Abstract
Studies about hypoxia-induced oxidative stress in human health disorders take advantage from the use of unicellular eukaryote models. A widely extended model is the fermentative yeast Saccharomyces cerevisiae. In this paper, we describe an overview of the molecular mechanisms induced by a decrease in oxygen availability and their interrelationship with the oxidative stress response in yeast. We focus on the differential characteristics between S. cerevisiae and the respiratory yeast Kluyveromyces lactis, a complementary emerging model, in reference to multicellular eukaryotes.
1. Introduction
Interest in hypoxic and oxidative stress studies is increasing in recent years, mostly in relation to aging or diseases such as neurodegenerative disorders or cancer. These processes in human cell lines show a very complex regulation, and therefore the availability of simple models is extremely useful. Yeasts have demonstrated to be suitable unicellular eukaryotic models since, in addition to generalized culture facilities, global “omic” analysis is fully developed and molecular mechanisms are generally well conserved. For instance, although obviously without nervous system, most of the molecular signaling pathways and the proteins involved in human neurological diseases are functionally conserved in yeasts [1]. Besides, functional characterization of human genes is sometimes achieved by means of their heterologous expression in mutant yeasts.
Most studies about the hypoxic and oxidative stress responses and their connections have been carried out hitherto on Saccharomyces cerevisiae, a yeast model with a predominantly fermentative metabolism [2]. In the same way, S. cerevisiae mutants have been frequently used as research models in aging [3] and in human pathologies [4]. Frequently, the mechanisms discovered with this yeast proved to be conserved in multicellular eukaryotes. However, human tissues such as the neuronal network have oxidative metabolism, and therefore the use of alternative yeast models has been suggested [5]. We analyze Kluyveromyces lactis from the perspective of an alternative eukaryote model in these studies or similar studies, since this yeast has a predominantly respiratory metabolism.
Molecular mechanisms that support the metabolic differences between S. cerevisiae and K. lactis and the specific responses to hypoxia or oxidative stress have been studied. Redox metabolism is a key differential point between S. cerevisiae and K. lactis, both thiol-redox and NAD(P)H-redox reactions. K. lactis is characterized, opposite to S. cerevisiae, by a higher glucose flow through the pentose phosphate pathway (PPP) than through glycolysis [6] and as a consequence by a higher production of NADPH in the cytosol. In fact, one of the molecular keys supporting the difference in the Crabtree phenotype (inhibition of respiration by fermentation) of the two yeast species lies in the mechanisms involved in the re-oxidation of the NADPH [7, 8]. A significant part of this reoxidation is carried out in K. lactis by mitochondrial external alternative dehydrogenases (NDEs), which use NADPH, the enzymes of S. cerevisiae being NADH-specific. Unlike S. cerevisiae, transcription of NDEs genes in K. lactis is not regulated by the carbon source. Since NDEs may compete with alcohol dehydrogenases for the cytosolic NADH, their repression at high glucose concentrations, as it occurs in S. cerevisiae, correlates with an increase of reoxidation of glycolytic NADH by the alcohol dehydrogenases and therefore with the prevalence of aerobic fermentation and the Crabtree-positive phenotype [9]. Interestingly, NDEs have been reported to influence ROS production and life span in S. cerevisiae [10, 11]. Moreover, the NADPH-dependent pathways of response to oxidative stress also contribute to NADPH reoxidation. In S. cerevisiae, they play a main role together with glutamate dehydrogenase and also operate, although to a lesser extent, in K. lactis, [8, 12, 13].
In this paper, we focus on the complex interdependence of multiple mechanisms, which arise as a consequence of the decrease of oxygen availability and on the responses elicited to compensate this stress. A general overview of all the subjects analyzed is shown in Figure 1. Along the text, special reference is made on the differences found between S. cerevisiae and K. lactis, looking for the potential advantages and disadvantages of these models in reference to each other and in comparison to multicellular eukaryotes.
Figure 1.
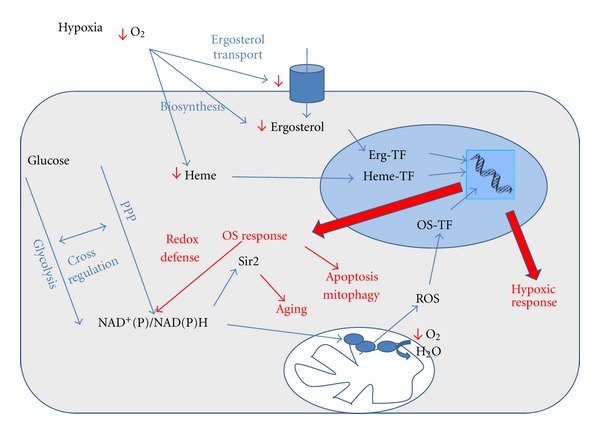
A panorama of the multiple connections between hypoxia, metabolic rerouting, oxidative stress response, and cell defense mechanisms. Erg: ergosterol; PPP: pentoses phosphate pathway; TF: transcriptional factors.
The first intracellular signal sensing low levels of oxygen is the heme content. The biosynthesis of heme includes enzymes that directly use oxygen as electron acceptor during the catalysis, and, besides, several steps are regulated by oxygen availability. Other important pathway that directly uses oxygen is the biosynthesis of ergosterol, which is also regulated by oxygen availability. The intake of ergosterol from the media through the membrane is also regulated by oxygen levels. Downstream in these sensing strategies is heme and ergosterol dependent transcriptional factors, which act in the nucleus to regulate the transcription of more than 100 genes, those conditioning the “hypoxic response” and improving the use of the low levels of oxygen. Hypoxia signals the activation of mechanisms that regulate the transcription of genes involved in the oxidative stress response. Simultaneously, the decrease in oxygen levels causes a complete rerouting of nutrients through different metabolic pathways. This principally affects glucose and other sugars, which can follow fermentative or respiratory transformations and, in turn, condition the systems of redox exchange between cytoplasm and mitochondria and the mechanisms that produce ROS. Reoxidation of reduced NAD(P)H also has regulatory effects on the diverse metabolic routes that need the oxidized coenzyme forms to function. ROS also elicit other mechanisms of cell defense, including reoxidation of NAD(P)H and life span adjustment, programmed cell death, and mitophagy (Figure 1).
2. The Hypoxic Response in K. lactis
During hypoxia, it is advantageous for cells to adapt the pattern of gene expression in order to improve oxygen utilization. The hypoxic response is well documented in the model yeast Saccharomyces cerevisiae, whose cells sense oxygen via the levels of heme and sterols [14–17]. The response of S. cerevisiae to hypoxia produces increased expression of genes related to ergosterol synthesis, cell wall composition, and glycolytic genes and reduced expression of components of the respiratory chain, ATP synthesis, and the citric acid cycle [14, 18, 19]. However, this knowledge is not directly applicable to other yeasts, even to those close-related in phylogeny, which became evident in the last years with the advances in the study of the hypoxic response in K. lactis and other yeasts. It has been proposed that a whole-genome duplication (WGD) contributed to yeast evolution from strict aerobes to facultatives/fermentatives [20–22]. Functional specialization between duplicated genes explains the existence in S. cerevisiae of homologous genes (COX5a/COX5b; CYC1/CYC7; HYP2/ANB1; AAC1/AAC2/AAC3) differentially expressed in aerobic and hypoxic conditions [14, 23–25]. Kluyveromyces lactis does not present duplication of genes with specialized aerobic, hypoxic transcription, but the unique copy is regulated by oxygen availability [26]. Probably, K. lactis and S. cerevisiae are diverged from one common ancestor yeast previously to the WGD event and this could explain the multiple differences observed when comparing the response to hypoxia in both yeasts, as explained below.
Although K. lactis is unable to grow under strictly anoxic conditions [27], probably due to the absence of expression of genes related to the import of sterols in this condition [28], this yeast ferments sugars and grows in hypoxic conditions defined as oxygen availability below 1% of fully aerobic levels [7, 29]. Several reports of genes upregulated during hypoxia in K. lactis have been published. A K. lactis heme-deficient strain, obtained by KlHEM1 disruption, was used to assess the functional significance of heme-directed regulation in K. lactis; KlHEM13, encoding the coproporphyrinogen oxidase (EC 1.3.3.3), an oxygen-requiring enzyme that catalyzes the sixth step of heme biosynthesis, was the first hypoxic gene functionally characterized in this yeast [30, 31]. Other gene from the heme biosynthetic pathway, KlHEM1, is upregulated during hypoxia in K. lactis [32]. The KlPDC1 gene, encoding for the enzyme pyruvate decarboxylase, is also induced by hypoxia [33]. After the completion of the Génolevures sequencing project [34], the availability of the complete sequence of the K. lactis genome allowed the design of specific DNA arrays containing selected DNA probes putatively related to the aerobic-hypoxic response by their similarity to the orthologs in S. cerevisiae [26]. The nature of the hypoxic transcriptional response in K. lactis, as revealed by using these arrays, differed notably from S. cerevisiae, but confirmed the existence of hypoxic upregulated genes in K. lactis such as KlOYE2 (KLLA0A09075g), KlGSH1 (KLLA0F14058g) and KlOLE1 [26]. Besides KlOLE1, the transcription of other lipid biosynthetic genes like KlERG1, KlFAS1, and KlATF1 is also induced by hypoxia [35].
