mother cell can undergo a mitotic event and originate daughter cells before senescence [36]. RLS is accurately measured by moving and counting small daughters away from the mothers via microscopic manipulation and simulates aging of mitotically active mammalian cells [35, 37, 38]. On the other hand, CLS defines the length of time nondividing yeast cells remain viable [39] thus simulating aging of the postmitotic mammalian cells [40, 41]. This viability is assessed by cells reentering the cell cycle after transfer from the depleted medium or water to nutrient complete-medium [35]. In CLS, two types of metabolic yeast cells can be studied: postdiauxic or stationary phase cells. Both metabolic state cells are grown in synthetic complete (SDC) medium but while postdiauxic cells are kept in the culture medium, the stationary phase ones are transferred to water (extreme calorie restriction) after 3 days of growth. Some protocols for postdiauxic cells could also use cells grown in YPD (yeast extract, peptone, dextrose) medium [35, 42] instead of SDC medium.
The two paradigms of aging in yeast, CLS and RLS, have become useful tools to compare the aging process in proliferating and nonproliferating cells as well as to serve as models to study the mechanics of the aging process in mitotic and postmitotic cells of multicellular organisms [38].
The yeast S. cerevisiae divides by budding and therefore undergoes asymmetrical cell division, with the mother cell retaining more volume than the daughter cell. In this asymmetric division mother cells retain most of the age-associated damage, thus sacrificing individual replicative capacity while daughter cells retain full replicative potential [43, 44]. One of the aging factors affecting RLS is the accumulation of extrachromosomal ribosomal DNA circles (ERCs) [45]. These circular DNA molecules are self-replicating units formed in the nucleus by homologous recombination between adjacent rDNA repeats which segregate asymmetrically to the mother-cell nucleus during cell division. During each division ERCs replicate leading to an exponential accumulation in the mother cell and consequently to cell senescence [44, 45]. This finding in yeasts came in large part from the study of important age-related proteins called sirtuins. Sirtuins are NAD+-dependent protein deacetylases involved in chromatin silencing and known to mediate longevity in yeast, nematodes, flies, and mammals [40, 44, 46]. Deletion of SIR2 decreases RLS and its overexpression increases RLS showing that Sir2p mediates RLS in yeast most probably by regulating rDNA recombination and ERCs formation [47]. ERCs appear to be an aging factor specific to yeast, although without relevance in nondividing yeast cells (CLS) and so far without a role in aging of multicellular eukaryotes [46, 48].
Another factor known to decrease longevity both in CLS as RLS is accumulation of oxidative damage due to the production of reactive oxygen species (ROS). Deletion of the yeast antioxidant defense enzymes superoxide dismutases (SOD), reduces significantly CLS [39] and RLS [49]. However, overexpression of cytosolic (SOD1) and mitochondrial (SOD2) superoxide dismutases increased longevity of nondividing cells [50] while it decreased RLS. Although studies show several similarities but also major differences between CLS and RLS mechanistic regulation, these two models are interconnected as RLS decreases in chronologically aged cells [48]. In addition, both aging models are regulated by the nutrient-signaling kinases, as screenings for long-lived mutants identified the same gene mutations in both paradigms [27, 51–53].
3. Glucose-Signaling Pathways Involved in Yeast Longevity
The most common dietary regimes used to study the interaction between nutrient-signaling pathways and longevity include: dietary restriction (DR) in which the intake of nutrients, but not necessarily calories, is reduced without causing malnutrition; calorie restriction (CR), a regime in which only calories are reduced without compromising other nutrients, for instance amino acids and vitamins [54, 55].
In yeast, when studying both replicative lifespan (RLS) and chronological lifespan (CLS), several results correlating environmental growth conditions and longevity emerged. Many studies, including those using calorie restriction (CR), showed that reducing the glucose or amino acids concentrations of the culture media is sufficient to increase replicative and chronological lifespan [56–60]. The composition of culture media has proven to be an extrinsic factor affecting chronological lifespan but this is still giving rise to different interpretations on longevity regulation.
The manipulation of nutrient-signaling pathways for the study of aging regulating mechanisms, as previously mentioned, can be accomplished by genetic manipulations of key components of these pathways or by dietary (DR) and calorie restriction (CR). In yeast, the vast majority of protocols for CR are based on the decrease of the glucose concentration in the medium from the standard 2% to 0.5 or 0.05%. The latter (0.05% glucose) is considered extreme calorie restriction as well as the one achieved by transferring cells grown in 2% glucose to water [35, 61].
