Abstract
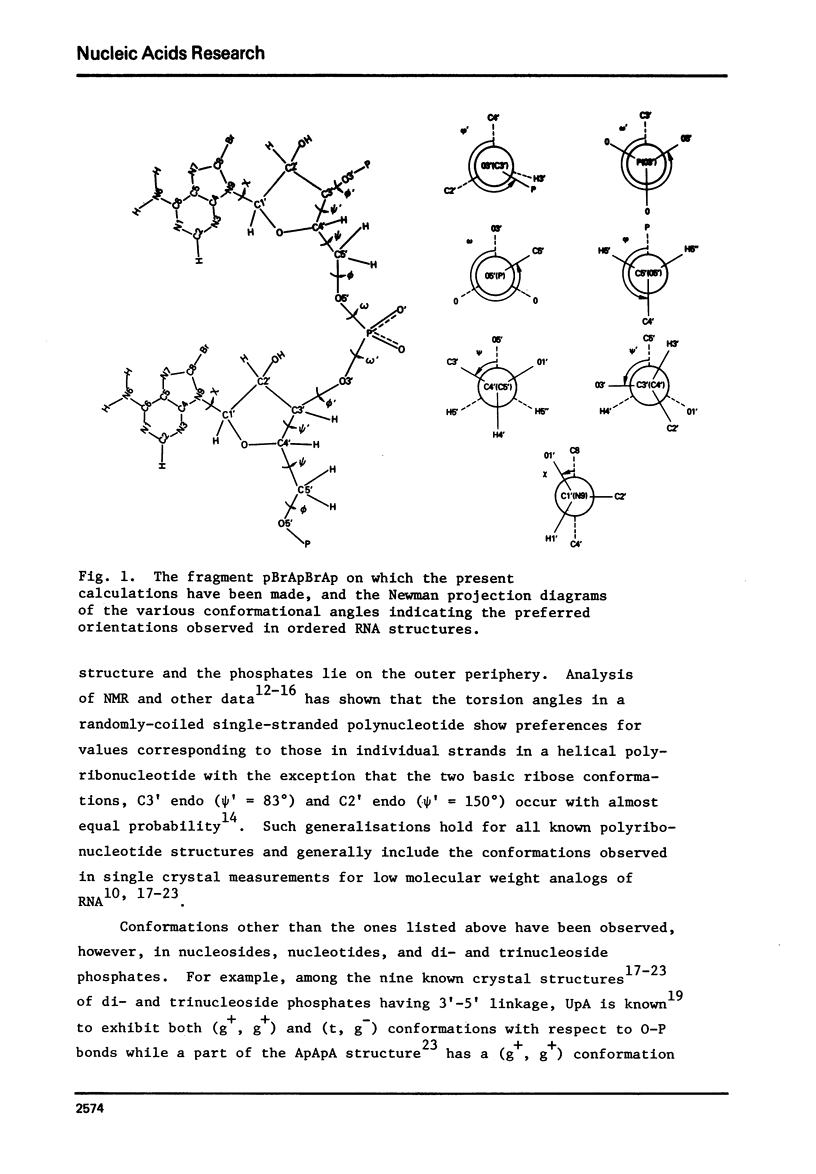
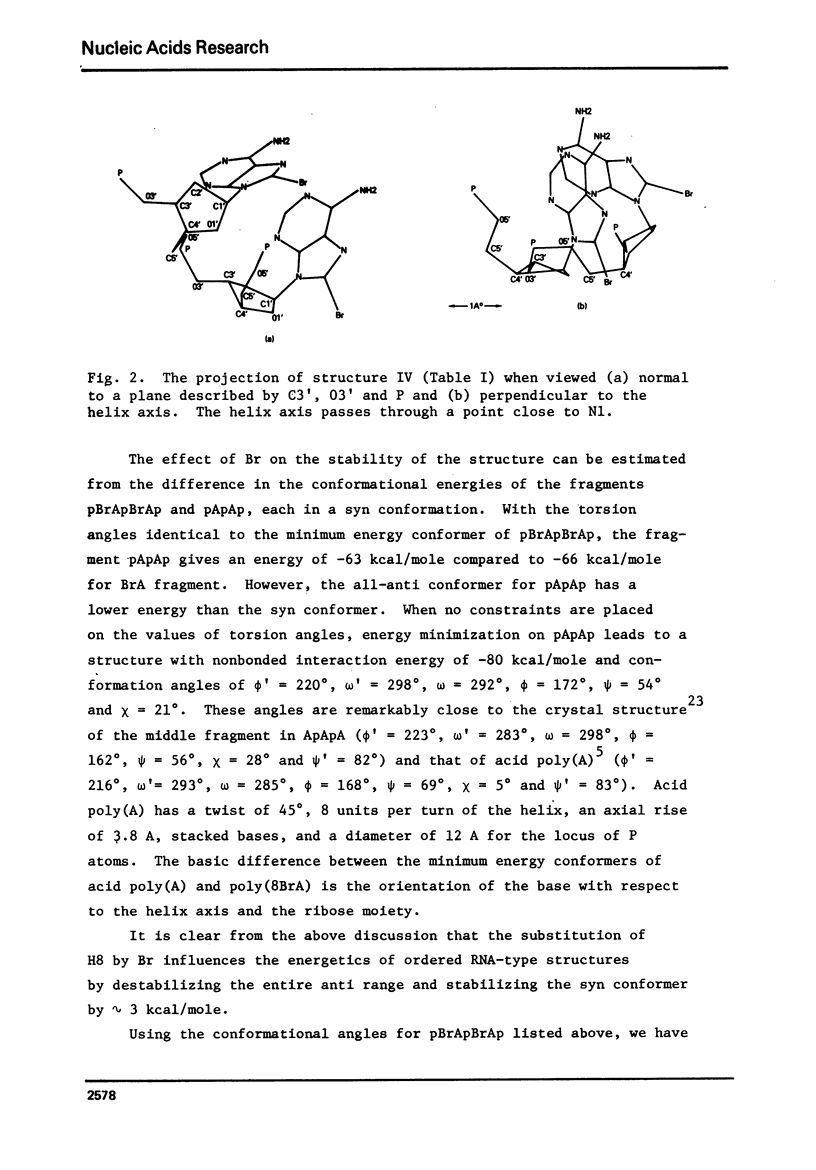
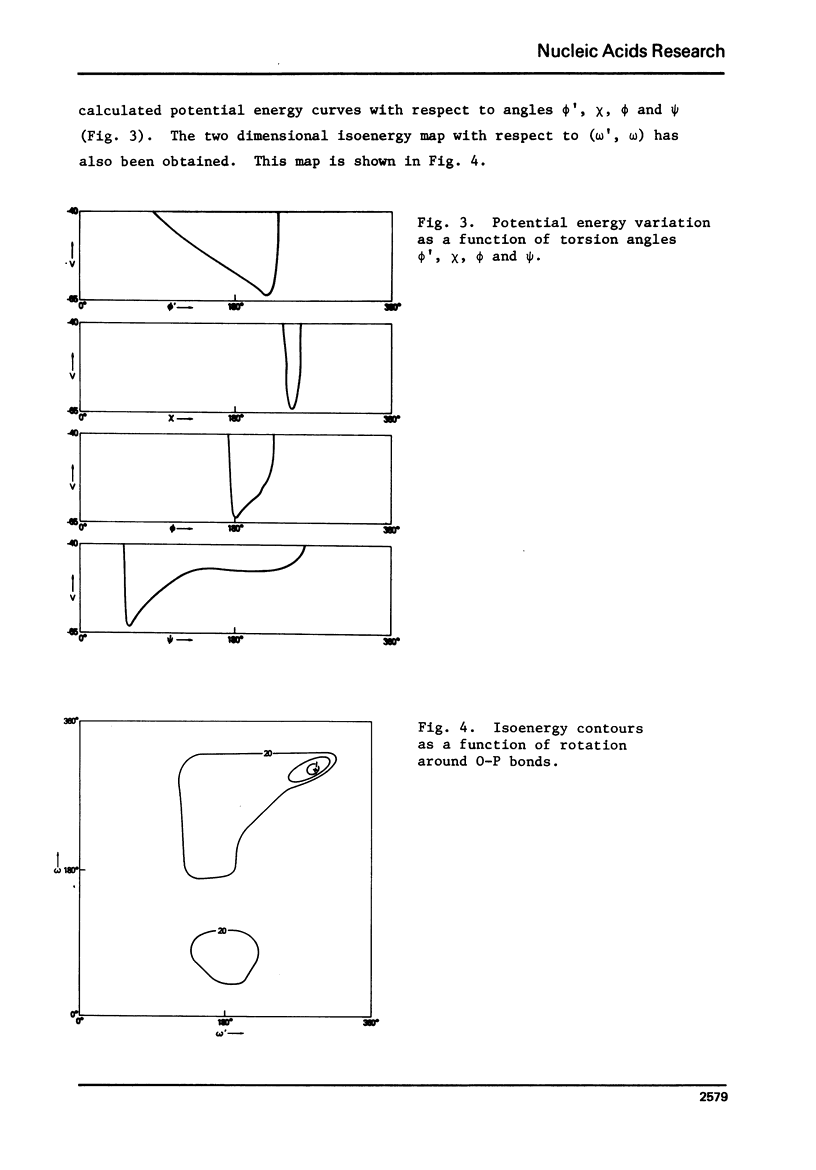
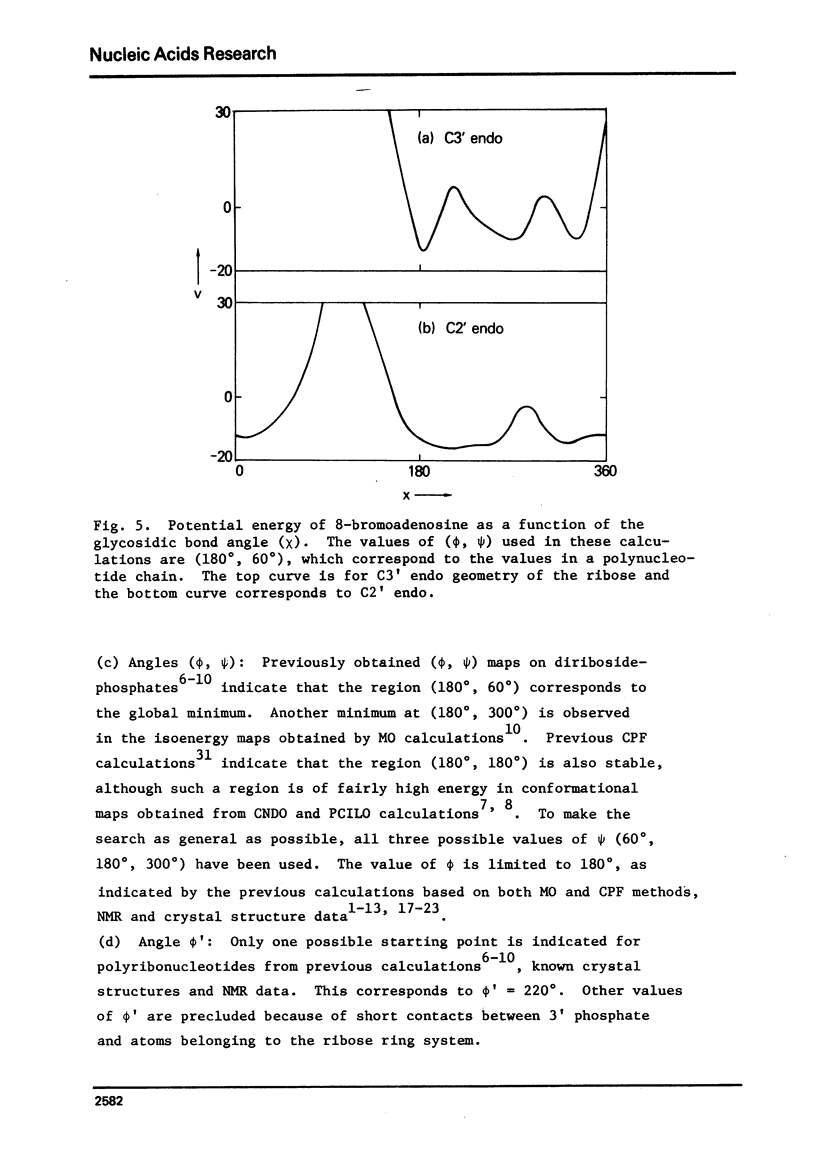
Poly 8-bromoadenylic acid [poly(BBrA)] is the only known all-syn polynucleotide. It shows a helix-coil transition with a melting curve centred around 55 degrees C. Energy calculations based on classical potential functions have been used to explore the three-dimensional structure of this polymer in helix and random coil. It is concluded that the ordered state is a helix of two parallel strands with a two-fold rotation axis, and the duplex is stabilised by hydrogen bonds involving N1 and H6. Each strand has a conformation with C3' endo geometry, phi' = 216 degrees, omega' = 280 degrees, omega = 294 degrees, phi = 179 degrees, chi = 243 degrees and psi = 57 degrees. Such a conformation leads to approximately 8 nucleotide units per turn of the helix and an axial rise of 3.9A degrees. The structure of poly(8BrA) has been compared with that of the related polymer poly(A) which forms a double helical structure in acidic conditions with bases in the anti conformation and with interstrand hydrogen-bonds between N7 and H6. This is the first time that a specific geometrical model of a novel polynucleotide structure has been produced initially by potential energy calculations, though such calculations on a number of known structures have been reported previously.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J Am Chem Soc. 1973 Apr 4;95(7):2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W., Dover S. D., Fuller W., Hodgson A. R. Structures of synthetic polynucleotides in the A-RNA and A'-RNA conformations: x-ray diffraction analyses of the molecular conformations of polyadenylic acid--polyuridylic acid and polyinosinic acid--polycytidylic acid. J Mol Biol. 1973 Dec 5;81(2):107–122. doi: 10.1016/0022-2836(73)90183-6. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W., Dover S. D. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun. 1972 Sep 26;48(6):1392–1399. doi: 10.1016/0006-291x(72)90867-4. [DOI] [PubMed] [Google Scholar]