In S. cerevisiae, adaptation to hypoxia requires the transcriptional induction or derepression of multiple genes organized in regulons controlled by specific transcriptional regulators. Considering that in K. lactis the hypoxic response exists but the target genes are not coincident and are not equally regulated, the question arose about the functionality of sensors and transcriptional regulators. The principal sensors in the yeast response to hypoxia are heme and ergosterol. The heme biosynthetic pathway is well conserved in different organisms throughout evolution [36], and this is also true between S. cerevisiae and K. lactis. Both yeasts have eight highly homologous genes necessary for the biosynthesis of heme. For the three genes of the heme biosynthetic pathway characterized in K. lactis to date (KlHEM1, KlHEM12,and KlHEM13), functional equivalence with their S. cerevisiae homologs has been confirmed experimentally by cross-complementation [31, 32, 37]. As it happens in S. cerevisiae, the transcriptional regulation of KlHEM12 is not a key point for regulation of heme synthesis in K. lactis [38] and its transcriptional regulation in different carbon sources [38] is also similar to that reported for its homolog in S. cerevisiae [39, 40]. However, notable differences exist in the regulation of the other two characterized genes. In S. cerevisiae, the expression of HEM1 is constitutive [41], but in K. lactis the first step of the heme biosynthesis is under double-feedback regulation by heme, at the level of gene transcription [37] and mitochondrial import of the protein [42]. Although in S. cerevisiae the aerobic repression of HEM13 is mediated by the transcriptional regulator Rox1p, diverse data indicate that the mechanism is different in K. lactis and a similar repressor does not operate [43, 44].
In S. cerevisiae, intracellular levels of heme regulate the activity of the transcriptional regulator Hap1 [14]. The CCAAT-binding complex Hap2/3/4/5, an evolutionarily conserved multimeric transcriptional activator in eukaryotes [45], is also necessary for the activation of many genes involved in respiratory metabolism [45], although its direct activation by heme has not been demonstrated. Targets of Hap1 include genes required for respiration and for controlling oxidative damage [46–48] and also the aerobic repressor Rox1. When the oxygen levels drop, heme does not bind to Hap1. Then, the interaction with Ssa1, Ydj1, and Sro9 maintains to Hap1 inactive [49, 50]. As a result, ROX1 is not expressed and no longer represses aerobic expression of genes involved in the hypoxic response. Moreover, Mot3 collaborates in the repression exerted by Rox1 in target promoters [51], and Ixr1 has been related to the hypoxic response of S. cerevisiae in cross-regulation with ROX1 [52–54]. Although several homologs to the components of the Hap2/3/4/5 complex have been cloned in K. lactis [55–57], the respiratory system of K. lactis, escapes from HAP2 control [56]. Contrary to data previously described for the homologous gene of S. cerevisiae, the function of the KlHAP1 gene does not affect growth in media with carbon sources used by fermentative or respiratory pathways in K. lactis and KlHap1 is not a transcriptional activator of the expression of genes related to respiration or sterol biosynthesis [58] but represses the expression of the major glucose transporter [59]. In a similar way, KlROX1 does not regulate the hypoxic response in K. lactis [60] and the KlROX1 promoter is not regulated by KlHap1 or KlRox1 in response to changes aerobiosis/hypoxia [44].
Parallel, Hap1-Rox1-independent, oxygen response pathways exist in yeast. For instance, in S. cerevisiae, the transcription of the hypoxic gene OLE1 depends on cytochrome c oxidase [61] and requires the transcription factor Mga2 that is functional in hypoxia [62]. In K. lactis, this regulatory circuit is also different and, although KlMGA2 shows homology to the MGA2 gene from S. cerevisiae, KlMga2does not regulate KlOLE1 hypoxic expression [35]. Sut1 and Sut2 are also involved in the transcriptional induction of hypoxic genes and in sterol uptake and synthesis in S. cerevisiae [63, 64].
Another sensing pathway includes the regulators of sterol biosynthesis Upc2 and Ecm22 [65]. Sterol depletion leads to activation of the paralogous genes UPC2 and ECM22 [16], which control expression of a subset of hypoxic genes. Both bind to a sequence motif known as the sterol regulatory element (SRE) in the promoters of their target genes, but Ecm22 is an aerobic repressor and Upc2 an anaerobic activator, which is upexpressed during hypoxia. They regulate expression of ergosterol biosynthesis genes and the DAN/TIR family of cell wall proteins [65–67]. This regulatory circuit remains unstudied in K. lactis, although analysis of the genome sequence shows that the hypoxic genes from the sterol biosynthetic pathway are conserved in the two yeasts [26].
Quantitation of the homology between proteins translated from verified or putative orthologous genes of K. lactis and S. cerevisiae reveals that, with few exceptions, the proteins related to biosynthetic routes producing intracellular changes in heme and ergosterol are more conserved than the transcriptional factors, which are regulated by their levels and produce the hypoxic response (Figure 2).
Figure 2.
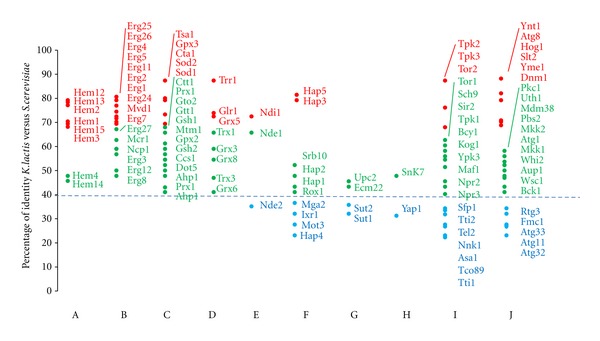
Homologies (percentage of identity calculated according to BLASTp in Génolevures) between K. lactis and S. cerevisiae proteins related to the pathways summarized in Figure 1. Red, 100–70% identity; green 69–40% identity; blue, <40% identity. (A) heme biosynthesis; (B) ergosterol biosynthesis and supply; (C) NAD(P)H consuming oxidative defense reactions; (D) other oxidative defense reactions; E, NAD(P)-dehydrogenases from the inner membrane of mitochondria; (F) heme/respiration-related transcriptional factors; (G) sterol-related transcriptional factors; (H) peroxide-related transcriptional factors; (I) life span-related proteins; (J) mitophagy-related proteins.
3. Oxidative Stress Response in K. lactis
The oxidative stress response in K. lactis is a mostly unexplored field. The number of published works is less than 1.5% compared to S. cerevisiae. Several studies based on comparative genomics have been performed, combining in silico and experimental approaches [26, 68, 69]. The search in the K. lactis genome of putative S. cerevisiae orthologs related to the oxidative stress response (genes coding for superoxide dismutases and their chaperones, catalases and peroxidases, proteins of the glutathione, and thioredoxin systems) has suggested that pathways and proteins are generally conserved, with a few exceptions mainly affecting gene redundancy or predicted subcellular location of the proteins [69]. These exceptions comprise the groups of genes encoding thioredoxin peroxidases, thioredoxin reductases, thioredoxins, glutaredoxins, and glutathione transferases (Figure 2). However, several functional differences affecting mainly connections between redox pathways related to carbohydrate metabolism, respiratory functions and oxidative stress response are observed [69]. In support of these results, a recent study about glutathione transferases, and synthetases in yeast species representatives of fermentative, respiratory, and oxidative metabolism [70] found significant differences in homology and predicted intracellular sorting.
Among the reported differences, it is remarkable to outline the role of the thiol-redox pathways, specifically glutathione reductase (GLR), the enzyme that catalyzes the interconversion of oxidized (GSSG) and reduced glutathione (GSH) using NADPH as reducing power. Whereas in S. cerevisiae the expression of GLR increases in response to oxidative stress produced after addition of peroxides by a Yap1-mediated mechanism, this effect is absent in K. lactis [26, 71, 72]. Surprisingly, both S. cerevisiae and K. lactis GLR depletion mutants are more sensitive to oxidative stress [12, 13]. In such K. lactis GLR mutants, increase in ROS production, catalase, and thioredoxin reductase (TRR) activities are observed and the expression of a pool of other antioxidant enzymes and oxidoreductases is also upregulated [73]. It is likely that TRR and other NADPH-dependent oxidoreductases might replace GLR in maintaining the GSH/GSSG ratio. In fact, purified K. lactis TRR shows GLR activity in vitro (our unpublished results). In support of this explanation, it has been reported that the thioredoxin-TRR system can reduce GSSG in S. cerevisiae [74]. Other reported functional differences affecting the OS defense enzymes of K. lactis and S. cerevisiae are regarding the mechanism of cation handling of the superoxide dismutase Sod1 [75] and the transcriptional regulation of the SOD1 gene after a shift to hypoxia [26, 76]. Also the transcriptional regulation of the genes encoding catalases [12, 26, 77, 78] and glutathione synthetases [26, 79] under aerobic/hypoxic conditions and under peroxide-treatment is different in the two yeasts. About comparative analyses between the transcriptional factors related to the oxidative stress response in S. cerevisiae and K. lactis, Yap1 and Snk7 share, respectively, 33% and 50% identities (Figure 2), and KlYap1 has been functionally characterized in relation to the oxidative stress response induced by metals and peroxides [80].
Several evidences support that in K. lactis the OS response has a regulatory role upon fermentation/respiration balance. Thus, there is a positive correlation between the increase of GLR activity and the glucose-6-phosphate-dehydrogenase activity (from PPP) when oxygen levels increase [12]. Besides, the glucose respiration rate, in K. lactis cells that metabolize all the monosaccharide through the PPP, increases upon GLR depletion and decreases upon GLR overexpression [13]. Proteome analysis reveals that there is a different response to H2O2-treatment, which is dependent on GLR in such a way that the expression of several enzymes of the glycolysis and the Krebs' cycle decreases in the wild-type strain, while enzymes of these pathways and the PPP increase in the GLR depleted mutant [73]. Other indirect evidence is that K. lactis GLR activity decreases in a Gcr1-mutant [13] being Gcr1 at the same time a positive transcriptional regulator of KlNDI1 [9]; KlNDI1 is the gene encoding the internal mitochondrial alternative dehydrogenase (NDI), the enzyme that replaces the respiratory chain complex I found in other eukaryotes [81]. GLR depletion mutants grow better in glucose than the wild type, overall when all the glucose is metabolized through the PPP, which might be explained by rerouting the oxidation of the NADPH produced in the PPP from GLR to NDEs, thus increasing ATP production in the respiratory chain [12, 13].