The first studies to report glucose as an agent affecting lifespan in yeast were conducted by Granot and coworkers who showed that addition of glucose to stationary-phase cells previously transferred to water leads to a reduction of CLS [62]. The authors further demonstrated that glucose, in the absence of other complementing nutrients, induces apoptotic cell death accompanied by an increase in ROS production [63]. Further studies in yeast have revealed that the major nutrient-signaling pathways TOR, SCH9, and Ras/AC/PKA are all involved in longevity regulation by glucose [6, 46, 58]. These pathways promote cell division and growth in response to nutrients while inhibiting the general stress response and autophagy. SCH9 was one of the first genes to be implicated in CLS [27]. Sch9p is the yeast closest homolog of the mammalian AKT/PKB and S6K, and its deletion leads to an increase in both CLS and RLS [27, 53]. Sch9p is a kinase that mediates PKA activation in the fermentable-growth-medium-(FGM)-induced pathway and also mediates many of the TOR complex 1 (TORC1) controlled processes [64–66]. Sch9p was first described as having a partially redundant role with PKA pathway, since deletion of SCH9 could be compensated by increased activity of PKA and vice versa [67], and later as a direct target of TORC1 regulation [65]. More recently, TORC1 was also identified as a target for regulating longevity in both CLS and RLS [51, 52]. The TOR pathway responds to nitrogen and carbon sources, mainly to control cell growth, through the regulation of processes such as translation initiation, ribosome biogenesis, mRNA and amino acid permeases stability, transcription of nutrient-regulated genes and stress response genes, and actin cytoskeleton organization and autophagy [68, 69]. Reduction of TORC1-Sch9p signaling was shown to promote longevity by increasing the expression of stress-response genes in a Rim15p-dependent manner [58], as RIM15 deletion reduced the lifespan extension of the long-lived sch9Δ cells [27]. Alternatively, a recent study proposes a Rim15p-independent mechanism for lifespan extension in reduced TORC1-Sch9p signaling [70]. This study shows that in tor1Δ cells, CLS is reduced if mitochondrial respiration is uncoupled. The authors suggest that during growth, mitochondrial ROS signaling downregulates both the mitochondrial membrane potential and ROS accumulation of stationary phase cells to promote their longevity [70]. This is in agreement with previous data showing that preadaptation to respiratory growth can also promote extension of CLS [71]. CR also promoted CLS extension by doubling the lifespan of the long-lived sch9Δ and tor1Δ cells by a Rim15p partially dependent mechanism. In fact, cells with a triple mutation in Rim15p downstream transcription factors (msn2Δ msn4Δ gis1Δ) do not display a reduction of CR promoted CLS extension when compared to the long-lived sch9Δ and tor1Δ cells suggesting the involvement of additional Rim15p independent transcriptional factors [58].
Another pathway involved in aging is the other major nutrient-signaling pathway Ras/AC/PKA, responsible for the link between glucose availability and the control of growth, proliferation, metabolism, stress resistance, and longevity [6, 27, 50, 72]. Deletion of RAS2 or a reduced activity of adenylate cyclase (Cyr1p), which is activated by the Ras proteins, causes lifespan extension and stress resistance [50]. Mutation in the CYR1 gene increases both RLS and CLS while deletion of RAS2 decreases RLS [53, 73]. Rim15p also mediates ras2Δ lifespan extension by enhancing cellular protection against oxidative stress through the activation of SOD2 [50], indicating that Rim15p is a common denominator of the pathways Ras/AC/PKA, Sch9p, and TOR. In addition, deletion of MSN2/4 in ras2Δ cells leads to lifespan reduction indicating that Msn2p/4p and Gis1p transcription factors controlled by Rim15p are also required for CLS extension. Nevertheless, the Rim15p downstream transcription factors (Msn2p, Msn4p, and Gis1p) appear to have different roles in sch9Δ and ras2Δ cells given that only the abrogation of GIS1, and not of MSN2/4, was shown to almost completely abolish the lifespan expansion of sch9Δ cells [27, 58]. Therefore, Sch9p and Ras2p seem to differentially modulate the common downstream effectors, which is also corroborated by the higher stress resistance and increased CLS exhibited by ras2Δ sch9Δ double knockout cells in comparison to the single deletion mutants [58, 74].
More recently, the correlation between glucose signaling, oxidative stress and aging was further addressed in a study showing that increasing glucose from the standard 2% to 10%, promotes a shortening of CLS accompanied by increased levels of intracellular superoxide anion (O2 ∙−), decreased levels of hydrogen peroxide (H2O2), reduced efficiency of stationary phase G0/G1 arrest, and activation of DNA damage [12]. On the other hand, CR by reducing glucose or by deletion of SCH9 or TOR1 extends CLS and diminishes superoxide anion levels promoting at the same time a more efficient G0/G1 arrest. These and other results point to superoxide levels as one of the key factors regulating aging [75], which is in agreement with the aforementioned results showing that reduction of signaling pathways leads to the activation of oxidative stress responses mediated by Rim15p [12]. Nevertheless, an alternative activation of oxidative stress responses independent of Rim15p [58] and mediated by H2O2 has been also reported [12]. Furthermore, high levels of H2O2, which respond to glucose in an inversely dose-dependent manner, promotes activation of SODs, leading to a reduction in superoxide anion levels and therefore to CLS extension [12, 76].
