Abstract
When growing on solid surfaces, yeast, like other microorganisms, develops organized multicellular populations (colonies and biofilms) that are composed of differentiated cells with specialized functions. Life within these populations is a prevalent form of microbial existence in natural settings that provides the cells with capabilities to effectively defend against environmental attacks as well as efficiently adapt and survive long periods of starvation and other stresses. Under such circumstances, the fate of an individual yeast cell is subordinated to the profit of the whole population. In the past decade, yeast colonies, with their complicated structure and high complexity that are also developed under laboratory conditions, have become an excellent model for studies of various basic cellular processes such as cell interaction, signaling, and differentiation. In this paper, we summarize current knowledge on the processes related to chronological aging, adaptation, and longevity of a colony cell population and of its differentiated cell constituents. These processes contribute to the colony ability to survive long periods of starvation and mostly differ from the survival strategies of individual yeast cells.
1. Introduction
The molecular basis of aging has been examined using numerous methods and organisms and is one of the main biological questions under investigation. A substantial amount of information on individual gene products, metabolic pathways, signal transduction cascades, environmental factors, and cellular mechanisms impacting aging is available, but a deeper insight into how all these components interact under specific conditions and how they contribute to the process of aging is still unavailable. In addition, aging is tightly linked with the process of adaptation, through which the cells adjust to a hostile environment and can reenter the proliferative state. With metazoans, this potentially leads to tumor formation and the subsequent death of the organism.
Yeast cells have been used for a long time as a tool for identifying the genes and pathways involved in basic cellular processes such as the cell cycle, aging, and stress response. Two types of lifespan, the replicative and the chronological, have been defined and studied in the yeast Saccharomyces cerevisiae [1]. While the replicative (mitotic) lifespan is characterized by the number of divisions that an individual yeast cell can undergo, the chronological (postmitotic) lifespan represents a period during which nondividing (stationary phase, G0) yeast cells remain viable. Various genes and pathways have been identified as being involved in both types of lifespan in yeast. For example, several nutrient sensing pathways affect the chronological lifespan in a way that their inactivation leads to a longevity phenotype. This was shown for the TOR (target of rapamycin), protein kinase A, and Sch9p pathways, as deletions of TOR1, RAS2, SCH9, or rapamycin treatment increase chronological lifespan in yeast [2–5].
Moreover, metabolic products can also modulate chronological lifespan. Acetic acid-induced medium acidification was shown to be a cause of accelerated aging in glucose-grown yeast cultures [1, 6]. Importantly, these mechanisms are evolutionary conserved from yeast to mammals, as rapamycin treatment induces longevity in yeast, worms, fruit flies, and mice [7], and lactate-induced acidification induces senescence in human tumor cells [8]. Various high-throughput screens performed to identify the genes responsible for long-living versus short-living yeast phenotypes led to the identification of large collections of genes, which highly differed between individual screens, however [4, 9]. These observations indicate that chronological lifespan is dependent on specific environmental conditions (e.g., nutrient sources), genetic background, and developmental program. Similarly, many recent results point to the conclusion that the properties of stationary-phase cells are also dependent on environmental conditions [10–15]. In other words, there are different types of chronologically aged (i.e., stationary-phase) yeast populations under different conditions. In addition, chronological lifespan analyses in liquid cultivations are complicated by the facts that cells are heterogeneous and mutual interactions between the cellular subpopulations are difficult to analyze.
Various findings suggested that different mechanisms and regulations can occur in liquid cultivations (where yeast cells behave as unicellular individuals) and in precisely structured multicellular colonies. During the past decade, studies on multicellular yeast colonies have developed in different directions and indicated that colonies represent a promising model for studying various aspects of microbial multicellularity. Thus, findings on colony communication, adaptation, and the differing longevity of cell subpopulations occupying different colony regions raised an attractive possibility of using colonies as a model for studies of processes such as stress defense, aging, adaptation, programmed cell death (PCD), and longevity, that is, of processes that are important for any organism and that are linked to each other. In addition, recent findings on the differentiation of S. cerevisiae laboratory strain smooth colonies and of structured biofilm colonies formed by natural isolates of S. cerevisiae showed that different developmental programs can be realized by yeast communities growing on solid media [10, 16]. These programs are dependent on both environmental factors such as medium composition, and, importantly, also on phenotypic switching between different colony types that is reversible and epigenetically regulated in some cases. Thus, very probably, processes and mechanisms related to aging and adaptation that have been already identified and that are discussed in this paper are only a fraction of the broad capabilities of yeast cells developing within differently organized colonies.
In this paper, we summarize current knowledge on aging-related processes that have been described in colonies of the two different types, those formed by laboratory strains and those formed by strains isolated from natural settings.