- Arnott S. The geometry of nucleic acids. Prog Biophys Mol Biol. 1970;21:265–319. doi: 10.1016/0079-6107(70)90027-1. [DOI] [PubMed] [Google Scholar]
- Davies D. B., Danyluk S. S. Nuclear magnetic resonance studies of 2'- and 3'-ribonucleotide structures in solution. Biochemistry. 1975 Feb 11;14(3):543–554. doi: 10.1021/bi00674a013. [DOI] [PubMed] [Google Scholar]
- Davies D. B., Danyluk S. S. Nuclear magnetic resonance studies of 5'-ribo- and deoxyribonucleotide structures in solution. Biochemistry. 1974 Oct 8;13(21):4417–4434. doi: 10.1021/bi00718a027. [DOI] [PubMed] [Google Scholar]
- Day R. O., Seeman N. C., Rosenberg J. M., Rich A. A crystalline fragment of the double helix: the structure of the dinucleoside phosphate guanylyl-3',5'-cytidine. Proc Natl Acad Sci U S A. 1973 Mar;70(3):849–853. doi: 10.1073/pnas.70.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg H., Felsenfeld G. Studies of the temperature-dependent conformation and phase separation of polyriboadenylic acid solutions at neutral pH. J Mol Biol. 1967 Nov 28;30(1):17–37. doi: 10.1016/0022-2836(67)90240-9. [DOI] [PubMed] [Google Scholar]
- Govil G. Conformational structure of polynucleotides around the O-P bonds: refined parameters for CPF calculations. Biopolymers. 1976 Nov;15(11):2303–2307. doi: 10.1002/bip.1976.360151119. [DOI] [PubMed] [Google Scholar]
- Govil G., Saran A. Quantum chemical studies on the conformational structure of nucleic acids. II. EHT and CNDO calculations on the puckering of D-ribose. J Theor Biol. 1971 Nov;33(2):399–406. doi: 10.1016/0022-5193(71)90073-7. [DOI] [PubMed] [Google Scholar]