4. Amino Acid Metabolism in the Regulation of the Yeast Chronological Lifespan
In nature, yeast cells enter a resting or quiescent state in the absence of favorable nutritional conditions. When inadequate carbon, nitrogen, sulfur, or phosphorus levels are sensed by yeast cells, growth ceases and cell cycle is arrested as a survival strategy. In natural environments, yeast are prototrophs capable of synthetizing most of their metabolites from simple carbon and nitrogen sources, whereas laboratory strains commonly have auxotrophic markers that confer a nutrient-limiting growth phenotype useful for genetic manipulation. These markers are usually genes involved in the biosynthesis of specific amino acids or nucleotides. Amino acids are important nutrients that can also be recycled by autophagy. This recycling process maintains amino acid homeostasis and is crucial for cell survival under nitrogen starvation leading to rapid loss of viability in autophagy-defective mutants [77] and therefore has been implicated in CLS regulation. Curiously, it was demonstrated that prototrophic and auxotrophic strains display different responses to nutrient starvation [78]. Starvation of “natural” nutrients leads to an arrest in G0/G1 cell cycle phase of prototrophics cells, while auxotrophic cells failed to arrest the cell cycle upon starvation of “supplemental” nutrients (auxotrophic nutrients) [78]. It was also observed that auxotrophic cells limited for leucine or uracil consume glucose at a much faster rate, exhausting it from the medium, than prototrophic cells limited for phosphate, sulfate, or ammonium that spare glucose [79]. These findings clearly reveal a failure of auxotrophic cells in regulating nutrient sensing in response to starvation of “supplemental” nutrients [80]. Furthermore, limiting levels of auxotrophy-complementing amino acids, in the growth medium, induce an early arrest in G2/M phase, negatively affecting chronological longevity and leading to a premature aging phenotype [81]. In accordance, reduction of total amino acid levels, including essential ones, in the medium also decreases CLS [82]. Starvation for leucine in nondividing leucine auxotrophic cells induces a rapid loss of viability [80]. Nevertheless, this phenotype is partially dependent on the carbon source present in the starvation medium but not in that used in the growth medium. For example the presence of ethanol/glycerol or galactose in the starvation medium increases CLS in contrast to starvation in glucose [80]. However, not all essential amino acids have the same effect on CLS. In fact, methionine starvation of methionine auxotrophic cells has no effect on viability [83]. Another study also reported that from the auxotrophic-complementing amino acids, lysine, histidine, and leucine, the latter has a more pronounced negative effect in CLS in both autophagy-competent and autophagy-deficient strains [84]. The authors pointed out that the enhanced sensitivity of yeast cells to leucine starvation is correlated to the high levels of leucine codon, the most frequent amino acid codon [84]. CLS is extended by the presence of nonessential amino acids, particularly isoleucine, and its precursors threonine and valine, via the general amino acid control (GAAC) pathway. The authors proposed a mechanism for CLS regulation by the branched side chain amino acids (BCAA) leucine, isoleucine and valine, in which low levels of these amino acids induce the GAAC pathway therefore shortening CLS and vice versa [84].
Starvation for nonessential amino acids was reported to extend RLS [56] and starvation for preferred amino acids such as asparagine- or glutamate- induced CLS extension in direct proportion to the nature of the amino acid removed [52].
5. Impact of Products of Fermentation in the Yeast Chronological Lifespan
Ethanol is the main product resulting from alcoholic fermentation and it is used as a carbon source during the diauxic shift and postdiauxic phase. Nevertheless, ethanol is known to negatively affect the metabolic activity of the yeast cells by inhibiting cell growth and fermentation. It is also known to cause among others the damage of cell membranes by increasing membrane fluidity [85, 86] and the inhibition of transport systems across the plasma membrane [87, 88]. The severity of the effects is dependent on the alcohol concentration and at high ethanol levels it results in cell death [89]. Recently, ethanol was described as an apoptotic inducer [90] and has also been implicated as an extrinsic factor in aging, significantly decreasing CLS of severely calorie restricted strains (CR in water), known for their lifespan extension in this condition [91]. In contrast to wild type cells, long-lived sch9Δ cells consume all the ethanol from the medium during chronological aging, further supporting ethanol as a modulator of aging [91].
A recent study on the genetic expression profile of long-lived tor1Δ, sch9Δ, and ras2Δ cells revealed an upregulation of genes involved in the metabolism of glycerol. In contrast to wild type cells that accumulate ethanol and rapidly deplete glycerol, those long-lived mutant cells accumulate glycerol whereas ethanol was early depleted. These observations suggest that inhibition of Tor1p/Sch9p mediates a metabolic switch from biosynthesis and release of ethanol to activation of glycerol biosynthesis and its consequent release [74]. Glycerol, unlike glucose and ethanol [58, 61, 91], does not promote aging or cell death and so this metabolic change extends CLS [74, 92].
In calorie restricted cells, ethanol is completely consumed before the beginning of viability decline. Conversely, noncalorie restricted cells were unable to completely consume ethanol before viability decline. The authors suggested a correlation between ethanol accumulation and loss of peroxisome function in noncalorie restricted cells since ethanol suppresses the synthesis of certain proteins localized to peroxisomes [93].