2. Cell Growth and Developmental Phases of Smooth Colonies Formed by S. cerevisiae Laboratory Strains
Yeast cells growing in shaken liquid media exhibit a typical grow profile. After lag phase (adaptation to growth medium), it begins with an exponential growth phase, which is characterized by rapid growth under conditions of nutrient abundance, followed by a diauxic phase, which is characterized by slower growth as a consequence of a limitation of the preferred nutrients and modulation of the cellular metabolism to utilize alternative nutrients. A typical example is the shift from fermentative to respiratory growth. These two phases of growth are followed by a stationary phase when the growth is significantly reduced or stopped. The duration of the respective phases differs depending on the type and amount of nutrient sources and on the yeast population doubling time. Similarly, early growth of smooth colonies of S. cerevisiae laboratory strains starts with an exponential growth phase. On complex glucose medium (YPD), this phase lasts about 42 hours with a doubling time that is independent of colony density and similar to the doubling time of cells growing in YPD liquid medium. The majority of the cells in a colony divide during this phase [17]. Later, the number of budding cells drops to about 15% and the colonies continue to grow more slowly. During this second “slow-growth” phase, the colony population starts to diversify. While cell growth continues mostly at the colony periphery stationary-phase cells appear in the colony center, as documented by the activation of the expression of the stationary-phase specific gene SSA3 in the colony center and growth-specific gene ACT1 at the colony margin. In addition to preferential cell growth at the colony periphery [17, 18], cells in the uppermost layers of the colony center continue to divide efficiently after 3 days of colony growth. Cells in the lower layers of the colony center enter a stationary phase at that time [10]. Slowly dividing cells are present in all marginal regions and in the central uppermost cell layers, even in older colonies (Figure 1(b)).
Figure 1.
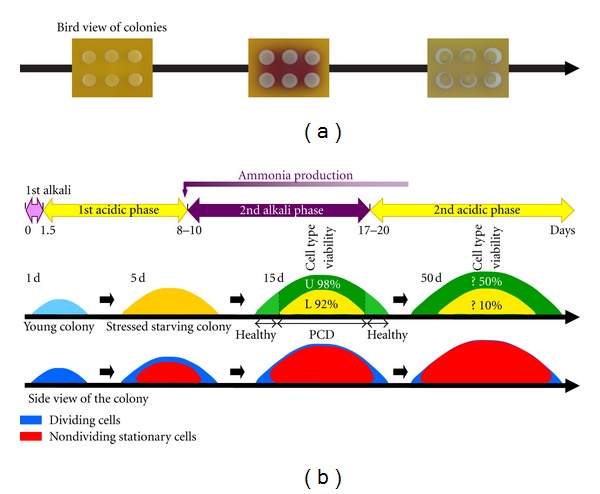
(a) Development of colonies formed by laboratory strain of S. cerevisiae. Colonies develop on complex glycerol agar with Bromocresol purple, pH dye indicator with pKa of 6.3, the color of which changes from yellow at acidic pH to purple at more alkali pH. (b) Upper part: process of colony differentiation to cells with specific metabolic properties localized in upper (U cells) and lower (L cells) layers of the colony center, respectively. Viability: percentage of colony-forming cells is given. PCD: programmed cell death. Lower part: presence of subpopulations of dividing and nondividing cells.
In contrast to the typical diauxic shift in liquid cultures caused by the change from fermentative to respiratory growth, the shift between the exponential and the slow-growth phase of colonies was independent of the carbon source used. Thus, colonies underwent these phases on both fermentative and respiratory medium, however, the exponential phase was shorter on fermentative (about 2 days) than on respiratory medium (about 3 days) where cells grow more slowly [17]. The slow growth phase continued even after 8 days of growth, while the diauxic slow growth in liquid cultures was almost stopped after 40 hours (or even earlier) [17]. These observations are in agreement with the finding that the biomass of S. cerevisiae giant colonies increases linearly when growing on complex respiratory medium during a relatively long interval from day 3 to day 16–18. In colonies older than 18 days the biomass accrual is reduced, but not stopped completely [18, 19]. This growth profile is probably a consequence of two factors: firstly, only a small subpopulation of cells located in specific colony regions undergo cell division and proliferate during colony growth, and secondly, nutrients only reach the cells by the slow process of diffusion, creating conditions of the slow continuous supply of low concentrations of nutrients.