- Govil G., Smith I. C. A carbon-13 magnetic resonance study of the helix-coil transition in polyuridylic acid. Biopolymers. 1973 Nov;12(11):2589–2598. doi: 10.1002/bip.1973.360121111. [DOI] [PubMed] [Google Scholar]
- Howard F. B., Frazier J., Miles H. T. Poly(8-bromoadenylic acid): synthesis and characterization of an all-syn polynucleotide. J Biol Chem. 1975 May 25;250(10):3951–3959. [PubMed] [Google Scholar]
- Howard F. B., Frazier J., Miles H. T. Poly-8-bromoadenylic acid. A helical, all-syn homopolynucleotide. J Biol Chem. 1974 May 10;249(9):2987–2990. [PubMed] [Google Scholar]
- Inners L. D., Felsenfeld G. Conformation of polyribouridylic acid in solution. J Mol Biol. 1970 Jun 14;50(2):373–389. doi: 10.1016/0022-2836(70)90199-3. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Ezra F. S., Kondo N. S., Sarma R. H., Danyluk S. S. Conformational properties of dinucleoside monophosphates in solution: dipurines and dipyrimidines. Biochemistry. 1976 Aug 10;15(16):3627–3639. doi: 10.1021/bi00661a034. [DOI] [PubMed] [Google Scholar]
- Olson W. K., Flory P. J. Spatial configuration of polynucleotide chains. II. Conformational energies and the average dimensions of polyribonucleotides. Biopolymers. 1972 Jan;11(1):25–56. doi: 10.1002/bip.1972.360110103. [DOI] [PubMed] [Google Scholar]
- Olson W. K. Syn-anti effects on the spatial configuration of polynucleotide chains. Biopolymers. 1973;12(8):1787–1814. doi: 10.1002/bip.1973.360120808. [DOI] [PubMed] [Google Scholar]
- Pullman B., Perahia D., Saran A. Molecular orbital calculations on the conformation of nucleic acids and their constituents. 3. Backbone structure of di- and polynucleotides. Biochim Biophys Acta. 1972 Apr 26;269(1):1–14. doi: 10.1016/0005-2787(72)90068-8. [DOI] [PubMed] [Google Scholar]