Yeasts alternative dehydrogenases of the mitochondrial inner membrane are also related to OS and are other point of connection with the response to metabolic changes produced by oxygen availability. The transcription of the two K. lactis genes encoding NDEs decreases when cells are under oxidative stress and the NADPH-related defense mechanisms are activated [9, 82]. Regarding K. lactis NDI, we have proved that the transcription of the KlNDI1 gene is induced in nonfermentable carbon sources through a process mediated by the factor Adr1 and that the expression of the gene did not decrease after an hypoxic shift [9]. The homologous S. cerevisiae enzyme has been more widely studied and its role in aging and ROS production has been reported [83]. Differences found in yeasts NDI are of clinical interest since they have been used in gene therapy of diseases [84] such as Parkinson's disease [85] or hereditary optic neuropathy [86, 87]. Heterologous expression of the S. cerevisiae NDI1 gene reduces the overproduction of ROS caused by mitochondrial complex I defects in multicellular eukaryotes [88].
These connections between thiol-redox OS reactions and carbohydrate metabolism described above in K. lactis are also supported in other organisms. Ralser et al. [89] discovered that S. cerevisiae cells with reduced activity of the key glycolytic enzyme triose-phosphate isomerase exhibit an increased resistance to the thiol-oxidizing reagent diamide. This phenotype is conserved in Caenorhabditis elegans and the underlying mechanism is based on a redirection of the metabolic flux from glycolysis to the PPP, altering the redox equilibrium of the cytoplasmic NADP(H) pool. Another key glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), is known to be inactivated in response to various oxidant treatments, and this causes a similar redirection of the metabolic flux [89].
4. The Hypoxic-Induced Oxidative Stress Response in K. lactis
A connection between the hypoxic and oxidative stress responses has been reported in the fermentative yeast S. cerevisiae. Several S. cerevisiae genes that are induced during hypoxia are related to the oxidative stress response. CUP1 and CUP2, which are necessary for the removal of superoxide radicals, are upregulated 11.6-fold during hypoxia in a Rox1 and Srb10-dependent mechanism [18]. Other genes related to oxidative stress (HSP12, FMP46, and GRE1) DNA repair (ALK1) or mitochondrial genome maintenance (MGM1)also increase their expression during hypoxia [18]. The level of mitochondrial and cytosolic protein carbonylation, the level of mitochondrial and nuclear DNA damage measured by 8-OH-dG modification, and the expression of SOD1, encoding superoxide dismutase, increases transiently during a shift to anoxia [76]. Besides, the specific proteins, which become carbonylated during the shift to anoxia, are the same proteins that become carbonylated during peroxidative stress. These results demonstrate that yeast cells exposed to anoxia experience transient oxidative stress and suggest that ROS generated could also signal the variation in oxygen levels and trigger the nuclear response to hypoxia affecting transcription [76]. However, the specific connection between ROS production, protein, or DNA modifications and transcriptional regulation has not yet been elucidated in yeasts. The question about whether mitochondrial or cytosolic proteins, which are specifically oxidized in cells exposed to anoxia, play a role in signaling pathways from the mitochondrion to the nucleus that function to induce hypoxic genes remains unanswered.
In K. lactis, after analyzing 30 genes related to oxidative stress, only two (KlGSH1 and KlOYE2) increased their expression after the hypoxic shift [26]. However, a whole-genome approach has not been carried out in this yeast and, therefore, a direct comparison of these data with those reported from S. cerevisiae is not accurate. An interesting observation, which suggests that also in K. lactis the hypoxic response might be triggered by ROS production, is that the hypoxic response is highly dependent on the relative flux of glucose through glycolysis or the pentose phosphate pathway (PPP). The predominant use of PPP versus glycolysis is accompanied by a higher expression of mitochondrial cytochrome c [7], which might be associated with the mitochondrial chain activation and changes in ROS production. Indeed, in a rag2 mutant, lacking phosphoglucose isomerase and committed to reroute the glucose-6-phosphate through PPP, in order to bypass the blocked glycolytic step, a more intense hypoxic response than the wild-type strain, and that affects the genes of heme metabolism and the oxidative stress response, is observed [26].
A new link between oxidative stress and hypoxia comes from the analysis of multiple functions attributed to transcriptional regulators initially characterized in the aerobic/hypoxic response. In S. cerevisiae, Hap1 not only acts as an aerobic activator but has a regulatory function during hypoxia [17, 90]. Hap1 also controls the expression of genes related to sterol biosynthesis [14, 91–94] and SOD2, involved in the oxidative stress response [95].
Although putative homologues of two of the principal regulators of the aerobic/hypoxic response in S. cerevisiae (Hap1 and Rox1) have been characterized in K. lactis, their sequence and function diverge notably from those described in S. cerevisiae [44, 58]. Remarkably, their functions in K. lactis are somehow related to the metal-induced oxidative-stress response. Deletion of KlHAP1 increases the resistance to oxidative stress or cadmium [58]. Moreover, the induction by 0.5 mM H2O2 of two genes related to the oxidative stress response, KlYAP1 and KlTSA1, is repressed by KlHap1p [58]. KlROX1 mediates the response to arsenate and cadmium [44]. KlRox1p binds to the KlYCF1 promoter, a gene related to cadmium detoxification, and causes its activation [44].
5. Adjustment of Cell Survival and the Oxidative Stress Response
Besides the transcriptional, proteomic, and metabolomic reorganization caused by the oxidative stress, ROS also elicit other mechanisms of cell defense, including life span adjustment, programmed cell dead, autophagy, and mitophagy. Mitochondria, being the major intracellular source of ROS, are involved in aging and life span regulation [96] Yeasts have been proved to be good models for studying these processes. In S. cerevisiae, a cross-regulation between glycolysis and PPP has been proposed in order to prevent oxidative stress when cells switch from anaerobic to oxidative metabolism. Low activity of the glycolytic enzyme pyruvate kinase causes accumulation of PEP and blocks the pathway diverting the glucose flux into the PPP [97]. This mechanism helps to balance the increased ROS production during oxidative metabolism [97]. Also in mammals, during oxidative damage in cancer cells, a similar redirection of metabolic fluxes contributes to ROS clearance [98]. Therefore, it is possible that the hypoxic and the oxidative stress responses, influenced by the reorganization of the utilization of different metabolic pathways, also contribute to modulate these cell defense mechanisms in yeasts and other cells.
Studies pioneered in S. cerevisiae by measuring life span have revealed several molecular mechanisms underlying cellular aging and which are well conserved in eukaryotes. Two basic experimental approaches have been applied in unicellular organisms. Replicative life span (RLS) is defined as the number of daughters a single cell produces during its life [99]. Chronological life span (CLS) is defined as the time a population of cells survive in stationary phase [100]. In S. cerevisiae, a Crabtree-positive yeast, calorie restriction by glucose limitation increases both RLS and CLS, a feature that coincides with increased cytochrome content, and NADH-cytochrome c reductase activity [101]. In K. lactis, a Crabtree-negative yeast for which glucose limitation does not promote an enhancement of the respiratory capacity [2], the increase in CLS by glucose limitation is not produced [102]. These results suggest that calorie restriction-dependent increase in longevity may be due to mitochondrial control and more particularly the regulation of oxidative phosphorylation activity.
An additional nexus between aging and the redox cell balance came from the discovery of sirtuins. They are NAD+-dependent enzymes and they belong to a highly conserved family of proteins that in yeasts, invertebrates, and mammals act in diverse functions related to longevity [103]. In S. cerevisiae, Sir2 is induced in cells treated with 4 mM H2O2 or 10 mM menadione [104] and these data suggest a connection between oxidative stress, Sir2 activation and longevity. The existence of complexes of Sir2 with other metabolic enzymes NAD+-dependent, like those formed with the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase Tdh3 or the alcohol dehydrogenase Adh1 [105], might indicate that the ratio NAD+/NADPH in the microenvironment surrounding Sir2 could act as its modulator. Recently, a third complex of Sir2 with Mdh1, the mitochondrial malate dehydrogenase, has been proposed based on multiple common network interactions involving also the proteins Rad53, Aat1, Fob1, and Hst1 [106], although not yet proved by physical interactions. Since Mdh1 is overexpressed under conditions of calorie restriction [107] and it causes extension of Sir2-dependent RLS [108], further investigation is promising. In K. lactis sirtuins or its regulators have not been studied but there is an ORF (KLLA0F14663g) with 62% identities to S. cerevisiae Sir2 (Figure 2). Considering the importance of sirtuins and their modulators in the etiology and treatment of human pathologies such as metabolic, cardiovascular, and neurodegenerative diseases or cancer [104, 109] and the similarities found between the high respiratory metabolism of K. lactis and certain human cell types like neurons [5], sirtuins and related proteins from K. lactis are good targets for study.
