Abstract
Many RNA structures are comprised of simple secondary structure elements linked by a few, critical, tertiary interactions. SHAPE chemistry has made interrogation of RNA dynamics at single-nucleotide resolution straightforward. However, de novo identification of nucleotides involved in tertiary interactions remains a challenge. Here we show that nucleotides that form non-canonical or tertiary contacts are detected by comparing information obtained using two SHAPE reagents, N-methylisatoic anhydride (NMIA) and 1-methyl-6-nitroisatoic anhydride (1M6). Nucleotides that react preferentially with NMIA exhibit slow local nucleotide dynamics and preferentially adopt the less common C2′-endo ribose conformation. Experiments and first-principle calculations show 1M6 reacts preferentially with nucleotides in which one face of the nucleobase allows an unhindered stacking interaction with the reagent. Differential SHAPE reactivities were used to detect non-canonical and tertiary interactions in four RNAs with diverse structures and to identify pre-formed non-canonical interactions in partially folded RNAs. Differential SHAPE reactivity analysis will enable experimentally concise, large-scale identification of tertiary structure elements and ligand binding sites in complex RNAs and in diverse biological environments.
RNA molecules are involved in nearly every aspect of cellular information transfer.1 Information is encoded both in the RNA primary sequence and in its three-dimensional structure. Higher-order RNA structures are typically comprised of secondary structure elements held together by a few key tertiary interactions2,3 including long-range stacking, loop-loop and loop-helix contacts, and pseudoknots. Regions of an RNA that contain significant tertiary structures ultimately have numerous important functional roles.
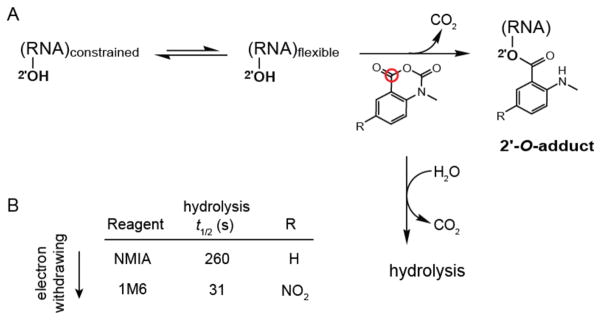
Nucleotides that participate in either base pairing or stable higher-order tertiary structure interactions can be detected by protection from solution-phase chemical probing reagents, whereas single-stranded and relatively unstructured elements are reactive.4 Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) has emerged as an especially informative approach for probing RNA structure and dynamics.4,5 SHAPE chemistry exploits the discovery that the reactivity of the ribose 2′-hydroxyl is highly sensitive to local nucleotide flexibility (Figure 1A). Flexible nucleotides sample many conformations, a few of which preferentially react with hydroxyl-selective, electrophilic reagents to form 2′-O-adducts (Figure 1A). The strong relationship between SHAPE reactivity and molecular motion6 makes it possible to use this chemistry to achieve accurate secondary structure predictions, to monitor RNA dynamics and folding, and to explore RNA-protein interactions.7–10
Figure 1.
RNA SHAPE chemistry. (A) Mechanism in the context of the concurrent hydrolysis reaction. The red circle denotes the reactive center of the reagent. (B) SHAPE reagents and hydrolysis half-lives.
However, it is not obvious based on the chemical reactivity of a nucleotide whether a given constraining interaction reflects a base pairing or tertiary interaction. Nucleotides involved in tertiary interactions often have unusual backbone or stacking geometries,3,11 adopt the syn conformation,12 or undergo conformational changes on slow timescales.8,9 There is an unmet need for RNA chemical probing technologies that identify nucleotides involved in tertiary interactions. Here we report an approach for rapidly “fingerprinting” RNA non-canonical and tertiary structures using structure-selective SHAPE chemical reactivities.
We initially screened potential SHAPE reagents for the ability to “fingerprint” RNA tertiary structure motifs using the aptamer domain of the TPP riboswitch in the ligand-bound state. The TPP riboswitch has been extensively characterized by crystallography,13,14 in-solution dynamics,15,16 and SHAPE chemistry.17 This RNA contains many tertiary structure features, especially at or near the ligand binding pocket, that are common to highly structured RNAs including base stacking, long-range docking interactions, and tight turns in the RNA backbone.