- Pullman B., Saran A. Quantum-mechanical studies on the conformation of nucleic acids and their constituents. Prog Nucleic Acid Res Mol Biol. 1976;18:215–322. doi: 10.1016/s0079-6603(08)60589-9. [DOI] [PubMed] [Google Scholar]
- RICH A., DAVIES D. R., CRICK F. H., WATSON J. D. The molecular structure of polyadenylic acid. J Mol Biol. 1961 Feb;3:71–86. doi: 10.1016/s0022-2836(61)80009-0. [DOI] [PubMed] [Google Scholar]
- Ramachandran G. N., Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg J. M., Seeman N. C., Kim J. J., Suddath F. L., Nicholas H. B., Rich A. Double helix at atomic resolution. Nature. 1973 May 18;243(5403):150–154. doi: 10.1038/243150a0. [DOI] [PubMed] [Google Scholar]
- Rubin J., Brennan T., Sundaralingam M. Crystal and molecular structure of a naturally occurring dinucleoside monophosphate. Uridylyl-(3'-5')-adenosine hemihydrate. Conformational "rigidity" of the nucleotide unit and models for polynucleotide chain folding. Biochemistry. 1972 Aug 1;11(16):3112–3128. doi: 10.1021/bi00766a027. [DOI] [PubMed] [Google Scholar]
- Sakore T. D., Sobell H. M. Crystal and molecular structure of a hydrogen-bonded complex containing adenine and hypoxanthine derivatives: 9-ethyl-8-bromoadenine-9-ethyl-8-bromohypoxanthine. J Mol Biol. 1969 Jul 14;43(1):77–87. doi: 10.1016/0022-2836(69)90080-1. [DOI] [PubMed] [Google Scholar]
- Sarma R. H., Lee C. H., Evans F. E., Yathindra N., Sundaralingam M. Probing the interrelation between the glycosyl torsion, sugar pucker, and the backbone conformation in C(8) substituted adenine nucleotides by 1H and 1H-(31P) fast Fourier transform nuclear magnetic resonance methods and conformational energy calculations. J Am Chem Soc. 1974 Nov 13;96(23):7337–7348. doi: 10.1021/ja00830a028. [DOI] [PubMed] [Google Scholar]
- Shefter E., Barlow M., Sparks R. A., Trueblood K. N. The crystal and molecular structure of a dinucleoside phosphate: beta-adenosine-2'-beta-uridine-5'-phosphoric acid. Acta Crystallogr B. 1969 May 15;25(5):895–908. doi: 10.1107/s0567740869003190. [DOI] [PubMed] [Google Scholar]