About the signaling pathways that affect life span in yeast (reviewed in [103]), the serine threonine kinases Tor1, Sch9, and PKA that control nutrient signaling pathways also regulate aging in yeasts, and their homologs in animals share conserved functions in aging. Depletion of Tor1 kinase increases both RLS and CLS in budding yeast, flies, and C. elegans. Likewise, PKA kinase activation lengthens life span in budding yeast and longevity in mice. The kinase S6K1, which is known to be related to the control of aging in mice, Drosophila, C. elegans, and the yeast Sch9 kinase that controls RLS and CLS in yeast represent other group or orthologous genes. Purification of TOR from yeast and human cells revealed that TOR can exist in at least two multiprotein complexes, termed TORC1 and TORC2 [110]. Interestingly, it has been found that superoxide anions regulate the TORC1 complex and its ability to bind the Fpr1-rapamycin complex [111], thus establishing another link between OS and aging. In S. cerevisiae, 15 genes are functionally related to TORC1 function and Tor, Sch9, and PKA signaling are interconnected (Table 1). In S. cerevisiae, the genes TPK1 (alias PKA1, SRA3; ORF, YJL164C), TPK2 (alias PKA2, YKR1, PKA3; ORF, YPL203W), and TPK3 (YKL166C) encode for three forms of the cAMP-dependent protein kinase catalytic subunit of the cyclic AMP-dependent protein kinase (PKA) and BCY1 (YIL033C) for the regulatory subunit. In K. lactis, homologous genes of main participants in these signaling pathways are present (Table 1) and homology (recorded using BLASTp in Génolevures at http://www.genolevures.org/) is summarized in Figure 2 The most remarkable observation is that in K. lactis, there is only one ORF (KLLA0B13948g), which encodes for a protein with 71% identity to S. cerevisiae Tor2 and 68% identity to S. cerevisiae Tor1. This opens a question about the existence and composition of two TORC complexes in K. lactis as previously reported in S. cerevisiae [110] and outlines this issue as a differential point to study in relation to divergences in life span signaling. Besides, the K. lactis proteins in this group with less than 40% identity to their S. cerevisiae counterparts are also good targets for further studies.
Table 1.
Putative main actors of aging and its signaling pathways in K. lactis and S. cerevisiae. Degree of identity (%) between homologs is indicated in Figure 2.
|
Protein |
ORF K. lactis |
ORF S. cerevisiae |
Function |
|---|---|---|---|
| Sir2 | YDL042C | KLLA0F14663g | NAD+ -dependent histone deacetylase |
| Tpk1 | YJL164C | KLLA0B12716g | PKA catalytic subunit |
| Tpk2 | YPL203W | KLLA0D03190g | PKA catalytic subunit |
| Tpk3 | YKL166C | KLLA0B07205g | PKA catalytic subunit |
| Bcy1 | YIL033C | KLLA0E04181g | PKA regulatory subunit |
| Ypk3 | YBR028C | KLLA0F24618g | An AGC kinase phosphorylated by cAMP-dependent protein kinase (PKA) in a TORC1-dependent manner |
| Asa1 | YPR085C | KLLA0D09086g | Subunit of the ASTRA complex involved in the stability or biogenesis of PIKK*s such as TORC1 |
| Tor1 | YJR066W | KLLA0B13948g | PIK-related protein kinase and rapamycin target, subunit of TORC1, a complex that controls growth in response to nutrients by regulating translation, transcription, ribosome biogenesis, nutrient transport, and autophagy, involved in meiosis |
| Tor2 | YKL203C | KLLA0B13948g | PIK-related protein kinase and rapamycin target, subunit of TORC1 and TORC2, a complex that regulates cell-cycle dependent polarization of the actin cytoskeleton, involved in meiosis |
| Nnk1 | YKL171W | KLLA0A06776g | Protein kinase, implicated in proteasome function, interacts with TORC1, Ure2, and Gdh2 |
| Tco89 | YPL180W | KLLA0E18855g | Subunit of TORC1 (Tor1 or Tor2-Kog1-Lst8-Tco89) |
| Kog1 | YHR186C | KLLA0A09471g | Subunit of TORC1, it may act as a scaffold protein to couple TOR and its effectors |
| Tti1 | YKL033W | KLLA0F25762g | Subunit of the ASTRA complex, involved in chromatin remodeling, telomere length regulator involved in the stability or biogenesis of PIKK*s such as TORC1 |
| Tti2 | YJR136C | KLLA0B04026g | Subunit of the ASTRA complex, involved in chromatin remodeling, telomere length regulator involved in the stability or biogenesis of PIKK*s such as TORC1 |
| Sch9 | YHR205W | KLLA0B03586g | AGC family protein kinase and functional ortholog of mammalian S6 kinase, phosphorylated by Tor1p and required for TORC1-mediated regulation of ribosome biogenesis, translation initiation, and entry into G0 phase, integrates nutrient signals and stress signals from sphingolipids to regulate life span |
| Maf1 | YDR005C | KLLA0E17535g | Negative regulator of RNA polymerase III, binds to the N-terminal domain of the Rpc160 subunit of Pol III to prevent closed-complex formation, localization and activity are regulated by phosphorylation, mediated by TORC1, protein kinase A, and Sch9 |
| Tel2 | YGR099W | KLLA0D15158g | Subunit of the ASTRA complex, involved in the stability or biogenesis of PIKK*s such as TORC1 |
| Sfp1 | YLR403W | KLLA0B03047g | Regulates transcription of ribosomal protein, response to nutrients and stress, G2/M transitions during mitotic cell cycle, and DNA-damage response and modulates cell size, regulated by TORC1 and Mrs6 prion |
| Npr2 | YEL062W | KLLA0D01067g | Npr2/3 complex mediates downregulation of TORC1 activity upon amino acid limitation |
| Npr3 | YHL023C | KLLA0F18238g | Npr2/3 complex mediates downregulation of TORC1 activity upon amino acid limitation |
*PIKK phosphoinositide 3-kinase related kinase.
Apoptosis is one type of programmed cell death (PCD) with great importance for the development and homeostasis of multicellular organisms. Basal apoptosis machinery exists also in yeast, unicellular fungus, and in some filamentous fungi [112]. Regarding the respiratory yeast K. lactis, once more the number of studies performed hitherto is very scarce [113]. A mutant in the essential gene KlLSM4, an ortholog to LSM4 of S. cerevisiae, which encodes an essential protein involved in both pre-mRNA splicing and mRNA decapping, shows phenotypic markers of apoptosis such as chromatin condensation, DNA fragmentation, accumulation of ROS, and increased sensitivity to different drugs. Mechanisms of Bax-induced [114, 115] and lactose-induced [116] cell death have also been described in K. lactis. We have recently investigated PCD in K. lactis, using the drugs staurosporine (STS) and phytosphingosine (PHS), which induce PCD in other organisms, and found that glutathione and GLR played an important role. While K. lactis seemed to be insensitive to STS, PHS induced PCD. The insensitivity of K. lactis to STS might be dependent upon the higher levels of GSH found in cells treated with STS. In human cells, PCD induced by STS causes GSH efflux, but GSH exporter proteins are absent in K. lactis. In addition, GLR appears to be involved in PHS-triggered PCD because cells lacking this enzyme are more resistant to the drug than the wild-type strain. Moreover, the addition of GSSG or GSH to the medium partially restores growth of the wild-type K. lactis strain on PHS [117].
The strictly regulated removal of oxidized structures is a universal stress response of eukaryotic cells that targets damaged or toxic components for vacuolar or lysosome degradation. Autophagy stands at the crossroad between cell survival and death. It promotes survival by degrading proteins and organelles damaged during oxidative stress, but it is also activated as a part of death programs, when the damage cannot be overcome. Evidence is accumulating that the cellular sites of ROS production and signaling (including mitochondria) may be primary targets of autophagy [118]. The surplus ROS damage the mitochondria themselves and the damaged mitochondria produce more ROS in a vicious circle, ultimately leading to mitochondrial DNA deletion, a form of the so-called petite-mutant phenotype [119]. Selective mitochondria autophagy is called mitophagy and contributes to the maintenance of mitochondrial quality by eliminating damaged or excess mitochondria [120]. Although little is known about the mechanism, glutathione influences mitophagy [121, 122]. The interplay between mitochondria and autophagy seems to be evolutionarily conserved from yeast to higher eukaryotes. Defects in one of these elements could simultaneously impair the other, resulting in risk increments for various human diseases [123]. Autophagy is associated with tumor genesis, neurodegenerative diseases, cardiomyopathy, Crohn's disease, fatty liver, type 2 diabetes, defense against intracellular pathogens, antigen presentation, and longevity [121, 122, 124].
Recent studies in yeast identified several mitophagy-related proteins, which have been characterized with regard to their function and regulation, allowing to compare the similarities and differences of this degradation process between yeast and mammalian cells [120]. Up to our knowledge, no studies at all about mitophagy or even autophagy have been published at present in K. lactis. We have performed a custom Blastp in Génolevures for the main actors of mitophagy [125] and its signaling pathways [126] in S. cerevisiae versus K. lactis and we found sequences with different degrees of similarity as shown in Table 2 and Figure 2. Further research is required to analyze if the sequence similarity corresponds to similarity of function or not. This is a new field of research in the respiratory yeast K. lactis. As occurs with other pathways, it is likely that functional differences exist according to fermentative or respiratory predominant metabolism in yeasts. K. lactis proteins in this group (Figure 2) are good targets for comparative studies in mitophagy.
Table 2.