Various data indicate that the entry of colony cell population to the slow growing/stationary phase is only the starting point of various metabolic and other changes that occur during subsequent colony development. Accordingly, cells within colonies survive much longer than a yeast population in shaken liquid cultivations. The number of colony forming cells (CFC) (i.e., the cells that regrow when replated on fresh media) decreased to about 2–5% after 10 days of cultivation in liquid complete synthetic glucose medium [20, 21] and to 10% and 30% after a 10-day cultivation in synthetic medium with ethanol and glycerol, respectively, as the carbon source [5]. Complex medium is much better for population survival, as 40–70% of the population survive after a 10-day cultivation in complex glucose medium (YPD) [22, 23], while respiratory complex medium shows the best results for long-term survival, as about 50–90% of cells are able to form colonies after 10 days in liquid glycerol complex medium [23, 24]. The population of the colony of a relatively long-living BY4742 strain grown on complex respiratory medium contained 90% CFC after 10 days of cultivation, which is almost double the value in liquid cultivations of the same strain grown in a similar medium [18, 19, 24]. Moreover, during prolonged cultivation the difference in survival between liquid cultivations and colonies becomes even more evident. Liquid cultures rapidly lose viability over time while a colonial population possesses a high fraction of CFC, even in very old colonies (75% on day 30, 40% on day 50, and 5% on day 135) [19]. Thus, the survival rate in any liquid cultivation does not reach the survival rate in colonies. In addition, it seems that cell survival within the colony is even better, as a subpopulation of resting cells resembling VBNC (“viable-but-non-culturable”) cells of bacteria [25] emerges within the colony after day 20 of its development [19]. Such data indicate that different types of stationary-phase and slow-growing cells are present within colonies (see also below).
3. Metabolic Reprogramming and Stress Defense of Smooth Colonies
In addition to developmental changes related to cell division and colony growth, metabolic reprogramming has been observed during the development of both microcolonies and giant colonies of S. cerevisiae grown on complex respiratory medium. These metabolic changes are accompanied by changes in external pH, which shifts from near to alkali to acidic and vice versa (Figure 1(a)) [26–29]. The so-called alkali phases are linked with the production of volatile ammonia that functions as a signal influencing colony metabolic reprogramming and cell differentiation. The first alkali phase starts early after giant colonies are inoculated and lasts until about 24–35 hours, that is, it approximately correlates with the exponential growth phase described above. Later, in parallel to colony entry to the slow growth phase [17], colonies start to intensively acidify their surroundings [26]. At day 8–10 of colony growth, the second alkali phase is initiated, that lasts until approximately day 17–20 [30] and is accompanied by expressive metabolic changes. Linear biomass accrual of colonies continues during this alkali phase. Only later, when colonies enter the second acidic phase, linear biomass accrual gradually decreases. The metabolic reprogramming that occurs during the second alkali phase includes the abrupt activation of some metabolic genes, including those of amino acid metabolism, peroxisomal functions (including fatty acid β-oxidation) and of some alternative branches of carbon metabolism (such as the methylglyoxylate cycle) [26]. Genes for various transporters including putative ammonium transporters (e.g., Ato1p) and transporters that may contribute to pH alterations (e.g., phosphate, sulphate and carboxylic acid transporters) are also strongly induced. In contrast, other cellular functions are gradually decreased, such as the expression of genes involved in mitochondrial oxidative respiration and those belonging to the group of environmental stress response genes [26]. Moreover, it was shown that the functionality of some of the stress defense enzymes such as cytosolic superoxide dismutase Sod1p, which is crucial for yeast longevity in liquid cultivations [31, 32], are dispensable in the healthy development and survival of a colony population [33]. These findings raised the intriguing possibility that chronologically aged colonies activate metabolic pathways that may allow them to exploit some previously released waste products and that are more economical than the metabolism of young colonies growing in nutrient abundance. This metabolism, paralleled with reduced mitochondrial phosphorylation, subsequently allows the cells to decrease the level of stress evoked by nutrient depletion within colonies and promotes population longevity. This adaptation, thus, subsequently makes stress eliminating systems (e.g., catalases and superoxide dismutases) unnecessary. Importantly, colonies formed by strains defective in ammonia signaling and in the activation of an adaptive metabolism decrease their vitality over long periods of starvation [33, 34]. The metabolic changes activated at the beginning of the second alkali phase partially persist in the second acidic phase and are supplemented by additional changes, the function of which is mostly unknown [30].
4. Smooth Colony Center-Margin Differentiation and PCD Contribute to Population Longevity
As indicated above, from the third day of colony growth on respiratory medium, the colony diversifies, and nondividing cells can be found in lower central colony areas (Figure 1(b)). Subsequently, the number of dividing cells gradually decreases and, for example, in 7-day old colonies dividing cells are present mostly at the colony margin and in the uppermost layer of the colony.
In addition to the localization of dividing/nondividing cells into distinct colony areas, colony stratification and the differentiation of chronologically aged nondividing cells has been discovered to occur later in aging colonies. Such differentiation is in terms of metabolic differences, stress-related characteristics and the survival of specifically localized colony cell subpopulations. In the late first acidic phase, nutrients seem to be exhausted and a level of reactive oxygen species (ROS) starts to increase in cells randomly distributed within the colony, including infant cells located at the colony margin. In parallel with the transition to the second alkali phase, the colony became differentiated both “horizontally” and “vertically” (and independently of whether cells are dividing or not).