Two reagents, N-methylisatoic anhydride (NMIA) and 1-methyl-6-nitroisatoic anhydride (1M6), proved especially promising. NMIA, one of the first reagents used in the SHAPE approach,18 reacts slowly with RNA and can be used to identify nucleotides that undergo local conformational changes on slow timescales (Figure 1B).8 These nucleotides are usually in the relatively rare C2′-endo conformation and, in some cases, govern the folding of entire RNA domains.9 The second reagent, 1M6, differs from NMIA by a single nitro (–NO2) group on the double ring system (Figure 1B). This modification changes the chemical behavior of 1M6 in two ways relative to that of NMIA. Addition of the electron-withdrawing group increases the electrophilicity of the reactive center (Figure 1A, red circle), and consequently 1M6 reacts more rapidly than NMIA. Second, the –NO2 group significantly changes the electronic distribution of the reagent ring system which, we will show below, allows 1M6 to stack with RNA nucleobases.
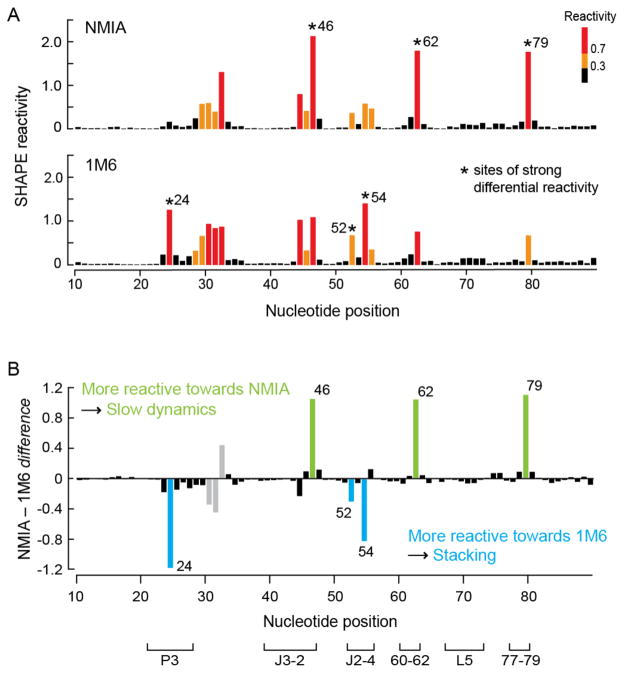
When the folded, ligand-bound TPP riboswitch was allowed to react with NMIA, the observed reactivities agreed with the known structure for the ligand-bound TPP riboswitch (Figures 2A and S1A). When this RNA was treated with 1M6, the overall SHAPE reactivity profile was very similar to that for NMIA (Figures 2A and S1B). In particular, all base-paired nucleotides were unreactive and many single-stranded nucleotides exhibited similar reactivity towards both reagents. Critically, a few nucleotides exhibited strongly enhanced reactivity towards one of the two reagents (Figure 2A, asterisks). SHAPE chemistry is quantitative; therefore, reagent-specific reactivities can be identified by simply subtracting one profile from another. After excluding nucleotides that participate in crystal contacts or that have poorly-defined electron densities in the previously determined crystal structure (Figure 2B, gray columns), we identified six nucleotides that exhibited statistically significant differential reactivities to NMIA versus 1M6 (Figure 2B).
Figure 2.
SHAPE analysis of the ligand-bound state of the TPP riboswitch. (A) Absolute SHAPE reactivities resulting from reaction with NMIA (top) and 1M6 (bottom). Columns are colored by nucleotide reactivities. Asterisks indicate sites of strong differential reactivity. (B) Differential SHAPE reactivities calculated by subtracting the 1M6 profile from that of NMIA. Columns corresponding to nucleotides that exhibit statistically significant differential reactivity (absolute reactivity difference ≥ 0.3 SHAPE units and a p-value < 0.05, calculated using the Student’s t-test) are colored in green (for NMIA) and blue (for 1M6). Gray columns represent nucleotides with differential reactivity that are involved in crystal contacts or have poorly-defined electron density.