- Suck D., Manor P. C., Germain G., Schwalbe C. H., Weimann G., Saenger W. X-ray study of helix, loop and base pair stacking in trinucleoside diphosphate ApApA. Nat New Biol. 1973 Dec 12;246(154):161–165. doi: 10.1038/newbio246161a0. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Seeman N. C., Kim S. H., Berman H. M. Crystal structure of a naturally occurring dinucleoside phoaphate: uridylyl 3',5'-adenosine phosphate model for RNA chain folding. J Mol Biol. 1972 May 28;66(3):403–421. doi: 10.1016/0022-2836(72)90423-8. [DOI] [PubMed] [Google Scholar]
- Tavale S. S., Sobell H. M. Crystal and molecular structure of 8-bromoguanosine and 8-bromoadenosine, two purine nucleosides in the syn conformation. J Mol Biol. 1970 Feb 28;48(1):109–123. doi: 10.1016/0022-2836(70)90222-6. [DOI] [PubMed] [Google Scholar]
- Tewari R., Nanda R. K., Govil G. Quantum chemical studies on the conformational structure of nucleic acids. IV. Calculation of backbone structure by CNDO method. J Theor Biol. 1974 Jul;46(1):229–239. doi: 10.1016/0022-5193(74)90149-0. [DOI] [PubMed] [Google Scholar]
- Tewari R., Nanda R. K., Govil G. Spatial configuration of single-stranded polynucleotides. Calculations of average dimensions and Nmr coupling constants. Biopolymers. 1974;13(10):2015–2035. doi: 10.1002/bip.1974.360131007. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976 Sep 23;263(5575):289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- Yathindra N., Sundaralingam M. Backbone conformations in secondary and tertiary structural units of nucleic acids. Constraint in the phosphodiester conformation. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3325–3328. doi: 10.1073/pnas.71.9.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]


