Abstract
Background:
Predict (www.predict.nhs.uk) is an online, breast cancer prognostication and treatment benefit tool. The aim of this study was to incorporate the prognostic effect of HER2 status in a new version (Predict+), and to compare its performance with the original Predict and Adjuvant!.
Methods:
The prognostic effect of HER2 status was based on an analysis of data from 10 179 breast cancer patients from 14 studies in the Breast Cancer Association Consortium. The hazard ratio estimates were incorporated into Predict. The validation study was based on 1653 patients with early-stage invasive breast cancer identified from the British Columbia Breast Cancer Outcomes Unit. Predicted overall survival (OS) and breast cancer-specific survival (BCSS) for Predict+, Predict and Adjuvant! were compared with observed outcomes.
Results:
All three models performed well for both OS and BCSS. Both Predict models provided better BCSS estimates than Adjuvant!. In the subset of patients with HER2-positive tumours, Predict+ performed substantially better than the other two models for both OS and BCSS.
Conclusion:
Predict+ is the first clinical breast cancer prognostication tool that includes tumour HER2 status. Use of the model might lead to more accurate absolute treatment benefit predictions for individual patients.
Keywords: breast cancer, HER2, prognostic model, HER2
Accurate prognostication is essential when selecting the most appropriate adjuvant therapy following surgery for early breast cancer. A number of predictive models are now available to help estimate the survival for individual patients, including the Nottingham Prognostic Index (NPI), Adjuvant and Predict, and to date, these have all been based on known pathological prognostic factors including tumour size, tumour grade and lymph node status. The NPI, first described in1982 (Haybittle et al, 1982), has been prospectively validated (Todd et al, 1987; D'Eredita et al, 2001) and more recently updated (Blamey et al, 2007a) to provide accurate survival estimates following breast cancer surgery, including individual survival estimates (Blamey et al, 2007b). The introduction of Adjuvant!, a web-based (www.adjuvantonline.com) prognostication tool, in 2001 (Ravdin et al, 2001) went a step further by also providing absolute treatment benefits for hormone therapy and chemotherapy by applying risk reductions from the Early Breast Cancer Trialists Collaborative Group (1998a, 1998b) to breast cancer-specific mortality estimates based on the Surveillance Epidemiology and End Results Program (www.seer.cancer.gov). This model has been validated in case cohorts from British Columbia (Olivotto et al, 2005), the Netherlands (Mook et al, 2009) and the United Kingdom (Campbell et al, 2009).
Prognostication is becoming more sophisticated, and additional prognostic and predictive factors need to be considered in any current prognostic and treatment benefit model. There is a growing body of evidence to show that screen detection confers an additional survival benefit beyond stage shift and also reduces the risk of systemic recurrence when compared with symptomatic cancers of a similar stage (Joensuu et al, 2004; Shen et al, 2005). Recent studies have shown that the majority of the survival advantage associated with breast screening can be explained by this shift to an earlier stage at diagnosis, and more favourable prognostic factors (Dawson et al, 2009), but approximately 25% of the survival advantage is still unexplained (Wishart et al, 2008). These findings were recently confirmed in a data set from the Netherlands in which screen detection was associated with a 26 and 38% reduction in all-cause and breast cancer-specific mortality, respectively (Mook et al, 2011). The authors from this paper concluded that method of detection should be taken into account when estimating individual prognosis.
Predict is an online prognostication and treatment benefit tool developed in the United Kingdom, which is based on 5694 women diagnosed in East Anglia from 1999 to 2003 (Wishart et al, 2010). The model includes mode of detection and provides 5- and 10-year survival estimates as well as treatment benefit predictions at both time points, and it has been validated in an independent case cohort from the United Kingdom and more recently in a British Columbia data set wherein it was also compared with Adjuvant! (Wishart et al, 2011). This comparison showed that Predict and Adjuvant! provide accurate overall and breast cancer-specific survival (BCSS) estimates that were comparable. However, the Predict estimates did not utilise the mode of detection component of the model as the mode of detection was not available for the British Columbia cohort. Moreover, Adjuvant! does not take account of the mode of detection. Predict was slightly better calibrated for breast cancer-specific mortality with predicted deaths being within 3.4% of observed deaths compared with 6.7% for Adjuvant!. Both models showed good discrimination with similar area under the receiver-operator characteristic curve (AUC; 0.723 vs 0.727 for Predict and Adjuvant, respectively). Predict is now available online at www.predict.nhs.uk.