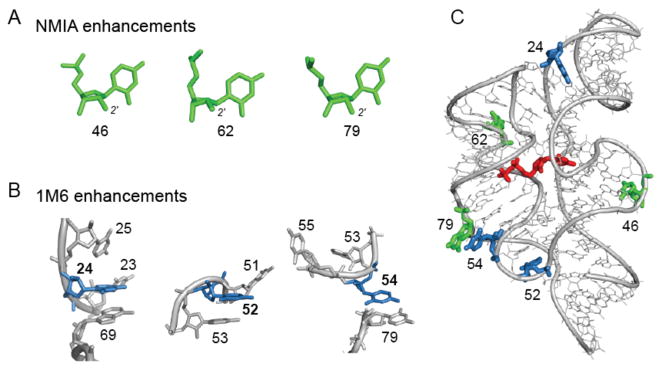
Each of the three nucleotides that reacted preferentially with NMIA (Figure 3A) adopts the relatively rare C2′-endo conformation, consistent with previous studies.8 However, the mechanism by which nucleotides might react preferentially with 1M6 has not been previously explored. The three nucleotides that reacted preferentially with 1M6 are located in diverse local structural environments but share the characteristic that one face of the nucleobase is available for π-π stacking interactions with a small molecule like 1M6 (Figure 3B). This conformation is unusual because, both in A-form helices and in most highly folded RNAs, base-base stacking is nearly fully saturated.2,3 Only a few nucleotides in special structural contexts – especially at bulges, turns and the termini of some helices – form “one-sided” stacking interactions.
Figure 3.
Conformations and structural contexts for nucleotides exhibiting differential reactivities in the ligand-bound state of the TPP riboswitch. (A) Sites of enhanced reactivity toward NMIA correspond to nucleotides in the C2′-endo ribose conformation. (B) Sites of 1M6 enhancement reflect one-sided stacking conformations. (C) NMIA (green) and 1M6 (blue) enhancements superimposed on a three-dimensional model for the TPP riboswitch aptamer domain with the bound ligand shown in red (2gdi).
We hypothesized that the –NO2 substituent polarizes the two-ring system to stabilize the 1M6-nucleobase stacking interaction. We evaluated this model experimentally by varying the electron-withdrawing ability of the ring functional group of the reagent from a methyl group (slightly electron-donating), to bromine (moderately electron-withdrawing), to a nitro group (strongly electron-withdrawing) (Figures 1B & S2A). The SHAPE reactivities of the “one-sided” stacking nucleotide C24 increased monotonically with increasing electron-withdrawing ability of the reagent substituent as reflected by the Hammett coefficient19 for each functional group (Pearson’s linear r = 0.97; Figure S2B). In contrast, this trend was not observed for A45, which is also reactive towards SHAPE reagents but forms stacking interactions on both sides of the adenine base. These reactivity patterns are consistent with the formation of increasingly favorable reagent-nucleobase stacking interactions,20 which are possible at C24 but not A45.
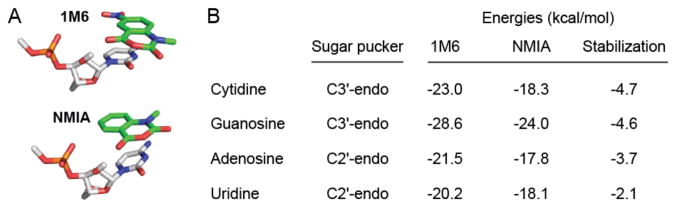
Since the effect of electron-withdrawing groups on the stacking interaction and resulting SHAPE reactivity is quantitative, we estimated representative electronic contributions associated with this interaction from first principles for the four RNA nucleotide types (Figure 4). Complexes formed between 1M6 and the four RNA nucleotides were −2 to −5 kcal/mol more stable than those formed with NMIA (Figure 4B). These values are significant when compared to the approximate net stabilization energy of a two base pair stack of 2–3 kcal/mol21 and likely reflect the upper limit for favorable interaction. Favorable stacking appears to enhance 1M6 reactivity by increasing the local reagent concentration at nucleotides where one face is available for the one-sided stacking interaction.
Figure 4.
Electronic structure calculations for the 1M6- and NMIA-nucleotide complex stabilities. (A) The most stable stacking conformations for the cytidine-1M6 and -NMIA complexes. (B) Representative open-faced stacking complex energies and net stabilization energy as a function of nucleotide, ribose conformation, and reagent.