Acetic acid is a byproduct of fermentative metabolism in yeast accumulating in the medium during fermentation of glucose to ethanol and is also one factor described to affect CLS [94, 95]. After sugar is depleted in 2% glucose standard conditions, a shift in metabolism occurs from fermentation to respiration and the metabolization of ethanol also leads to the production and accumulation of acetic acid. Acetic acid is a well-known inducer of apoptotic cell death leading to ROS production [96, 97]. In a recent study, Burtner and coworkers identified acetic acid as an important extracellular factor affecting CLS in SDC medium [94]. The authors showed that cells grown for 48 hours under extreme calorie restriction conditions (0.05% glucose concentration), known to extend CLS, rapidly loss viability if transferred to cell-free supernatants of 2% standard glucose-depleted medium, indicating that cell-extrinsic aging factors were present in the SDC depleted medium [94]. Although several other organic acids also accumulate in the culture medium during chronological aging, only acetic acid was identified as being sufficient to cause chronological aging [94]. In the same study it was also shown that buffering of aging cultures to pH 6 is sufficient to increase CLS, neutralizing the toxic effect of acetic acid. Actually, the acetate anion is not readily taken up from the environment by glucose-grown yeast cells, but the protonated acetic acid can cross the plasma membrane resulting in intracellular acidification [98]. This negative effect of acetic acid in CLS was diminished by mutational inactivation of conserved signaling pathways, namely deletion of SCH9 and RAS2, conferring resistance via unknown mechanisms [94]. SCH9 and RAS2 mutant cells are known to have a more frequent growth arrest in G1 phase when compared to the wild-type, promoted by the reduction in growth signaling in these mutants [99]. In accordance, nutrient-depleted stationary phase cells are continuously subjected to acetic acid-induced growth signals, even in the absence of glucose, that promote cell cycle progression and consequently replication stress due to the lack of favorable conditions [100]. These and other results show that acetic acid, as glucose, activates Sch9p and RAS pathways and seems to mediate cell death by promoting the accumulation of superoxide anion (O2 ∙−) in consequence of downregulation of SODs and other oxidative stress defenses by the activated pathways [12]. The long-lived ade4Δ cells (Ade4p is involved in the purine de novo biosynthetic pathway) do not accumulate acetic acid in the culture medium when compared to the wild type cells, while the short-lived atg16Δ cells (Atg16p is involved in the autophagic process) accumulate acetic acid at higher concentrations than the wild type cells, inversely correlating the amount of acid release from cells and the extension of CLS [101]. Buffering the growth media to pH 6.0 of the short-lived atg16Δ cells and the wild type strain, also dramatically increase CLS to the same levels obtained for the CR growth condition and for the long-lived ade4Δ cells, indicating that pH neutralized the toxic effects of acetic acid. Overall the results demonstrate that acetic acid can have an important impact on CLS through a cell extrinsic mechanism that is dependent on media pH.
6. Ammonium Toxicity in Aging Yeast Cells
Ammonium (NH4 +) toxicity has been well described in animals and plant systems [102]. In yeast, NH4 + is commonly used as nitrogen source for growth and it is usually not toxic having a central role in nitrogen metabolism both in degradative and biosynthetic pathways [103]. In yeast, nitrogen sources, prior to their use, need to be converted into glutamate and glutamine. However not all nitrogen sources are equally preferred and yeast can select the nitrogen sources through nitrogen catabolite repression (NCR) mechanism also known as nitrogen discrimination pathway (NDP). This pathway enables yeast to repress genes that code for proteins required for the use of poor nitrogen sources, when in the presence of sufficient quantities of rich nitrogen sources like glutamine [104].
Production of ammonia in yeast colonies has even been described as a mechanism of protection from cell death during colony development [105]. An excess of ammonium was found to be toxic for S. cerevisiae, under potassium limitation, resulting in amino acid excretion similar to the detoxifying mechanism found in mammals [106]. The authors described that ammonium toxicity in yeast is related to a “leak current” of ammonium ions that enter the cell through potassium channels, in limiting potassium conditions, and this influx causes an excess of internal ammonium that becomes toxic for the cell. To cope with this ammonium excess, cells excrete amino acids possibly through the Ssy1p-Ptr3p-Ssy5p (SPS)-system of amino acid transporters, which were found to be strongly upregulated in this condition, or by directly excreting ammonium via the Ato (Ammonium Transporter Outward) transporters [106].
Recently, we have reported that ammonium is toxic for aging cells and acts as an extrinsic factor affecting CLS [107]. In this study, it was shown that decreasing the concentration of NH4 + in the culture medium increases yeast CLS in amino acid restriction conditions. In contrast, when the initial (NH4)2SO4 concentration in the culture medium, either with or without restriction of amino acids, was increased from 0.5% to 1%, there was a decrease in cell survival, demonstrating that the toxic effects of ammonium are correlated with its concentration in the culture medium. Moreover, after transferring cultured cells to extreme calorie restriction conditions in water, the addition of ammonium drastically decreases the CLS, indicating that ammonium alone could also induce loss of cell viability as observed in culture media, and providing, for the first time, a role for ammonium in chronological longevity regulation [107].