When examined in the horizontal direction, central chronologically aging cells maintain higher stress-defense enzyme activities [30, 33] and a fraction of these cells undergoes PCD (Figure 1(b)) [18, 33]. On the other hand, relatively young infant cells at the colony margin activate the adaptive metabolism and acquire the phenotype of healthy cells with a low ROS level. This center-margin diversification is dependent on alkalization and ammonia signaling, as colonies that fail to enter the alkali phase and produce ammonia contain dying cells spread throughout the colony, including the margin region [18, 33]. Interestingly, the removal of central cells from a differentiated colony diminished accrual at the colony margin [18]. These data showed that a kind of PCD occurs within the central regions of an aged colony. PCD is regulated in parallel with medium alkalization and ammonia production, and is an important factor in the subsequent longevity and development of cells inhabiting more propitious areas at the colony margin.
Potentially, dying cells of chronologically aged central areas may release some compounds that may serve as nutrients for late colony growth. When compared with accidental (or regulated) cell lysis, PCD exhibiting apoptosis-like features prevents the release of various lytic enzymes (proteases and other hydrolases) that could destroy healthy cells located in the vicinities of dying cells. As shown by confocal microscopy, cells are tightly attached within laboratory yeast colonies [35]. Thus, PCD in colonies could have a similar purpose as the apoptosis of a fraction of the cells during the development of a real multicellular organism. On the other hand, while apoptosis in multicellular organisms depends on a group of evolutionary conserved effectors and pathways, PCD observed within the colonies is independent of the presence of the yeast metacaspase Mca1p and the yeast homologue of apoptosis-inducing factor Aif1p, which implies that programmed dying in yeast colonies is regulated independently of these evolutionary conserved factors. As alternative pathways involved in PCD in yeast are proposed [36, 37], these could regulate cell dying in yeast colonies. Such regulated dying contributes to the longevity of the whole colony by providing nutrients released by the death of one subpopulation to the benefit of the other subpopulations.
5. Differentiation of Chronologically Aged Cells in Distinct Smooth Colony Layers
A confocal microscopy study on the localization of GFP-labeled transporter Ato1p showed that the central colony population is not homogeneous and that Ato1p is exclusively produced in the upper cell layers of the colony, that is, this protein starts to be produced both in the colony margin and in upper central colony areas during colony entry to the second alkali phase [35]. The localization of Ato1p-producing cells in the upper layer of the colony center corresponds to the localization of one of the two major cell subpopulations revealed by a recent study [10]. This study provided a complex view of the properties of these two cell subpopulations, the upper and the lower, that form the colony center. These subpopulations are formed by the differentiation of cells of late acidic-phase colonies (approximately 7-day-old colonies), that is, by the differentiation of a population composed mostly of chronologically aged cells with only a negligible fraction of slowly dividing cells in most upper colony regions. Colony stratification develops over about the next 3 days and both layers (upper and lower) are fully developed in colonies more than 10 days old. Thus, the population of chronologically aged stressed cells of the colony center differentiates into two prominent subpopulations that apparently differ from each other as well as from their ancestors (Figure 1(b)).
Cells in the upper colony layer (U cells) gain a high resistance to stress and exhibit a longevity phenotype, while cells in the lower layers (L cells) are sensitive to stress and die more rapidly [10]. This simple comparison would imply that U and L cells could resemble so called Q (quiescent) and NQ (nonquiescent) cells, respectively, described in stationary liquid cultures [38, 39]. However, transcriptomic, microscopic, and biochemical analyses revealed that the situation within a colony is more complicated. U cells, although resistant and long-living, are metabolically active cells that activate various metabolic pathways controlled by an unusual combination of nutrient sensing regulators. On the one hand, U cells activate pathways (including TOR pathway) that are usually active under nutrient-rich conditions. On the other hand, the activation of a group of amino acid metabolic genes indicates that the general amino acid control (GCN) pathway with transcription factor Gcn4p, a master regulator of amino acid biosynthetic genes, is active in U cells. Both TOR and GCN pathways monitor the availability of amino acids [40], but the GCN pathway usually opposes TOR pathway and is activated under conditions of nutrient shortage [41]. Thus, the expression of genes involved in glycolysis, pentose shunt, and translation indicates the active consumption of nutrients by U cells, but the expression of amino acid metabolic genes usually induced by amino acid starvation [41] indicates some degree of nutrient limitation in U cells. U cells also store glycogen and activate autophagy; both of these features are also found in cells entering the stationary phase. A surprising feature of U cells is the downregulation of the respiratory function of their mitochondria, which are swollen and contain only a few cristae. All of the above characteristics imply that U cells develop a unique metabolism that contributes to their longevity and stress resistance. This metabolism is dissimilar to that of Q cells identified in stationary liquid cultivations by Allen et al. [38]. These authors isolated two subpopulations of aged cells in stationary liquid cultures and named them Q cells and NQ cells. Q cells are highly respiring cells [42] and do not exhibit prominent features of U cells such as the activation of amino acid metabolic genes. Thus, similarly to the previous indications of the differing properties of cell populations from colonies and cells from liquid cultivations [33, 43], the mechanisms of aging and related metabolic reprogramming also seem to significantly differ in these two different yeast life-styles. What are the reasons for such expressive differences? In contrast to the mixed population of Q and NQ cell types within a liquid culture, cell subpopulations within colonies possess specific positions, can mutually interact with their neighbors, and can form gradients of nutrients and signaling compounds that affect remote colony subpopulations. Hence, in contrast to a liquid population, the colony behaves more like an organized primitive multicellular organism containing specialized mutually interacting cell types.