In sum, each of the three nucleotides with enhanced reactivity to NMIA has an unusual ribose geometry that either stabilizes the RNA fold (Figure 3C, nucleotide 79) or is adjacent to the ligand-binding pocket (Figure 3C, nucleotides 46 and 62). Each of the three nucleotides with enhanced reactivity towards 1M6 form either a long-range stacking interaction (Figure 3C, nucleotides 24 and 54) or lie at a turn in the backbone (Figure 3C, nucleotide 52). These differential reactivities thus comprise an RNA non-canonical and tertiary structure “fingerprint” that highlights nucleotides that form relatively rare, but structurally critical, interactions.
To examine the generality of differential SHAPE reactivity analysis, we probed three additional RNAs with well-characterized structures: the adenine and lysine riboswitches and the RNase P specificity domain8,22 (Figure S3). Differential reactivities in the adenine riboswitch occurred at the ligand-binding pocket and in the region of a crucial loop-loop interaction (Figure S3A, nucleotides 47 and 63, respectively). In the lysine riboswitch, the three differentially reactive nucleotides are located in the P2a-L2 turn motif that is important for stabilizing the folded riboswitch structure23 (Figure S3B, nucleotides 38, 40, and 52). The RNase P specificity domain had many nucleotides with significant differential reactivities (Figure S3C, nucleotides 130 and 194 are involved in long-range interactions, nucleotide 142 is in a sarcinricin motif, and nucleotide 186 is adjacent to a T-loop motif24).
In the four RNAs analyzed in this study, 18 nucleotides displayed statistically significant reagent-specific differential reactivities (Figures 3 and S3). Ten nucleotides displayed preferential reactivity with NMIA and presumably experience slow local dynamics8 (Figures 3 and S3, emphasized in green). Of these ten nucleotides, nine are in the C2′-endo ribose conformation (nucleotide 180 in the RNase P RNA is in the C3′-endo conformation). Eight nucleotides reacted preferentially with 1M6 (Figures 3 and S3, emphasized in blue) and seven represent clear examples in which one face of the nucleobase is accessible for a stacking interaction; the remaining 1M6 preferentially-reactive nucleotide (position 142 in the RNase P RNA) forms a partial one-sided stacking conformation. These 18 nucleotides were detected out of approximately 470 RNA nucleotides probed, and the strong correlation between reactivity and location in structurally distinctive regions highlights the ability of differential SHAPE reactivity analysis to identify unique and important nucleotide dynamics and conformations on a large scale.
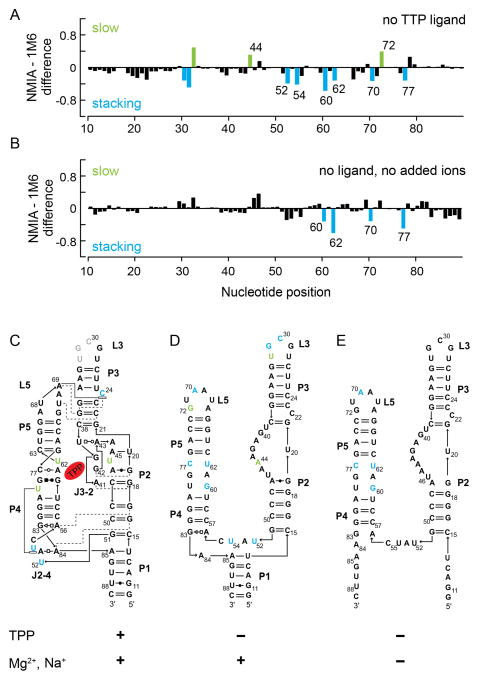
We next examined the TPP riboswitch aptamer domain in the less structured, ligand-free state. These experiments were motivated, in part, by studies suggesting that the J3-2 region of the TPP binding pocket may exist in a pre-formed state14,15,25 and that the ligand-free states of many riboswitches have significant structure.25,26 Two features were immediately evident when we compared the “fingerprint” of the ligand-bound state of the TPP riboswitch with that of the ligand-free state (compare Figures 2B and 5A). First, the magnitudes of the reagent-specific differential reactivities in the ligand-free state were much smaller than those in the ligand-bound state, consistent with a model in which local tertiary structure motifs in the ligand-bound state form transiently or less stably in the less structured ligand-free state. Second, there were more nucleotides with differential reactivities in the ligand-free state, especially 1M6 enhancements, in comparison to the ligand-bound RNA state (Figures 2B and 5A, numbered positions). This observation suggests that the TPP riboswitch samples diverse, yet structured, microstates in the absence of ligand.