Putative main actors of mitophagy and its signaling pathways in K. lactis and S. cerevisiae. Degree of identity (%) between homologs is indicated in Figure 2.
| Protein | ORF K. lactis |
ORF S. cerevisiae |
Function |
|---|---|---|---|
| Atg1 | KLLA0C17160g | YGL180W | Autophagy-dedicated protein serine/threonine kinase |
| Atg11 | KLLA0B12133g | YPR049C | Cytoplasm-to-vacuole targeting (Cvt) pathway and peroxisomal degradation (pexophagy) |
| Atg32 | KLLA0A00660g | YIL146C | Mitochondrial receptor specific to mitophagy |
| Atg33 | KLLA0A02695g | YLR356W | Detects or presents aged mitochondria for degradation at the stationary phase |
| Atg8 | KLLA0E20593g | YBL078C | Component of autophagosomes and Cvt vesicles |
| Aup1 | KLLA0F06985g | YCR079W | Mitochondrial protein phosphatase |
| Bck1 | KLLA0F14190g | YJL095W | MAP kinase kinase kinase acting in the protein kinase C signaling pathway |
| Dnm1 | KLLA0F12892g | YLL001W | Dynamin-related GTPase |
| Fmc1 | KLLA0F04081g | YIL098C | Assembly at high temperature of mitochondrial ATP synthase |
| Hog1 | KLLA0F20053g | YLR113W | MAP kinase involved in osmoregulation |
| Mdm38 | KLLA0B11748g | YOL027C | Mitochondrial distribution and morphology |
| Mkk1 | KLLA0D07304g | YOR231W | MAP kinase kinase acting in the protein kinase C signaling pathway |
| Mkk2 | KLLA0D07304g | YPL140C | MAP kinase kinase acting in the protein kinase C signaling pathway |
| Pbs2 | KLLA0E15313g | YJL128C | MAP kinase kinase in the osmosensing signal-transduction pathway |
| Pkc1 | KLLA0E06447g | YBL105C | Protein kinase C |
| Rtg3 | KLLA0E06513g | YBL103C | Transcription factor to activate the retrograde (RTG) and TOR pathways |
| Slt2 | KLLA0B11902g | YHR030C | MPK1 MAP kinase |
| Uth1 | KLLA0E14939g | YKR042W | Regulator outer membrane protein |
| Whi2 | KLLA0F15972g | YOR043W | Full activation of the general stress response |
| Wsc1 | KLLA0D14377g | YOR008C | Sensor transducer of the stress-activated PKC1-MPK1 kinase pathway |
| Yme1 | KLLA0E06711g | YPR024W | Protease catalytic subunit for degradation of unfolded or misfolded mitochondrial gene products |
| Ynt1 | KLLA0C06534g | YDR394W | Subunit of the 26S proteasome |
6. Conclusions and Perspectives
K. lactis is proposed as a respiratory eukaryote model, complementary to the fermentative S. cerevisiae, for the study of the pathways of hypoxia-induced oxidative stress. The experimental studies carried so far reveal that there are many differences in all the steps analysed from a comparative perspective, even when high homology exists between the acting proteins from the two yeasts. Some of these differences are briefly summarized in Table 3, although they are probably much wider than here exposed and they will increase with future studies. Besides, in silico analysis reveals that transcriptional factors and several actors from the cell-defense response (life span and mitophagy) are among the poorly homologous proteins, and therefore those become good candidates for functional characterization.
Table 3.
Main differences reported hitherto between K. lactis and S. cerevisiae, two alternative unicellular eukaryote models for hypoxic and oxidative stress responses.
| K. lactis | S. cerevisiae | |
|---|---|---|
| Crabtree effect | Negative | Positive |
| Glucose catabolism in aerobic conditions | Mainly respiratory | Mainly fermentative |
| Ratio PPP/glycolysis for glucose catabolism | High | Low |
| Reoxidation of NADPH from PPP | Mainly by mitochondrial alternative external dehydrogenases | Mainly by cytosolic NADPH oxidoreductases |
| Catabolic repression of respiration | Low | High |
| Respiratory capacity | Unlimited | Limited |
| Petite phenotype | Positive in specific mutant genetic backgrounds | Positive |
| Caloric restriction increases longevity | No | Yes |
| Aerobic/hypoxic gene pairs | Absent | Present |
| Upregulated by hypoxia | Genes related to ergosterol synthesis, cell wall composition, and glycolytic genes. OS genes: CUP1 and CUP2, HSP12, FMP46 and GRE1, and SOD1. |
Genes from the heme biosynthetic pathway, pyruvate decarboxylase, and lipid biosynthesis. OS genes: KlOYE2, KlGSH1. This response is highly dependent on the relative flux of glucose through glycolysis or PPP |
| Transcriptional regulators Hap1 and Rox1 | Not related to heme-mediated oxygen response | Related to heme-mediated oxygen response |
Many yeast genes related to the hypoxic, oxidative, and cell-defense responses are related to human diseases [127]. Although most of the studies performed hitherto about K. lactis physiology are focused on the respiro-fermentative metabolism, and much less is known about other pathways, there are representative examples of differences between K. lactis and S. cerevisiae that might be of interest for their applications in therapy of human health disorders and in the potential use of K. lactis as a model for this research. Among the potential genes or proteins of interest, SOD1 is homolog of the human gene involved in amyotrophic lateral sclerosis [128]. NDI is involved in gene therapy of complex I defects [88, 129] and is important in neurological diseases [84–86]. Several genes of ergosterol biosynthesis are targets to look for pharmacological drugs (anticholesterol, antifungal, anticancer, etc.) [125]. Diamine oxidases and catalases have been used as therapeutic approaches for the treatment of inflammatory bowel diseases, intestinal cancers, or pseudoallergic reactions [130]. Hereditary coproporphyria (HCP), an autosomal dominant acute hepatic porphyria, results from mutations in the gene that encodes coproporphyrinogen III oxidase [131]. Sirtuins have been associated to diabetes type 2 [132] and Huntington disease [104] as well as cardiopathies or cancer [109].
Neither S. cerevisiae nor other currently used models, even multicellular, manifest the complex set of alterations associated to each health disorder in humans. This makes necessary the combination of the information obtained from several models, as representative as possible of the diversity of human cell types (S. cerevisiae, K. lactis, and others), in order to advance in puzzling out the molecular basis of the diseases and in developing new preventive and therapeutic tools.
Acknowledgments
The authors thank all their colleagues, who along these years contributed to the study of the yeast hypoxic and oxidative stress responses in the authors' laboratory. This work was supported by Grant no. BFU2009-08854 from MICINN (Spain) and FEDER. General support for the laboratory during 2008–2011 was provided by Xunta de Galicia (Consolidación C.E.O.U.2008/008) and by FEDER.
References
- 1.Bharadwaj P, Martins R, Macreadie I. Yeast as a model for studying Alzheimer’s disease. FEMS Yeast Research. 2010;10(8):961–969. doi: 10.1111/j.1567-1364.2010.00658.x. [DOI] [PubMed] [Google Scholar]
- 2.Breunig KD, Bolotin-Fukuhara M, Bianchi MM, et al. Regulation of primary carbon metabolism in Kluyveromyces lactis . Enzyme and Microbial Technology. 2000;26(9-10):771–780. doi: 10.1016/s0141-0229(00)00170-8. [DOI] [PubMed] [Google Scholar]
- 3.Lepperdinger G, Berger P, Breitenbach M, et al. The use of genetically engineered model systems for research on human aging. Frontiers in Bioscience. 2008;13:7022–7031. doi: 10.2741/3207. [DOI] [PubMed] [Google Scholar]
- 4.Petranovic D, Nielsen J. Can yeast systems biology contribute to the understanding of human disease? Trends in Biotechnology. 2008;26(11):584–590. doi: 10.1016/j.tibtech.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Ocampo A, Barrientos A. From the bakery to the brain business: developing inducible yeast models of human neurodegenerative disorders. BioTechniques. 2008;45(4):7–14. doi: 10.2144/000112746. [DOI] [PubMed] [Google Scholar]
- 6.Blank LM, Lehmbeck F, Sauer U. Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Research. 2005;5(6-7):545–558. doi: 10.1016/j.femsyr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 7.González Siso MI, Freire Picos MA, Cerdán ME. Reoxidation of the NADPH produced by the pentose phosphate pathway is necessary for the utilization of glucose by Kluyveromyces lactis rag2 mutants. FEBS Letters. 1996;387(1):7–10. doi: 10.1016/0014-5793(96)00390-0. [DOI] [PubMed] [Google Scholar]
- 8.Tarrío N, Becerra M, Cerdán ME, Siso MIG. Reoxidation of cytosolic NADPH in Kluyveromyces lactis . FEMS Yeast Research. 2006;6(3):371–380. doi: 10.1111/j.1567-1364.2005.00021.x. [DOI] [PubMed] [Google Scholar]
- 9.Tarrío N, Díaz Prado S, Cerdán ME, González Siso MI. The nuclear genes encoding the internal (KlNDI1) and external (KlNDE1) alternative NAD(P)H:ubiquinone oxidoreductases of mitochondria from Kluyveromyces lactis . Biochimica et Biophysica Acta. 2005;1707(2-3):199–210. doi: 10.1016/j.bbabio.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Davidson JF, Schiestl RH. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae . Molecular and Cellular Biology. 2001;21(24):8483–8489. doi: 10.1128/MCB.21.24.8483-8489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacioglu E, Demir AB, Koc A. Identification of respiratory chain gene mutations that shorten replicative life span in yeast. Experimental Gerontology. 2012;47(2):149–153. doi: 10.1016/j.exger.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Tarrío N, García-Leiro A, Cerdán ME, González-Siso MI. The role of glutathione reductase in the interplay between oxidative stress response and turnover of cytosolic NADPH in Kluyveromyces lactis . FEMS Yeast Research. 2008;8(4):597–606. doi: 10.1111/j.1567-1364.2008.00366.x. [DOI] [PubMed] [Google Scholar]
- 13.García-Leiro A, Cerdán ME, González-Siso MI. A functional analysis of Kluyveromyces lactis glutathione reductase. Yeast. 2010;27(7):431–441. doi: 10.1002/yea.1760. [DOI] [PubMed] [Google Scholar]