Interestingly, upper cells in 8-day-old colonies of diploid S. cerevisiae strains grown on sporulation acetate medium exhibit efficient meiosis and sporulation [44]. Sporulating cells are located in the interior of an unusual stalk-like structure formed by yeast cells growing from a cavity in the agar medium [45, 46]. The process of sporulation is tightly connected with limited nutrient availability and produces highly stress-resistant spores capable of long-term survival. Sporulation is thus one of the possible developmental programs after entering nutrient-limiting conditions, being an alternative to stationary-phase state.
As indicated above, U cells seem to activate metabolic pathways that allow them to consume nutrients, possibly including sugars, although they have developed from chronologically aged stressed cells of acidic-phase colonies, which grow on complete respiratory medium without glucose [10]. Features of the other subpopulation of a stratified colony, the L cells, could explain such a paradox. In contrast to U cells, L cells exhibit typical features of starving cells including a high level of ROS. These cells are able to effectively consume oxygen and they activate various degradative mechanisms, including proteasomal and vacuolar functions. The hypothesis that L cells could release nutrients to feed U cells has been therefore postulated and supported by the discovery of extracellular amino acids within a differentiated colony [10]. In addition to amino acids originating from protein degradation, carbohydrates arising from gluconeogenesis (activated in L cells) or partial cell wall degradation could be released from L cells.
6. Yeast Laboratory-Strain Colonies: A Model for Studies of Different Types of Chronologically Aged Cells
The above-mentioned discoveries indicate that the colonies of S. cerevisiae laboratory strains are an excellent model for studies of chronological aging. At least three different types of chronologically aged cells have been identified in these colonies: stressed cells of acidic-phase colonies, which are ancestors of the two other cell types: U cells (adapted, “healthy” chronologically aged cells of the upper colony layer) and L cells (cells with an enhanced stressed phenotype of the lower colony part) (Figure 1(b)). Interestingly, similar metabolic features to those exhibited by U cells (i.e., activation of glycolysis and decrease in mitochondrial respiration) were identified in early stationary populations of long-living deletants tor1Δ, sch9Δ and ras2Δ when compared to the wild type [5], indicating that the longevity phenotype is linked to specific metabolic changes. In the colonial model, however, this longevity phenotype is achieved not by a genetic manipulation, but rather in a “natural” way, that is, the process of aging and the formation of these different types of chronologically aged cells occurs during spontaneous aging and starvation of the entire colony population. The aging of colonial populations occurs within a precisely defined multicellular structure that exhibits the features of a primitive multicellular organism. The existence of the three major chronologically aged cell subpopulations enables the analysis of not only temporal but also spatial changes occurring during chronological aging and analyses of the interactions and signaling among the two aged U and L subpopulations including the formation and effects of gradients of metabolites and signaling compounds. It is also important that such types of chronologically aged cell subpopulations can be quickly isolated as relatively homogeneous populations, which enables the acquisition of strong conclusions based on measured data.
7. Structured Biofilm Colonies of S. cerevisiae Natural Strains
In contrast to most S. cerevisiae laboratory strains that form smooth colonies with the typical architecture described above, various natural S. cerevisiae isolates form biofilm colonies that are often noticeably structured [47]. These colonies resemble those formed by so-called nonconventional yeasts, such as those of the Candida and Cryptococcus genera. Studies of the prominent characteristics of structured S. cerevisiae colonies revealed that these colonies possess some features typical of yeast biofilms [16]. Such features include the production of a protective low-permeable extracellular matrix (ECM), the presence of adhesins, and production of specifically localized multidrug resistance transporters (MDR). In contrast to the above-mentioned laboratory strain colonies, relatively little is known about the aging of these structured colonies. However, a recent study on structured colony ultrastructure and differentiation indicated that the mechanisms regulating the transition from cell growth to stationary phase differ between smooth and structured colonies. In contrast to smooth colonies, where dividing cells are localized to the margin and to the uppermost colony layer (Figure 1(b)), dividing and stationary-phase cells are located differently within structured colonies [16] (Figure 2). Up to approximately 35 hours of growth, the whole of a structured colony is composed of dividing cells. This timing approximately correlates with the exponential growth phase of smooth colonies. Starting at about 40 hours of growth, a layer of stationary-phase cells appears at the top of the colony. As nutrients are spread very efficiently throughout the structured colonies [16], such a cell transition to the stationary phase is very likely unrelated to nutrient shortage, as colonies still grow in nutrient abundance. A 72-hour-old colony is composed of aerial and root structures with dividing cells detectable at precise locations in the colony interior and at the tips of the roots invading the agar (Figure 2). In contrast to smooth colonies, cells in the uppermost layers over the whole aerial part of the structured colony occur in stationary phase and new cell generations are formed within the internal colony cavity, where they are protected against the environment by various mechanisms including MDR transporters and the ECM.