Cells starved for auxotrophic-complementing amino acids are particularly sensitive to ammonium-induced cell death [107]. Death induced by ammonium in these cells is mediated through the regulation of the evolutionary conserved pathways PKA, TOR, and SCH9 and is accompanied by an initial apoptotic cell death followed by a fast secondary necrosis. Autophagy, which has been described as essential for cell survival during nitrogen starvation and regulating amino acid homeostasis [84], does not seem to have a role in ammonium-induced cell death.
The ammonium effects were also not dependent on its metabolism as activity of enzymes involved in the metabolism of ammonium showed no correlation with ammonium toxicity and the use of a nonmetabolizable analog produced the same outcome as ammonium [107]. Even though, ammonium signaling is capable of activating the PKA pathway in agreement with previous results showing that the addition of ammonium, in nitrogen starvation medium, directly signals PKA activation through Mep2p [108]. In contrast, in aging yeast, although Mep2p is involved in ammonium-induced death it does not appear to have a major role in PKA activation. Tor1p and Sch9p were shown to be necessary for ammonium-induced PKA activation in amino acid-starvation conditions as deletion of TOR1 and SCH9 resulted in a decrease of PKA activation. Ammonium action on both pathways, resulting in the over-activation of PKA and TOR pathways and inhibition of Sch9p, culminates in the shortening of CLS [107].
Altogether results support that ammonium induces cell death in aging cultures through the regulation of evolutionary conserved pathways. They also show that the study of ammonium toxicity in yeast aging may be a powerful system to understand longevity regulation in multicellular organisms.
7. Final Remarks
Yeast has emerged as one of the most important model organisms to study the environmental and genetic factors affecting longevity, and its exploitation has made huge contributions to the progress in understanding aging. Although some aspects of aging in yeast are specific to this organism, many of the most important features reveled in yeast proved to be evolutionarily conserved in higher eukaryotic organisms. The two paradigms of aging in yeast, chronological and replicative lifespans, are useful tools to compare the aging process in proliferating and nonproliferating cells and to study the aging process in mitotic and postmitotic cells. The pathways controlling yeast lifespan occur through complex signaling cascades, allowing cells to stimulate proliferation in optimal conditions and also to induce cell cycle arrest and enter into a quiescent state in nutrient exhaustion conditions. In the present paper, we focused on the particular case of the lifespan in yeast and its modulation triggered by extrinsic culture medium factors. A scheme illustrating the current scenario on the regulation of CLS by different nutrient/energy signalling pathways in yeast described herein is presented in Figure 1. Major advances in this research field have come from dietary regimes that have been shown to increase longevity in organisms ranging from yeast to mammals. Altogether, data presented clearly establishes that the carbon and the nitrogen sources as well as the products of fermentation, are among the main extrinsic factors modulating yeast chronological longevity. Loss of cell viability induced by these environmental factors in aging cultures is regulated through evolutionary conserved pathways and their study can provide key insights into pathways that modulate aging in mammals, being a powerful system to understand longevity regulation in multicellular organisms.
Figure 1.
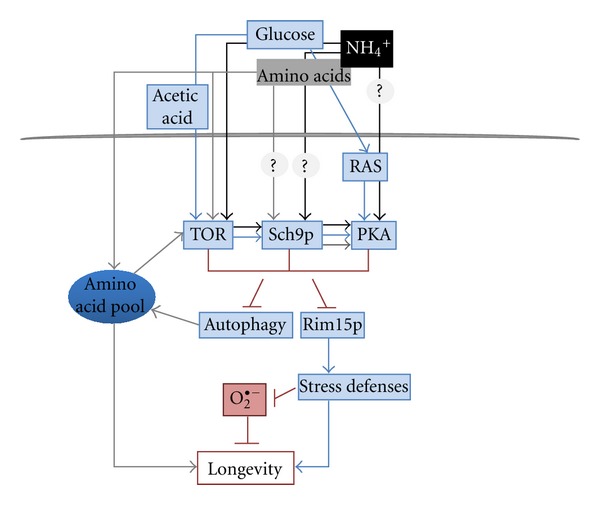
Scheme illustrating the current scenario on the regulation of chronological lifespan (CLS) in yeast by the different nutrient/energy signalling pathways described in the present paper (the detailed description and abbreviations are given in the text).
Authors' Contribution
M. J. Sousa and C. Leão contributed equally to this work.
Acknowledgments
This work was supported by Fundação para a Ciência e Tecnologia (FCT), Portugal Grant PTDC/AGR-ALI/102608/2008. J. Santos received a fellowship from FCT (SFRH/BD/33314/2008).