- 14.Zitomer RS, Lowry CV. Regulation of gene expression by oxygen in Saccharomyces cerevisiae . Microbiological Reviews. 1992;56(1):1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies BSJ, Wang HS, Rine J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Molecular and Cellular Biology. 2005;25(16):7375–7385. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies BSJ, Rine J. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae . Genetics. 2006;174(1):191–201. doi: 10.1534/genetics.106.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickman MJ, Winston F. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Molecular and Cellular Biology. 2007;27(21):7414–7424. doi: 10.1128/MCB.00887-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becerra M, Lombardía-Ferreira LJ, Hauser NC, Hoheisel JD, Tizon B, Cerdán ME. The yeast transcriptome in aerobic and hypoxic conditions: Effects of hap1, rox1, rox3 and srb10 deletions. Molecular Microbiology. 2002;43(3):545–555. doi: 10.1046/j.1365-2958.2002.02724.x. [DOI] [PubMed] [Google Scholar]
- 19.Kwast KE, Lai LC, Menda N, James DT, Aref S, Burke PV. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: Functional roles of Rox1 and other factors in mediating the anoxic response. Journal of Bacteriology. 2002;184(1):250–265. doi: 10.1128/JB.184.1.250-265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387(6634):708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 21.Keogh RS, Seoighe C, Wolfe KH. Evolution of gene order and chromosome number in Saccharomyces, Kluyveromyces and related fungi. Yeast. 1998;14(5):443–457. doi: 10.1002/(SICI)1097-0061(19980330)14:5<443::AID-YEA243>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 22.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae . Nature. 2004;428(6983):617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 23.Betina S, Gavurnikova G, Haviernik P, Sabova L, Kolarov J. Expression of the AAC2 gene encoding the major mitochondrial ADP/ATP carrier in Saccharomyces cerevisiae is controlled at the transcriptional level by oxygen, heme and HAP2 factor. European Journal of Biochemistry. 1995;229(3):651–657. doi: 10.1111/j.1432-1033.1995.tb20510.x. [DOI] [PubMed] [Google Scholar]
- 24.Gavurníková G, Šabova L, Kiššová I, Haviernik P, Kolarov J. Transcription of the AAC1 gene encoding an isoform of mitochondrial ADP/ATP carrier in Saccharomyces cerevisiae is regulated by oxygen in a heme-independent manner. European Journal of Biochemistry. 1996;239(3):759–763. doi: 10.1111/j.1432-1033.1996.0759u.x. [DOI] [PubMed] [Google Scholar]
- 25.Sabová L, Gavurníková G, Kolarov J. Regulation of AAC isogenes encoding mitochondrial ADP/ATP translocator in the yeast Saccharomyces cerevisiae . Folia Microbiologica. 1996;41(1):124–126. doi: 10.1007/BF02816370. [DOI] [PubMed] [Google Scholar]
- 26.Blanco M, Núñez L, Tarrío N, et al. An approach to the hypoxic and oxidative stress responses in Kluyveromyces lactis by analysis of mRNA levels. FEMS Yeast Research. 2007;7(5):702–714. doi: 10.1111/j.1567-1364.2007.00233.x. [DOI] [PubMed] [Google Scholar]
- 27.Kiers J, Zeeman AM, Luttik M, et al. Regulation of alcoholic fermentation in batch and chemostat cultures of Kluyveromyces lactis CBS, 2359. Yeast. 1998;14(5):459–469. doi: 10.1002/(SICI)1097-0061(19980330)14:5<459::AID-YEA248>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 28.Snoek IS, Steensma HY. Why does Kluyveromyces lactis not grow under anaerobic conditions? Comparison of essential anaerobic genes of Saccharomyces cerevisiae with the Kluyveromyces lactis genome. FEMS Yeast Research. 2006;6(3):393–403. doi: 10.1111/j.1567-1364.2005.00007.x. [DOI] [PubMed] [Google Scholar]
- 29.Fontanesi F, Viola AM, Ferrero I. Heterologous complementation of the Klaac null mutation of Kluyveromyces lactis by the Saccharomyces cerevisiae AAC3 gene encoding the ADP/ATP carrier. FEMS Yeast Research. 2006;6(3):414–420. doi: 10.1111/j.1567-1364.2005.00011.x. [DOI] [PubMed] [Google Scholar]
- 30.González-Domínguez M, Freire-Picos MA, Ramil E, Guiard B, Cerdán ME. Heme-mediated transcriptional control in Kluyveromyces lactis . Current Genetics. 2000;38(4):171–177. doi: 10.1007/s002940000146. [DOI] [PubMed] [Google Scholar]
- 31.Blanco M, Becerra M, González-Siso MI, Cerdán ME. Functional characterization of KlHEM13, a hypoxic gene of Kluyveromyces lactis . Canadian Journal of Microbiology. 2005;51(3):241–249. doi: 10.1139/w04-133. [DOI] [PubMed] [Google Scholar]
- 32.Núñez L, Rodríguez-Torres A, Cerdán ME. Regulatory elements in the KlHEM1 promoter. Biochimica et Biophysica Acta. 2008;1779(2):128–133. doi: 10.1016/j.bbagrm.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Camattari A, Bianchi MM, Branduardi P, Porro D, Brambilla L. Induction by hypoxia of heterologous-protein production with the K1PDC1 promoter in yeasts. Applied and Environmental Microbiology. 2007;73(3):922–929. doi: 10.1128/AEM.01764-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman D, Durrens P, Beyne E, Nikolski M, Souciet JL. Génolevures: comparative genomics and molecular evolution of hemiascomycetous yeasts. Nucleic Acids Research. 2004;32:D315–D318. doi: 10.1093/nar/gkh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Micolonghi C, Ottaviano D, di Silvio E, Damato G, Heipieper H, Bianchi MM. A dual signaling pathway 5 for the hypoxic expression of lipid genes, dependent on the glucose sensor Rag4, is revealed by the analysis of KlMGA2 gene in Kluyveromyces lactis. Microbiology. 2012;158:1734–1744. doi: 10.1099/mic.0.059402-0. [DOI] [PubMed] [Google Scholar]
- 36.Wyckoff EE, Phillips JD, Sowa AM, Franklin MR, Kushner JP. Mutational analysis of human uroporphyrinogen decarboxylase. Biochimica et Biophysica Acta. 1996;1298(2):294–304. doi: 10.1016/s0167-4838(96)00148-3. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Dominguez M, Mendez-Carro C, Cerdan ME. Isolation and characterization of the KlHEM1 gene in Kluyveromyces lactis . Yeast. 1997;13(10):961–971. doi: 10.1002/(SICI)1097-0061(199708)13:10<961::AID-YEA156>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 38.Núñes L, González-Siso I, Becerra M, Cerdán ME. Functional characterisation and transcriptional regulation of the KIHEM12 gene from Kluyveromyces lactis . Current Genetics. 2004;46(3):147–157. doi: 10.1007/s00294-004-0517-5. [DOI] [PubMed] [Google Scholar]
- 39.Chelstowska A, Zoladek T, Garey J, Kushner J, Rytka J, Labbe-Bois R. Identification of amino acid changes affecting yeast uroporphyrinogen decarboxylase activity by sequence analysis of hem12 mutant alleles. The Biochemical Journal. 1992;288(3):753–757. doi: 10.1042/bj2880753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diflumeri C, Larocque R, Keng T. Molecular analysis of HEM6 (HEM12) in Saccharomyces cerevisiae, the gene for uroporphyrinogen decarboxylase. Yeast. 1993;9(6):613–623. doi: 10.1002/yea.320090608. [DOI] [PubMed] [Google Scholar]
- 41.Keng T, Guarente L. Constitutive expression of the yeast HEM1 gene is actually a composite of activation and repression. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(24):9113–9117. doi: 10.1073/pnas.84.24.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Dominguez M, Freire-Picos MA, Cerdan ME. Haem regulation of the mitochondrial import of the Kluyveromyces lactis 5-aminolaevulinate synthase: an organelle approach. Yeast. 2001;18(1):41–48. doi: 10.1002/1097-0061(200101)18:1<41::AID-YEA654>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 43.Blanco M. KlHEM13, un gen hypóxico en la biosíntesis de hemo [Ph.D. thesis] Universidad de A Coruña; 2006. [Google Scholar]
- 44.Torres AM, Maceiras ML, Belmonte ER, Naveira LN, Calvo MB, Cerdan ME. KlRox1p contributes to yeast resistance to metals and is necessary for KlYCF1 expression in the presence of cadmium. Gene. 2012;497(1):27–37. doi: 10.1016/j.gene.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 45.McNabb DS, Pinto I. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae . Eukaryotic Cell. 2005;4(11):1829–1839. doi: 10.1128/EC.4.11.1829-1839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Creusot F, Verdière J, Gaisne M, Slonimski PP. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. Journal of Molecular Biology. 1988;204(2):263–276. doi: 10.1016/0022-2836(88)90574-8. [DOI] [PubMed] [Google Scholar]
- 47.Pfeifer K, Kim KS, Kogan S, Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Hach A. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cellular and Molecular Life Sciences. 1999;56(5-6):415–426. doi: 10.1007/s000180050442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hon T, Hach A, Lee HC, Cheng T, Zhang L. Functional analysis of heme regulatory elements of the transcriptional activator Hap1. Biochemical and Biophysical Research Communications. 2000;273(2):584–591. doi: 10.1006/bbrc.2000.2995. [DOI] [PubMed] [Google Scholar]
- 50.Hon T, Lee HC, Hach A, et al. The Hsp70-Ydj1 molecular chaperone represses the activity of the heme activator protein Hap1 in the absence of heme. Molecular and Cellular Biology. 2001;21(23):7923–7932. doi: 10.1128/MCB.21.23.7923-7932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klinkenberg LG, Mennella TA, Luetkenhaus K, Zitomer RS. Combinatorial repression of the hypoxic genes of Saccharomyces cerevisiae by DNA binding proteins Rox1 and Mot3. Eukaryotic Cell. 2005;4(4):649–660. doi: 10.1128/EC.4.4.649-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castro-Prego R, Lamas-Maceiras M, Soengas P, Carneiro I, González-Siso I, Cerdán ME. Regulatory factors controlling transcription of Saccharomyces cerevisiae IXR1 by oxygen levels: a model of transcriptional adaptation from aerobiosis to hypoxia implicating ROX1 and IXR1 cross-regulation. Biochemical Journal. 2010;425(1):235–243. doi: 10.1042/BJ20091500. [DOI] [PubMed] [Google Scholar]