Figure 5.
Differential SHAPE reactivities as a function of the extent of stable RNA structure. Differential SHAPE reactivity analysis of the (A) ligand-free and (B) ligand-free, no added ion (no Mg2+/Na+) states of the TPP riboswitch. Nucleotides that showed significant reagent-specific enhancements (as defined in Figure 2) are highlighted. Reagent enhancements superimposed on secondary structure models for the (C) ligand-bound, (D) ligand-free, and (E) no added ion states of the TPP riboswitch.
We superimposed the differential reactivities onto secondary structure models for the ligand-bound (Figure 5C) and ligand-free states (Figure 5D). The differential reactivities in the ligand-free state occurred predominantly at nucleotides in two specific structural contexts: (i) at or adjacent to the TPP ligand-binding pocket (Figure 5D, nucleotides 44, 60, 62, and 77) and (ii) at nucleotides that form key tertiary structure motifs in the fully folded, ligand-bound structure (nucleotides 52, 54, 70, and 72). The differential reactivity data thus support a model in which a population of ligand-free RNA molecules adopts a TPP binding pocket structure similar to the ligand-bound RNA (Figures 5C and 5D, compare J3-2 and nucleotides 60–62 and 77–79). A preformed ligand-binding pocket likely facilitates rapid recognition and binding of the ligand, consistent with fluorescence experiments that show fast TPP binding and ligand-induced riboswitch folding.15
Finally, we probed the ligand-free TPP riboswitch in the absence of added monovalent or divalent ions and generated an experimentally supported secondary structure model7 for the RNA in this state. The major structural difference in the presence versus absence of ions occurred in the P1 helix. In the absence of ions, nucleotides in the P1 helix were reactive towards SHAPE reagents, suggesting that this helix does not form stably (Figure 5E). We observed four statistically significant differential reactivities in the ion-free TPP RNA (Figures 5B and 5E, nucleotides 60, 62, 70, and 77). All four nucleotides that exhibited enhanced reactivity towards 1M6 were also observed in the ligand-free state (+Mg2+) of the TPP riboswitch. These observations support a model in which the P4–P5 side of the TPP ligand binding pocket is partially pre-structured, independent of ionic environment.
Differential SHAPE reactivity probing of the ligand-bound, ligand-free and low-ion states of the TPP riboswitch demonstrated that, as the extent of higher-order structure was systematically decreased, the magnitude and number of reagent-specific differential reactivities also decreased. Nonetheless, even in apparently loosely structured states, the TPP riboswitch RNA samples specific local conformations that allow for favorable, potentially transient, tertiary structure interactions involved in ligand binding.
Differential SHAPE analysis uses a dual-reagent detection scheme that takes advantage of two distinct, fundamental features of local RNA structure: (1) some nucleotides are constrained in a special structural environment such that they become reactive on slow timescales and (2) although the vast majority of nucleotides in folded RNAs have fully “saturated” base stacking interactions, a few nucleotides form unusual conformations that leave one face of the nucleobase accessible for a binding interaction. To a first-order approximation, these two features are selectively detectable by enhanced reactivity towards NMIA and 1M6, respectively. The longer lifetime of NMIA in solution, prior to degradation by hydrolysis, provides a longer window for nucleotides experiencing slow nucleotide dynamics to achieve a SHAPE-reactive conformation8 (Figure 1). Enhanced reactivity towards 1M6 reflects preferential reagent binding via stacking at accessible nucleobase sites, a model supported by direct visualization in high-resolution structures (Figure 3B), the correlation between strength of electron-withdrawing substituent and SHAPE reactivity (Figure S2), and high-level density functional theory calculations (Figure 4).