References
- 1.Gómez JMG. Aging in bacteria, immortality or not-a critical review. Current Aging Science. 2010;3(3):198–218. [PubMed] [Google Scholar]
- 2.Greer EL, Brunet A. Signaling networks in aging. Journal of Cell Science. 2008;121(4):407–412. doi: 10.1242/jcs.021519. [DOI] [PubMed] [Google Scholar]
- 3.Narasimhan SD, Yen K, Tissenbaum HA. Converging pathways in lifespan regulation. Current Biology. 2009;19(15):R657–R666. doi: 10.1016/j.cub.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontana L, Partridge L, Longo VD. Extending healthy life span-from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longo VD. Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells. Neurobiology of Aging. 1999;20(5):479–486. doi: 10.1016/s0197-4580(99)00089-5. [DOI] [PubMed] [Google Scholar]
- 6.Longo VD. The Ras and Sch9 pathways regulate stress resistance and longevity. Experimental Gerontology. 2003;38(7):807–811. doi: 10.1016/s0531-5565(03)00113-x. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Molecular and Cellular Biology. 1999;19(1):216–228. doi: 10.1128/mcb.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118(1):75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366(6454):461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 10.Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384(6604):p. 33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- 11.Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141(7):2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- 12.Weinberger M, Mesquita A, Caroll T, et al. Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging. 2010;2(10):709–726. doi: 10.18632/aging.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120(4):449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131(16):3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 15.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 16.Bjedov I, Toivonen JM, Kerr F, et al. Mechanisms of life span extension by rapamycin in the fruit fly drosophila melanogaster. Cell Metabolism. 2010;11(1):35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge L, Alic N, Bjedov I, Piper MDW. Ageing in Drosophila: the role of the insulin/Igf and TOR signalling network. Experimental Gerontology. 2011;46(5):376–381. doi: 10.1016/j.exger.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapahi P, Chen D, Rogers AN, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metabolism. 2010;11(6):453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown-Borg HM. Hormonal regulation of longevity in mammals. Ageing Research Reviews. 2007;6(1):28–45. doi: 10.1016/j.arr.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartke A, Brown-Borg H. Life extension in the dwarf mouse. Current Topics in Developmental Biology. 2004;63:189–225. doi: 10.1016/S0070-2153(04)63006-7. [DOI] [PubMed] [Google Scholar]
- 21.Kojima T, Kamei H, Aizu T, et al. Association analysis between longevity in the Japanese population and polymorphic variants of genes involved in insulin and insulin-like growth factor 1 signaling pathways. Experimental Gerontology. 2004;39(11-12):1595–1598. doi: 10.1016/j.exger.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Van Heemst D, Beekman M, Mooijaart SP, et al. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4(2):79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 23.Levine AJ, Feng Z, Mak TW, You H, Jin S. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes and Development. 2006;20(3):267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 24.Harrison DE, Strong R, Sharp ZD, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selman C, Tullet JMA, Wieser D, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326(5949):140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in saccharomyces cerevisiae. Science. 2000;289(5487):2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 27.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292(5515):288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 28.Yan L, Vatner DE, O’Connor JP, et al. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130(2):247–258. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 29.Enns LC, Morton JF, Treuting PR, et al. Disruption of protein kinase A in mice enhances healthy aging. PLoS ONE. 2009;4(6) doi: 10.1371/journal.pone.0005963.e5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerits N, Kostenko S, Shiryaev A, Johannessen M, Moens U. Relations between the mitogen-activated protein kinase and the cAMP-dependent protein kinase pathways: comradeship and hostility. Cellular Signalling. 2008;20(9):1592–1607. doi: 10.1016/j.cellsig.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Dechant R, Peter M. Nutrient signals driving cell growth. Current Opinion in Cell Biology. 2008;20(6):678–687. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Borras C, Daniel M, Raul L, et al. RasGrf1 deficiency delays in mice. Aging. 2011;3(3):262–276. doi: 10.18632/aging.100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wittinghofer A, Nassar N. How Ras-related proteins talk to their effectors. Trends in Biochemical Sciences. 1996;21(12):488–491. doi: 10.1016/s0968-0004(96)10064-5. [DOI] [PubMed] [Google Scholar]
- 34.Mirisola MG, Longo VD. Conserved role of Ras-GEFs in promoting aging: from yeast to mice. Aging. 2011;3(4):340–343. doi: 10.18632/aging.100320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2(2):73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 36.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183(4677):1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 37.Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiology and Molecular Biology Reviews. 2003;67(3):376–399. doi: 10.1128/MMBR.67.3.376-399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dilova I, Easlon E, Lin SJ. Calorie restriction and the nutrient sensing signaling pathways. Cellular and Molecular Life Sciences. 2007;64(6):752–767. doi: 10.1007/s00018-007-6381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae: mitochondrial production of toxic oxygen species in vivo. Journal of Biological Chemistry. 1996;271(21):12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 40.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126(2):257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 41.MacLean M, Harris N, Piper PW. Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast. 2001;18(6):499–509. doi: 10.1002/yea.701. [DOI] [PubMed] [Google Scholar]