- 53.Castro-Prego R, Lamas-Maceiras M, Soengas P, et al. Ixr1p regulates oxygen-dependent HEM13 transcription. FEMS Yeast Research. 2010;10(3):309–321. doi: 10.1111/j.1567-1364.2010.00616.x. [DOI] [PubMed] [Google Scholar]
- 54.Vizoso-Vazquez A, Lamas-Maceiras M, Becerra M, Gonzalez-Siso MI, Rodriguez-Belmonte E, Cerdan ME. Ixr1p and the control of the Saccharomyces cerevisiae hypoxic response, Applied Microbiology and Biotechnology. 2012;94(1):173–184. doi: 10.1007/s00253-011-3785-2. [DOI] [PubMed] [Google Scholar]
- 55.Mulder W, Scholten IHJM, De Boer RW, Grivell LA. Sequence of the HAP3 transcription factor of Kluyveromyces lactis predicts the presence of a novel 4-cysteine zinc-finger motif. Molecular and General Genetics. 1994;245(1):96–106. doi: 10.1007/BF00279755. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen C, Bolotin-Fukuhara M, Wésolowski-Louvel M, Fukuhara H. The respiratory system of Kluyveromyces lactis escapes from HAP2 control. Gene. 1995;152(1):113–115. doi: 10.1016/0378-1119(94)00684-k. [DOI] [PubMed] [Google Scholar]
- 57.Bourgarel D, Nguyen CC, Bolotin-Fukuhara M. HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis . Molecular Microbiology. 1999;31(4):1205–1215. doi: 10.1046/j.1365-2958.1999.01263.x. [DOI] [PubMed] [Google Scholar]
- 58.Lamas-Maceiras M, Núñez L, Rodríguez-Belmonte E, González-Siso MI, Cerdán ME. Functional characterization of KlHAP1: a model to foresee different mechanisms of transcriptional regulation by Hap1p in yeasts. Gene. 2007;405(1-2):96–107. doi: 10.1016/j.gene.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 59.Bao WG, Guiard B, Fang ZA, et al. Oxygen-dependent transcriptional regulator hap1p limits glucose uptake by repressing the expression of the major glucose transporter gene RAG1 in Kluyveromyces lactis . Eukaryotic Cell. 2008;7(11):1895–1905. doi: 10.1128/EC.00018-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fang ZA, Wang GH, Chen AL, et al. Gene responses to oxygen availability in Kluyveromyces lactis: an insight on the evolution of the oxygen-responding system in yeast. PLoS ONE. 2009;4(10) doi: 10.1371/journal.pone.0007561.e7561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwast KE, Burke PV, Staahl BT, Poyton RO. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(10):5446–5451. doi: 10.1073/pnas.96.10.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang Y, Vasconcelles MJ, Wretzel S, Light A, Martin CE, Goldberg MA. MGA2 is involved in the low-oxygen response element-dependent hypoxic induction of genes in Saccharomyces cerevisiae . Molecular and Cellular Biology. 2001;21(18):6161–6169. doi: 10.1128/MCB.21.18.6161-6169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ness F, Bourot S, Régnacq M, Spagnoli R, Bergès T, Karst F. SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. European Journal of Biochemistry. 2001;268(6):1585–1595. [PubMed] [Google Scholar]
- 64.Régnacq M, Alimardani P, El Moudni B, Bergès T. Sut1p interaction with Cyc8p(Ssn6p) relieves hypoxic genes from Cyc8p-Tup1p repression in Saccharomyces cerevisiae . Molecular Microbiology. 2001;40(5):1085–1096. doi: 10.1046/j.1365-2958.2001.02450.x. [DOI] [PubMed] [Google Scholar]
- 65.Vik Å, Rine J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae . Molecular and Cellular Biology. 2001;21(19):6395–6405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abramova NE, Cohen BD, Sertil O, Kapoor R, Davies KJA, Lowry CV. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae . Genetics. 2001;157(3):1169–1177. doi: 10.1093/genetics/157.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abramova N, Sertil O, Mehta S, Lowry CV. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae . Journal of Bacteriology. 2001;183(9):2881–2887. doi: 10.1128/JB.183.9.2881-2887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becerra M, Tarrío N, González-Siso MI, Cerdán ME. Genome-wide analysis of Kluyveromyces lactis in wild-type and rag2 mutant strains. Genome. 2004;47(5):970–978. doi: 10.1139/g04-039. [DOI] [PubMed] [Google Scholar]
- 69.González-Siso MI, García-Leiro A, Tarrío N, Cerdán ME. Sugar metabolism, redox balance and oxidative stress response in the respiratory yeast Kluyveromyces lactis . Microbial Cell Factories. 2009;8, article 46 doi: 10.1186/1475-2859-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koleva DI, Petrova VY, Kujumdzieva AV. Comparison of enzymatic antioxidant defence systems in different metabolic types of yeasts. Canadian Journal of Microbiology. 2008;54(11):957–963. doi: 10.1139/w08-093. [DOI] [PubMed] [Google Scholar]
- 71.Lee J, Godon C, Lagniel G, et al. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. The Journal of Biological Chemistry. 1999;274(23):16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 72.Tarrío N, Díaz Prado S, Cerdán ME, González Siso MI. Isolation and characterization of two nuclear genes encoding glutathione and thioredoxin reductases from the yeast Kluyveromyces lactis . Biochimica et Biophysica Acta. 2004;1678(2-3):170–175. doi: 10.1016/j.bbaexp.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 73.García-Leiro A, Cerdán ME, González-Siso MI. Proteomic analysis of the oxidative stress response in Kluyveromyces lactis and effect of glutathione reductase depletion. Journal of Proteome Research. 2010;9(5):2358–2376. doi: 10.1021/pr901086w. [DOI] [PubMed] [Google Scholar]
- 74.Tan SX, Greetham D, Raeth S, Grant CM, Dawes IW, Perrone GG. The thioredoxin-thioredoxin reductase system can function in vivo as an alternative system to reduce oxidized glutathione in Saccharomyces cerevisiae . Journal of Biological Chemistry. 2010;285(9):6118–6126. doi: 10.1074/jbc.M109.062844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raimondi S, Zanni E, Talora C, Rossi M, Palleschi C, Uccelletti D. SOD1, a new Kluyveromyces lactis helper gene for heterologous protein secretion. Applied and Environmental Microbiology. 2008;74(23):7130–7137. doi: 10.1128/AEM.00955-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dirmeier R, O’Brien KM, Engle M, Dodd A, Spears E, Poyton RO. Exposure of yeast cells to anoxia induces transient oxidative stress: implications for the induction of hypoxic genes. Journal of Biological Chemistry. 2002;277(38):34773–34784. doi: 10.1074/jbc.M203902200. [DOI] [PubMed] [Google Scholar]
- 77.Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) The EMBO Journal. 1996;15(9):2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 78.Izawa S, Ikeda K, Ohdate T, Inoue Y. Msn2p/Msn4p-activation is essential for the recovery from freezing stress in yeast. Biochemical and Biophysical Research Communications. 2007;352(3):750–755. doi: 10.1016/j.bbrc.2006.11.100. [DOI] [PubMed] [Google Scholar]
- 79.Sugiyama KI, Izawa S, Inoue Y. The Yap1p-dependent induction of glutathione synthesis in heat shock response of Saccharomyces cerevisiae . The Journal of Biological Chemistry. 2000;275(20):15535–15540. doi: 10.1074/jbc.275.20.15535. [DOI] [PubMed] [Google Scholar]
- 80.Billard P, Dumond H, Bolotin-Fukuhara M. Characterization of an AP-1-like transcription factor that mediates an oxidative stress response in Kluyveromyces lactis . Molecular and General Genetics. 1997;257(1):62–70. doi: 10.1007/s004380050624. [DOI] [PubMed] [Google Scholar]
- 81.Gonzalez-Siso MI, Cerdan ME. Mitochondrial alternative NAD(P)H-dehydrogenases and respiro-fermentative metabolism in yeast. In: Gonzalez-Siso MI, Cerdan ME, editors. Complex I and Alternative Dehydrogenases. Kerala, India: Transworld Research Network; 2007. pp. 69–84. [Google Scholar]
- 82.Tarrío N, Cerdán ME, Siso MIG. Characterization of the second external alternative dehydrogenase from mitochondria of the respiratory yeast Kluyveromyces lactis . Biochimica et Biophysica Acta. 2006;1757(11):1476–1484. doi: 10.1016/j.bbabio.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 83.Li W, Sun L, Liang Q, Wang J, Mo W, Zhou B. Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Molecular Biology of the Cell. 2006;17(4):1802–1811. doi: 10.1091/mbc.E05-04-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marella M, Byoung BS, Matsuno-Yagi A, Yagi T. Mechanism of cell death caused by complex I defects in a rat dopaminergic cell line. Journal of Biological Chemistry. 2007;282(33):24146–24156. doi: 10.1074/jbc.M701819200. [DOI] [PubMed] [Google Scholar]
- 85.Marella M, Seo BB, Yagi T, Matsuno-Yagi A. Parkinson’s disease and mitochondrial complex I: a perspective on the Ndi1 therapy. Journal of Bioenergetics and Biomembranes. 2009;41(6):493–497. doi: 10.1007/s10863-009-9249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park JS, Li YF, Bai Y. Yeast NDI1 improves oxidative phosphorylation capacity and increases protection against oxidative stress and cell death in cells carrying a Leber’s hereditary optic neuropathy mutation. Biochimica et Biophysica Acta. 2007;1772(5):533–542. doi: 10.1016/j.bbadis.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marella M, Seo BB, Thomas BB, Matsuno-Yagi A, Yagi T. Successful amelioration of mitochondrial optic neuropathy using the yeast NDI1 gene in a rat animal model. PLoS ONE. 2010;5(7) doi: 10.1371/journal.pone.0011472.e11472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yagi T, Seo BB, Nakamaru-Ogiso E, et al. Can a single subunit yeast NADH dehydrogenase (Ndi1) remedy diseases caused by respiratory complex I defects? Rejuvenation Research. 2006;9(2):191–197. doi: 10.1089/rej.2006.9.191. [DOI] [PubMed] [Google Scholar]