In sum, differential SHAPE analysis makes it possible to rapidly identify nucleotides involved in key non-canonical and tertiary interactions in RNA, essentially independent of RNA size, complexity, and dynamics. This approach takes advantage of the insight that reactivity differences between two hydroxyl-selective, nucleic acid probes can reveal new information, complementary to that obtained from either reagent alone. Nucleotides detected using differential SHAPE always mapped to nucleotides involved in or adjacent to important tertiary structure interactions. This approach detects the local dynamic and structural differences that are characteristic of important tertiary structure motifs and yields information complementary to hydroxyl radical footprinting,27 which detects tertiary interactions by virtue of their ability to shield nucleotides from reaction with a solvent-based probe.
We anticipate that differential SHAPE reactivity analysis and the resulting RNA tertiary structure “fingerprints” will prove especially useful in ongoing efforts to improve secondary and tertiary prediction, for identification of ligand and protein binding sites, and in large-scale searches for tertiary interactions in complex RNAs.
Supplementary Material
Acknowledgments
This work was supported by grants from the NSF (MCB-0919666) and NIH (GM064803) to K.M.W. We are grateful to Alexander Serganov for providing electron density maps and crystal packing information for the riboswitch RNAs and to Shubin Liu for assistance during early phases of the electronic structure calculations.
Footnotes
Supporting Information: Experimental methods and three figures. Available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sharp PA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]; (x) Darnell EJ. RNA: Life’s Indispensable Molecule. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2011. [Google Scholar]
- 2.Leontis NB, Lescoute A, Westhof E. Curr Opin Struct Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butcher SE, Pyle AM. Acc Chem Res. 2011 doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 4.Weeks KM. Curr Opin Struct Biol. 2010;20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weeks KM, Mauger DM. Acc Chem Res. 2011 doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. J Am Chem Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deigan KE, Li TW, Mathews DH, Weeks KM. Proc Natl Acad Sci U S A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. J Am Chem Soc. 2008;130:8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortimer SA, Weeks KM. Proc Natl Acad Sci U S A. 2009;106:15622–15627. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan CD, Weeks KM. Biochemistry. 2010;49:5418–5425. doi: 10.1021/bi100267g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holbrook SR. Annu Rev Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]; (x) Stombaugh J, Zirbel CL, Westhof E, Leontis NB. Nucleic Acids Res. 2009;37:2294–2312. doi: 10.1093/nar/gkp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sokoloski JE, Godfrey SA, Dombrowski SE, Bevilacqua PC. RNA. 2011;17:1775–1787. doi: 10.1261/rna.2759911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]; (x) Thore S, Leibundgut M, Ban N. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]; (x) Kulshina N, Edwards TE, Ferre-D’Amare AR. RNA. 2010;16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards TE, Ferre-D’Amare AR. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Lang K, Rieder R, Micura R. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baird NJ, Ferre-D’Amare AR. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]; (x) Ali M, Lipfert J, Seifert S, Herschlag D, Doniach S. J Mol Biol. 2010;396:153–165. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steen KA, Malhotra A, Weeks KM. J Am Chem Soc. 2010;132:9940–9943. doi: 10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 19.Hansch C, Leo A. Substituent constants for correlation analysis in chemistry and biology. Wiley-Interscience; NY: 1979. [DOI] [PubMed] [Google Scholar]
- 20.Hunter CA, Lawson KR, Perkins J, Urch CJ. J Chem Soc Perkin Trans. 2001;2:651–669. [Google Scholar]; (x) Mignon P, Loverix S, Steyaert J, Geerlings P. Nucleic Acids Res. 2005;33:1779–1789. doi: 10.1093/nar/gki317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersheim M, Turner DH. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 22.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; (x) Serganov A, Huang L, Patel DJ. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]; (x) Krasilnikov AS, Yang X, Pan T, Mondragon A. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 23.Blouin S, Lafontaine DA. RNA. 2007;13:1256–1267. doi: 10.1261/rna.560307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krasilnikov AS, Mondragon A. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montange RK, Batey RT. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 26.Haller A, Souliere MF, Micura R. Acc Chem Res. 2011 doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- 27.Brenowitz M, Chance MR, Dhavan G, Takamoto K. Curr Opin Struct Biol. 2002;12:648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]; (x) Tullius TD, Greenbaum JA. Curr Opin Chem Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.