- 42.Fabrizio P, Longo VD. Chronological aging-induced apoptosis in yeast. Biochimica et Biophysica Acta. 2008;1783(7):1280–1285. doi: 10.1016/j.bbamcr.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. Journal of Cell Biology. 1994;127(6):1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annual Review of Cell and Developmental Biology. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91(7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 46.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464(7288):513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes and Development. 1999;13(19):2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashrafi K, Sinclair D, Gordon JI, Guarente L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(16):9100–9105. doi: 10.1073/pnas.96.16.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wawryn J, Krzepiłko A, Myszka A, Biliński T. Deficiency in superoxide dismutases shortens life span of yeast cells. Acta Biochimica Polonica. 1999;46(2):249–253. [PubMed] [Google Scholar]
- 50.Fabrizio P, Liou LL, Moy VN, et al. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163(1):35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaeberlein M, Powers RW, Steffen KK, et al. Cell biology: regulation of yeast replicative life span by TOR and Sch9 response to nutrients. Science. 2005;310(5751):1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 52.Powers RW, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes and Development. 2006;20(2):174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mechanisms of Ageing and Development. 2005;126(4):491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Goldberg AA, Richard VR, Kyryakov P, et al. Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes. Aging. 2010;2(7):393–414. doi: 10.18632/aging.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piper MDW, Bartke A. Diet and aging. Cell Metabolism. 2008;8(2):99–104. doi: 10.1016/j.cmet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 56.Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB Journal. 2000;14(14):2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- 57.Kaeberlein M, Burtner CR, Kennedy BK. Recent developments in yeast aging. PLoS Genetics. 2007;3(5):p. e84. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei M, Fabrizio P, Hu J, et al. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genetics. 2008;4(1):p. e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biology. 2004;2(9) doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin SJ, Kaeberlein M, Andalis AA, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418(6895):344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 61.Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6(5):649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 62.Granot D, Snyder M. Glucose induces cAMP-independent growth-related changes in stationary-phase cells of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(13):5724–5728. doi: 10.1073/pnas.88.13.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Granot D, Levine A, Dor-Hefetz E. Sugar-induced apoptosis in yeast cells. FEMS Yeast Research. 2003;4(1):7–13. doi: 10.1016/S1567-1356(03)00154-5. [DOI] [PubMed] [Google Scholar]
- 64.Roosen J, Engelen K, Marchal K, et al. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Molecular Microbiology. 2005;55(3):862–880. doi: 10.1111/j.1365-2958.2004.04429.x. [DOI] [PubMed] [Google Scholar]
- 65.Urban J, Soulard A, Huber A, et al. Sch9 Is a Major Target of TORC1 in Saccharomyces cerevisiae. Molecular Cell. 2007;26(5):663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 66.Crauwels M, Donaton MCV, Pernambuco MB, Winderickx J, De Winde JH, Thevelein JM. The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable-growth-medium-induced (FGM) pathway. Microbiology. 1997;143(8):2627–2637. doi: 10.1099/00221287-143-8-2627. [DOI] [PubMed] [Google Scholar]
- 67.Toda T, Cameron S, Sass P, Wigler M. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes & Development. 1988;2(5):517–527. doi: 10.1101/gad.2.5.517. [DOI] [PubMed] [Google Scholar]
- 68.Crespo JL, Hall MN. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 2002;66(4):579–591. doi: 10.1128/MMBR.66.4.579-591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Virgilio C, Loewith R. The TOR signalling network from yeast to man. International Journal of Biochemistry and Cell Biology. 2006;38(9):1476–1481. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 70.Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metabolism. 2011;13(6):668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mechanisms of Ageing and Development. 2006;127(9):733–740. doi: 10.1016/j.mad.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 72.Thevelein JM, De Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Molecular Microbiology. 1999;33(5):904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 73.Sun J, Kale SP, Childress AM, Pinswasdi C, Jazwinski SM. Divergent roles of RAS1 and RAS2 in yeast longevity. Journal of Biological Chemistry. 1994;269(28):18638–18645. [PubMed] [Google Scholar]
- 74.Wei M, Fabrizio P, Madia F, et al. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genetics. 2009;5(5) doi: 10.1371/journal.pgen.1000467.e1000467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fabrizio P, Battistella L, Vardavas R, et al. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. Journal of Cell Biology. 2004;166(7):1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mesquita A, Weinberger M, Silva A, et al. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(34):15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Letters. 1993;333(1-2):169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 78.Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Molecular Biology of the Cell. 2004;15(9):4089–4104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brauer MJ, Huttenhower C, Airoldi EM, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Molecular Biology of the Cell. 2008;19(1):352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(19):6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gomes P, Sampaio-Marques B, Ludovico P, Rodrigues F, Leão C. Low auxotrophy-complementing amino acid concentrations reduce yeast chronological life span. Mechanisms of Ageing and Development. 2007;128(5-6):383–391. doi: 10.1016/j.mad.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 82.Murakami CJ, Burtner CR, Kennedy BK, Kaeberlein M. A method for high-throughput quantitative analysis of yeast chronological life span. Journals of Gerontology A. 2008;63(2):113–121. doi: 10.1093/gerona/63.2.113. [DOI] [PubMed] [Google Scholar]