- 89.Ralser M, Wamelink MM, Kowald A, et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. Journal of Biology. 2007;6(4, article 10) doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hon T, Hee CL, Hu Z, Iyer VR, Zhang L. The heme activator protein Hap1 represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae . Genetics. 2005;169(3):1343–1352. doi: 10.1534/genetics.104.037143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kennedy MA, Barbuch R, Bard M. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae . Biochimica et Biophysica Acta. 1999;1445(1):110–122. doi: 10.1016/s0167-4781(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 92.Turi TG, Loper JC. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14α-demethylase (ERG11) Journal of Biological Chemistry. 1992;267(3):2046–2056. [PubMed] [Google Scholar]
- 93.Jensen-Pergakes K, Guo Z, Giattina M, Sturley SL, Bard M. Transcriptional regulation of the two sterol esterification genes in the yeast Saccharomyces cerevisiae . Journal of Bacteriology. 2001;183(17):4950–4957. doi: 10.1128/JB.183.17.4950-4957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong CM, Ching YP, Zhou Y, Kung HF, Jin DY. Transcriptional regulation of yeast peroxiredoxin gene TSA2 through Hap1p, Rox1p, and Hap2/3/5p. Free Radical Biology and Medicine. 2003;34(5):585–597. doi: 10.1016/s0891-5849(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 95.Pinkham JL, Wang Z, Alsina J. Heme regulates SOD2 transcription by activation and repression in Saccharomyces cerevisiae . Current Genetics. 1997;31(4):281–291. doi: 10.1007/s002940050207. [DOI] [PubMed] [Google Scholar]
- 96.Pan Y. Mitochondria, reactive oxygen species, and chronological aging: a message from yeast. Experimental Gerontology. 2011;46(11):847–852. doi: 10.1016/j.exger.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 97.Gruning NM, Rinnerthaler M, Bluemlein K, et al. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metabolism. 2011;14(3):415–427. doi: 10.1016/j.cmet.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anastasiou D, Poulogiannis G, Asara JM, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334(6060):1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183(4677):1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 100.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae . Aging Cell. 2003;2(2):73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 101.Sharma PK, Agrawal V, Roy N. Mitochondria-mediated hormetic response in life span extension of calorie-restricted Saccharomyces cerevisiae . Age. 2011;33(2):143–154. doi: 10.1007/s11357-010-9169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oliveira GA, Tahara EB, Gombert AK, Barros MH, Kowaltowski AJ. Increased aerobic metabolism is essential for the beneficial effects of caloric restriction on yeast life span. Journal of Bioenergetics and Biomembranes. 2008;40(4):381–388. doi: 10.1007/s10863-008-9159-5. [DOI] [PubMed] [Google Scholar]
- 103.Ralser M, Lehrach H. Building a new bridge between metabolism, free radicals and longevity. Aging. 2009;1(10):836–838. doi: 10.18632/aging.100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sorolla MA, Nierga C, Rodríguez-Colman MJ, et al. Sir2 is induced by oxidative stress in a yeast model of Huntington disease and its activation reduces protein aggregation. Archives of Biochemistry and Biophysics. 2011;510(1):27–34. doi: 10.1016/j.abb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 105.Gavin AC, Bösche M, Krause R, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 106.Ralser M, Michel S, Breitenbach M. Sirtuins as regulators of the yeast metabolic network. Frontiers in Pharmacology. 2012;3, article 32 doi: 10.3389/fphar.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee YL, Lee CK. Transcriptional response according to strength of calorie restriction in Saccharomyces cerevisiae . Molecules and Cells. 2008;26(3):299–307. [PubMed] [Google Scholar]
- 108.Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes and Development. 2008;22(7):931–944. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carafa V, Nebbioso A, Altucci L. Sirtuins and disease: the road ahead. Frontiers in Pharmacology. 2012;3, article 4 doi: 10.3389/fphar.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Molecular Cell. 2002;10(3):457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 111.Neklesa TK, Davis RW. Superoxide anions regulate TORC1 and its ability to bind Fpr1:rapamycin complex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):15166–15171. doi: 10.1073/pnas.0807712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hamann A, Brust D, Osiewacz HD. Apoptosis pathways in fungal growth, development and ageing. Trends in Microbiology. 2008;16(6):276–283. doi: 10.1016/j.tim.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 113.Mazzoni C, Mancini P, Verdone L, et al. A truncated form of KlLsm4p and the absence of factors involved in mRNA decapping trigger apoptosis in yeast. Molecular Biology of the Cell. 2003;14(4):721–729. doi: 10.1091/mbc.E02-05-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Poliaková D, Sokolíková B, Kolarov J, Šabová L. The antiapoptotic protein Bcl-XL prevents the cytotoxic effect of Bax, but not Bax-induced formation of reactive oxygen species, in Kluyveromyces lactis . Microbiology. 2002;148(9):2789–2795. doi: 10.1099/00221287-148-9-2789. [DOI] [PubMed] [Google Scholar]
- 115.Kost’anova-Poliakova D, Šabová L. Lactate utilization in mitochondria prevents Bax cytotoxicity in yeast Kluyveromyces lactis . FEBS Letters. 2005;579(23):5152–5156. doi: 10.1016/j.febslet.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 116.Lodi T, Donnini C. Lactose-induced cell death of β-galactosidase mutants in Kluyveromyces lactis . FEMS Yeast Research. 2005;5(8):727–734. doi: 10.1016/j.femsyr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 117.García-Leiro A, Fernandes AS, Castro A, Videira A, Cerdán ME, González-Siso MI. Glutathione: Biochemistry, Mechanisms of Action and Clinical Implications. New York, NY, USA: Nova Science; 2012. Glutathione and programmed cell death in Kluyveromyces lactis . [Google Scholar]
- 118.Minibayeva F, Dmitrieva S, Ponomareva A, Ryabovol V. Oxidative Stress-Induced Autophagy in Plants: The Role of Mitochondria. Plant Physiology and Biochemistry: PPB/Societe Francaise De Physiologie Vegetale; 2012. [DOI] [PubMed] [Google Scholar]
- 119.Kurihara Y, Kanki T, Aoki Y, et al. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. The Journal of Biological Chemistry. 2012;287(5):3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kanki T, Klionsky DJ, Okamoto K. Mitochondria autophagy in yeast. Antioxidants and Redox Signaling. 2011;14(10):1989–2001. doi: 10.1089/ars.2010.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deffieu M, Bhatia-Kiššová I, Salin B, Galinier A, Manon S, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. The Journal of Biological Chemistry. 2009;284(22):14828–14837. doi: 10.1074/jbc.M109.005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kiššová IB, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. Autophagy. 2009;5(6):872–873. doi: 10.4161/auto.9065. [DOI] [PubMed] [Google Scholar]
- 123.Okamoto K, Kondo-Okamoto N. Mitochondria and autophagy: critical interplay between the two homeostats. Biochimica Et Biophysica Acta. 2012;1820(5):595–600. doi: 10.1016/j.bbagen.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 124.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxidants and Redox Signaling. 2011;14(11):2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- 125.Bhatia-Kiššová I, Camougrand N. Mitophagy in yeast: actors and physiological roles. FEMS Yeast Research. 2010;10(8):1023–1034. doi: 10.1111/j.1567-1364.2010.00659.x. [DOI] [PubMed] [Google Scholar]
- 126.Hirota Y, Kang D, Kanki T. The physiological role of mitophagy: new insights into phosphorylation events. International Journal of Cell Biology. 2012;2012:8 pages. doi: 10.1155/2012/354914.354914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bolotin-Fukuhara M, Dumas B, Gaillardin C. Yeasts as a model for human diseases. FEMS Yeast Research. 2010;10(8):959–960. doi: 10.1111/j.1567-1364.2010.00693.x. [DOI] [PubMed] [Google Scholar]
- 128.Morimoto N, Miyazaki K, Kurata T, et al. Effect of mitochondrial transcription factor a overexpression on motor neurons in amyotrophic lateral sclerosis model mice. Journal of Neuroscience Research. 2012;90(6):1200–1208. doi: 10.1002/jnr.23000. [DOI] [PubMed] [Google Scholar]
- 129.Yagi T, Seo BB, Nakamaru-Ogiso E, et al. Possibility of transkingdom gene therapy for Complex I diseases. Biochimica et Biophysica Acta. 2006;1757(5-6):708–714. doi: 10.1016/j.bbabio.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 130.Calinescu C, Mondovi B, Federico R, Ispas-Szabo P, Mateescu MA. Carboxymethyl starch: chitosan monolithic matrices containing diamine oxidase and catalase for intestinal delivery. International Journal of Pharmaceutics. 2012;428(1-2):48–56. doi: 10.1016/j.ijpharm.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 131.Schmitt C, Gouya L, Malonova E, et al. Mutations in human CPO gene predict clinical expression of either hepatic hereditary coproporphyria or erythropoietic harderoporphyria. Human Molecular Genetics. 2005;14(20):3089–3098. doi: 10.1093/hmg/ddi342. [DOI] [PubMed] [Google Scholar]
- 132.Calabrese V, Cornelius C, Leso V, et al. Oxidative stress, glutathione status, sirtuin and cellular stress response in type 2 diabetes. Biochimica Et Biophysica Acta. 2012;1822(5):729–736. doi: 10.1016/j.bbadis.2011.12.003. [DOI] [PubMed] [Google Scholar]