Figure 2.
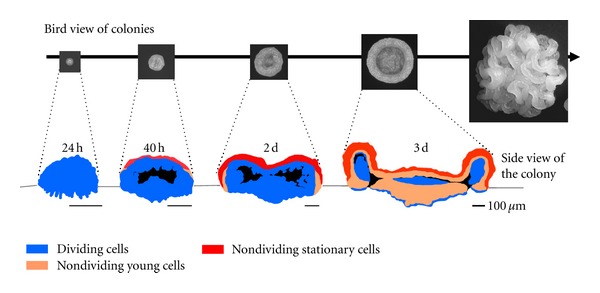
Development of colonies formed by wild strain of S. cerevisiae. Localization of dividing, young nondividing, and stationary cells within the colony is shown on vertical colony cross sections. Grey lines: surface of the agar.
When cultivated under laboratory conditions on rich nutrient media, the natural S. cerevisiae isolates forming structured colonies can domesticate, that is, they are capable of a phenotypic switch that results in smooth colony morphology. One can speculate that each of the colony life-styles (i.e., “biofilm-like” and “smooth”) is adapted to a particular environment. In natural settings with many threats, nutrient scarcity and ever changing conditions, the protection of internal cells is the priority for the population as a whole. Cells in the outer regions of the colony are exposed to the hostile environment and enter the stationary phase state, which is characterized by high resistance to environmental impacts, as soon as possible. These colonies also invest resources and energy in the formation of an ECM that can form a scaffold that keeps distances between the cells (or cell groups) and helps to form the internal colony cavity that provides sufficient space for dividing cell progeny. As shown in other microorganisms, ECMs can function as a sorptive sponge that sequesters organic molecules [48] and drugs [49]. Yeast ECMs also can have nutritional value [16, 50]. ECM in biofilm colony thus may play a dual function, a protective sequestration barrier and a nutrient pool for the cell progeny within the cavity. On the other hand, under stable conditions with rich initial nutrients such as those in the laboratory, the cells employ a different strategy. They do not produce an ECM scaffold, but rather form tightly packed colonies. Cells at the colony margin are those that can expand from the colony and reach nutrients in locations further away from the original colony position. In this case, stationary-phase cells are formed in the central colony regions, while cells at the margin are dividing. The question of whether the metabolic adaptive program identified in aged smooth colonies is also active in aged structured biofilm colonies remains to be elucidated.
8. Conclusions
Importantly, many genes and regulatory mechanisms are evolutionarily conserved so that the knowledge gained using primitive eukaryotes such as yeast can be often extrapolated to much more complex metazoans. Yeast multicellular colonies arising by the division of one or more of the original cells followed by a differentiation of nondividing cell population resemble multicellular organisms not only at the cellular level, but, to a limited extent, also at the level of tissue or even the whole organism. Similarly to metazoans, a significant part of the colony developmental program is likely to be determined genetically. On the other hand, environmental conditions can alter colony development and, in extreme cases, even induce a stable (genetic and/or epigenetic) modification of the developmental program. Colony “domestication,” as an example of such modification, leads to the shift from structured to smooth colony morphology. This colony morphology switch is reflected in internal colony organization and differentiation, as well as in distinct ways of transition of specifically localized cell subpopulations to the stationary phase. In addition, further development of chronologically aged colonies composed of mostly stationary-phase cells is linked to diversification of these cells to various subpopulations differing in characteristic properties and in their subsequent fate. Presumably, development of these subpopulations is coordinated and includes formation and function of gradients of metabolites and signaling compounds between the subpopulations. This variability of developmental programs and diversity of different types of stationary-phase cells makes yeast colonies a perspective model for investigation of different pathways and signaling compounds involved in chronological aging, some of which may have their counterparts in metazoans.
Acknowledgments
This work was supported by the Grant Agency of the Czech Republic (204/08/0718), the Ministry of Education (MSM0021620858), Charles University in Prague (UNCE 204013), and RVO 61388971.