- 83.Unger MW, Hartwell LH. Control of cell division in Saccharomyces cerevisiae by methionyl tRNA. Proceedings of the National Academy of Sciences of the United States of America. 1976;73(5):1664–1668. doi: 10.1073/pnas.73.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alvers AL, Fishwick LK, Wood MS, et al. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8(4):353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leão C, Van Uden N. Effects of ethanol and other alkanols on passive proton influx in the yeast Saccharomyces cerevisiae. Biochimica et Biophysica Acta. 1984;774(1):43–48. doi: 10.1016/0005-2736(84)90272-4. [DOI] [PubMed] [Google Scholar]
- 86.Swan TM, Watson K. Membrane fatty acid composition and membrane fluidity as parameters of stress tolerance in yeast. Canadian Journal of Microbiology. 1997;43(1):70–77. doi: 10.1139/m97-010. [DOI] [PubMed] [Google Scholar]
- 87.Cardoso H, Leão C. Sequential inactivation of ammonium and glucose transport in Saccharomyces cerevisiae during fermentation. FEMS Microbiology Letters. 1992;94(1-2):155–160. doi: 10.1016/0378-1097(92)90600-s. [DOI] [PubMed] [Google Scholar]
- 88.Leão C, Van Uden N. Effects of ethanol and other alkanols on the general amino acid permease of Saccharomyces cerevisiae. Biotechnology and Bioengineering. 1984;26(4):403–405. doi: 10.1002/bit.260260422. [DOI] [PubMed] [Google Scholar]
- 89.Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. Journal of Applied Microbiology. 2010;109(1):13–24. doi: 10.1111/j.1365-2672.2009.04657.x. [DOI] [PubMed] [Google Scholar]
- 90.Kitagaki H, Araki Y, Funato K, Shimoi H. Ethanol-induced death in yeast exhibits features of apoptosis mediated by mitochondrial fission pathway. FEBS Letters. 2007;581(16):2935–2942. doi: 10.1016/j.febslet.2007.05.048. [DOI] [PubMed] [Google Scholar]
- 91.Fabrizio P, Gattazzo C, Battistella L, et al. Sir2 blocks extreme life-span extension. Cell. 2005;123(4):655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 92.Fabrizio P, Hoon S, Shamalnasab M, et al. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genetics. 2010;6(7):1–14. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goldberg AA, Bourque SD, Kyryakov P, et al. Effect of calorie restriction on the metabolic history of chronologically aging yeast. Experimental Gerontology. 2009;44(9):555–571. doi: 10.1016/j.exger.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 94.Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8(8):1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pinto I, Cardoso H, Leão C, van Uden N. High enthalpy and low enthalpy death in Saccharomyces cerevisiae induced by acetic acid. Biotechnology and Bioengineering. 1989;33(10):1350–1352. doi: 10.1002/bit.260331019. [DOI] [PubMed] [Google Scholar]
- 96.Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Côrte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Molecular Biology of the Cell. 2002;13(8):2598–2606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ludovico P, Sousa MJ, Silva MT, Leão C, Côrte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001;147(9):2409–2415. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- 98.Casal M, Cardoso H, Leão C. Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology. 1996;142(6):1385–1390. doi: 10.1099/13500872-142-6-1385. [DOI] [PubMed] [Google Scholar]
- 99.Weinberger M, Feng L, Paul A, et al. DNA replication stress is a determinant of chronological lifespan in budding yeast. PloS one. 2007;2(1):p. e748. doi: 10.1371/journal.pone.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burhans WC, Weinberger M. Acetic acid effects on aging in budding yeast: are they relevant to aging in higher eukaryotes? Cell Cycle. 2009;8(14):2300–2302. doi: 10.4161/cc.8.14.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matecic M, Smith DL, Pan X, et al. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genetics. 2010;6(4) doi: 10.1371/journal.pgen.1000921.e1000921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.von Wirén N, Merrick M. Molecular Mechanisms Controlling Transmembrane Transport. Berlin, Germany : Springer; 2004. Regulation and function of ammonium carriers in bacteria, fungi, and plants; pp. 95–120. [Google Scholar]
- 103.Ter Schure EG, Van Riel NAW, Verrips CT. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiology Reviews. 2000;24(1):67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 104.Smets B, Ghillebert R, De Snijder P, et al. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Current Genetics. 2010;56(1):1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- 105.Váchová L, Palková Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. Journal of Cell Biology. 2005;169(5):711–717. doi: 10.1083/jcb.200410064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hess DC, Lu W, Rabinowitz JD, Botstein D. Ammonium toxicity and potassium limitation in yeast. PLoS Biology. 2006;4(11):p. e351. doi: 10.1371/journal.pbio.0040351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santos J, Sousa MJ, Leão C. Ammonium is toxic for aging yeast cells, inducing death and shortening of the chronological lifespan. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0037090.e37090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Van Nuland A, Vandormael P, Donaton M, et al. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Molecular Microbiology. 2006;59(5):1485–1505. doi: 10.1111/j.1365-2958.2005.05043.x. [DOI] [PubMed] [Google Scholar]