References
- 1.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464(7288):513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabrizio P, Liou LL, Moy VN, et al. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163(1):35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292(5515):288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 4.Powers RW, III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes and Development. 2006;20(2):174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei M, Fabrizio P, Madia F, et al. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genetics. 2009;5(5) doi: 10.1371/journal.pgen.1000467.e1000467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8(8):1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCormick MA, Tsai SY, Kennedy BK. TOR and ageing: a complex pathway for a complex process. Philosophical Transactions of the Royal Society B. 2011;366(1561):17–27. doi: 10.1098/rstb.2010.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leontieva OV, Blagosklonny MV. Yeast-like chronological senescence in mammalian cells: phenomenon, mechanism and pharmacological suppression. Aging. 2011;3(11):1078–1091. doi: 10.18632/aging.100402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matecic M, Smith DL, Pan X, et al. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genetics. 2010;6(4) doi: 10.1371/journal.pgen.1000921.e1000921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cap M, Stepanek L, Harant K, Vachova L, Palkova Z. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Molecular Cell. 2012;46(4):436–448. doi: 10.1016/j.molcel.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR. Yeast cells can access distinct quiescent states. Genes and Development. 2011;25(4):336–349. doi: 10.1101/gad.2011311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Zhang N, Hayes A, Panoutsopoulo K, Oliver SG. Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):3148–3153. doi: 10.1073/pnas.0308321100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boender LG, Almering MJ, Dijk M, et al. Extreme calorie restriction and energy source starvation in Saccharomyces cerevisiae represent distinct physiological states. Biochimica et Biophysica Acta. 2011;1813(12):2133–2144. doi: 10.1016/j.bbamcr.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Boender LG, van Maris AJ, de Hulster EA, et al. Cellular responses of Saccharomyces cerevisiae at near-zero growth rates: transcriptome analysis of anaerobic retentostat cultures. FEMS Yeast Research. 2011;11(8):603–620. doi: 10.1111/j.1567-1364.2011.00750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laporte D, Lebaudy A, Sahin A, et al. Metabolic status rather than cell cycle signals control quiescence entry and exit. The Journal of Cell Biology. 2011;192(6):949–957. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vachova L, Stovicek V, Hlavacek O, et al. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. The Journal of Cell Biology. 2011;194(5):679–687. doi: 10.1083/jcb.201103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meunier JR, Choder M. Saccharomyces cerevisiae colony growth and ageing: biphasic growth accompanied by changes in gene expression. Yeast. 1999;15(12):1159–1169. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1159::AID-YEA441>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 18.Váchová L, Palková Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. The Journal of Cell Biology. 2005;169(5):711–717. doi: 10.1083/jcb.200410064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palkova Z, Vachova L, Gaskova D, Kucerova H. Synchronous plasma membrane electrochemical potential oscillations during yeast colony development and aging. Molecular Membrane Biology. 2009;26(4):228–235. doi: 10.1080/09687680902893130. [DOI] [PubMed] [Google Scholar]
- 20.Fabrizio P, Battistella L, Vardavas R, et al. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae . The Journal of Cell Biology. 2004;166(7):1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabrizio P, Hoon S, Shamalnasab M, et al. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genetics. 2010;6(7) doi: 10.1371/journal.pgen.1001024.e1001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Lu Y, Qin LX, Bar-Joseph Z, Werner-Washburne M, Breeden LL. Budding yeast SSD1-V regulates transcript levels of many longevity genes and extends chronological life span in purified quiescent cells. Molecular Biology of the Cell. 2009;20(17):3851–3864. doi: 10.1091/mbc.E09-04-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLean M, Harris N, Piper PW. Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast. 2001;18(6):499–509. doi: 10.1002/yea.701. [DOI] [PubMed] [Google Scholar]
- 24.Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mechanisms of Ageing and Development. 2006;127(9):733–740. doi: 10.1016/j.mad.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Oliver JD. The viable but nonculturable state in bacteria. Journal of Microbiology. 2005;43(1):93–100. [PubMed] [Google Scholar]
- 26.Palková Z, Devaux F, Řičicová M, Mináriková L, Le Crom S, Jacq C. Ammonia pulses and metabolic oscillations guide yeast colony development. Molecular Biology of the Cell. 2002;13(11):3901–3914. doi: 10.1091/mbc.E01-12-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palková Z, Forstová J. Yeast colonies synchronise their growth and development. Journal of Cell Science. 2000;113(11):1923–1928. doi: 10.1242/jcs.113.11.1923. [DOI] [PubMed] [Google Scholar]
- 28.Palkova Z, Janderova B, Gabriel J, Zikanova B, Pospisek M, Forstova J. Ammonia mediates communication between yeast colonies. Nature. 1997;390(6659):532–536. doi: 10.1038/37398. [DOI] [PubMed] [Google Scholar]
- 29.Palkova Z, Vachova L. Ammonia signaling in yeast colony formation. International Review of Cytology. 2003;225:229–272. doi: 10.1016/s0074-7696(05)25006-4. [DOI] [PubMed] [Google Scholar]
- 30.Vachova L, Kucerova H, Devaux F, Ulehlova M, Palkova Z. Metabolic diversification of cells during the development of yeast colonies. Environmental Microbiology. 2009;11(2):494–504. doi: 10.1111/j.1462-2920.2008.01789.x. [DOI] [PubMed] [Google Scholar]
- 31.Gralla EB, Valentine JS. Null mutants of Saccharomyces cerevisiae Cu, Zn superoxide dismutase: characterization and spontaneous mutation rates. Journal of Bacteriology. 1991;173(18):5918–5920. doi: 10.1128/jb.173.18.5918-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo . The Journal of Biological Chemistry. 1996;271(21):12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 33.Čáp M, Váchová L, Palková Z. Yeast colony survival depends on metabolic adaptation and cell differentiation rather than on stress defense. The Journal of Biological Chemistry. 2009;284(47):32572–32581. doi: 10.1074/jbc.M109.022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Váchová L, Devaux F, Kučerová H, Řičicová M, Jacq C, Palková Z. Sok2p transcription factor is involved in adaptive program relevant for long term survival of Saccharomyces cerevisiae colonies. The Journal of Biological Chemistry. 2004;279(36):37973–37981. doi: 10.1074/jbc.M404594200. [DOI] [PubMed] [Google Scholar]
- 35.Váchová L, Chernyavskiy O, Strachotová D, et al. Architecture of developing multicellular yeast colony: spatio-temporal expression of Ato1p ammonium exporter. Environmental Microbiology. 2009;11(7):1866–1877. doi: 10.1111/j.1462-2920.2009.01911.x. [DOI] [PubMed] [Google Scholar]
- 36.Büttner S, Eisenberg T, Herker E, Carmona-Gutierrez D, Kroemer G, Madeo F. Why yeast cells can undergo apoptosis: death in times of peace, love, and war. The Journal of Cell Biology. 2006;175(4):521–525. doi: 10.1083/jcb.200608098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carmona-Gutierrez D, Eisenberg T, Büttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death and Differentiation. 2010;17(5):763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 38.Allen C, Büttner S, Aragon AD, et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. The Journal of Cell Biology. 2006;174(1):89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aragon AD, Rodriguez AL, Meirelles O, et al. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Molecular Biology of the Cell. 2008;19(3):1271–1280. doi: 10.1091/mbc.E07-07-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smets B, Ghillebert R, De Snijder P, et al. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae . Current Genetics. 2010;56(1):1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- 41.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annual Review of Microbiology. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 42.Davidson GS, Joe RM, Roy S, et al. The proteomics of quiescent and nonquiescent cell differentiation in yeast stationary-phase cultures. Molecular Biology of the Cell. 2011;22(7):988–998. doi: 10.1091/mbc.E10-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cap M, Vachova L, Palkova Z. How to survive within a yeast colony?: change metabolism or cope with stress? Communicative & Integrative Biology. 2010;3(2):198–200. doi: 10.4161/cib.3.2.11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piccirillo S, White MG, Murphy JC, Law DJ, Honigberg SM. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics. 2010;184(3):707–716. doi: 10.1534/genetics.109.113480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engelberg D, Mimran A, Martinetto H, et al. Multicellular stalk-like structures in Saccharomyces cerevisiae . Journal of Bacteriology. 1998;180(15):3992–3996. doi: 10.1128/jb.180.15.3992-3996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scherz R, Shinder V, Engelberg D. Anatomical analysis of Saccharomyces cerevisiae stalk-like structures reveals spatial organization and cell specialization. Journal of Bacteriology. 2001;183(18):5402–5413. doi: 10.1128/JB.183.18.5402-5413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Št’ovíček V, Váchová L, Kuthan M, Palková Z. General factors important for the formation of structured biofilm-like yeast colonies. Fungal Genetics and Biology. 2010;47(12):1012–1022. doi: 10.1016/j.fgb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 48.Decho AW. Microbial biofilms in intertidal systems: an overview. Continental Shelf Research. 2000;20(10-11):1257–1273. [Google Scholar]
- 49.Nett JE, Sanchez H, Cain MT, Andes DR. Genetic basis of Candida Biofilm resistance due to drug-sequestering matrix glucan. Journal of Infectious Diseases. 2010;202(1):171–175. doi: 10.1086/651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joubert LM, Wolfaardt GM, Botha A. Microbial exopolymers link predator and prey in a model yeast biofilm system. Microbial Ecology. 2006;52(2):187–197. doi: 10.1007/s00248-006-9063-7. [DOI] [PubMed] [Google Scholar]