The aim of this study was to incorporate the prognostic effect of HER2 status into Predict (hereafter referred to as Predict+), and to compare the 10-year survival estimates from Predict+ with Predict, Adjuvant! and the observed 10-year outcome from the British Columbia data set.
Methods
Prognostic effect of tumour HER2 status
Estimates for the prognostic effect of HER2 status were based on an analysis of data from the Breast Cancer Association Consortium (BCAC). Pathology data from 12 of these studies has been previously published (Blows et al, 2010) and the analysis was updated to include data from another 3 studies excluded from the previous publication because of missing data on basal markers that is not relevant for this analysis (Table 1). However, as the validation cohort for this study overlaps substantially with the British Columbia Cancer Agency (BCCA) case series published in Blows et al (2010), this case series was excluded from the BCAC data re-analysis. In addition, all patients diagnosed since 2004 were excluded, to exclude patients likely to have been treated with trastuzumab. In total, HER2 data were available for 10 179 cases (7278 ER-positive and 2931 ER-negative, Table 2) for whom data were also available for age at diagnosis, tumour size (⩽2, 2–4.9 and 5+ cm), tumour grade and nodal status. We estimated the hazard ratio for HER2-positive disease compared with HER2-negative disease using a Cox proportional hazards model stratified by study and adjusted for size, grade and nodal status. Separate regression models were used for ER-positive and ER-negative cases. As we have shown previously (Blows et al, 2010), the hazard ratio for HER2-positive disease decreases over time in women with ER-positive breast cancer, so the log hazard ratio was modelled to vary linearly with time. The effect of HER2 in women with ER-negative breast cancer is not time-dependent.
Table 1. Description of participating studies.
| Study | Country | Case ascertainment | Case definition | Age range (years) | References |
|---|---|---|---|---|---|
| Amsterdam Breast Cancer Study | The Netherlands | Hospital-based | All cases of operable, invasive cancer diagnosed from 1974–1994 in four Dutch hospitals. Familial non-BRCA1/2 cases <50 from the Clinical Genetic Centre at the Netherlands Cancer Institute | 23–50 | Schmidt et al (2007) |
| Bavarian Breast Cancer Case–Control Study | Germany | Hospital-based cases | Consecutive, unselected cases with invasive breast cancer recruited at the University Breast Centre, Franconia in Northern Bavaria during 2002–2006 | 27–86 | Fasching et al (2008) |
| Helsinki Breast Cancer Study | Finland | Hospital-based | (1) Consecutive cases (883) from the Department of Oncology, Helsinki University Central Hospital 1997–1998 and 2000, (2) consecutive cases (986) from the Department of Surgery, Helsinki University Central Hospital 2001–2004, (3) familial breast cancer patients (536) from the Departments of Oncology and Clinical Genetics, Helsinki University Central Hospital(1995) | 22–96 | Syrjakoski et al (2000); Kilpivaara et al (2005); Fagerholm et al (2008) |
| Jewish General Hospital | Canada | Hospital-based | Ashkenazi Jewish women diagnosed with non-metastatic, invasive breast cancer at Jewish General Hospital, Montreal, between 1980 and 1995 | 26–66 | Tischkowitz et al (2007) |
| Kuopio Breast Cancer Study | Finland | Women seen at Kuopio University Hospital between 1990 and 1995 because of breast lump, mammographic abnormality, or other breast symptom, who were found to have breast cancer | 23–92 | Hartikainen et al (2005) | |
| Mayo Clinic Breast Cancer Study | USA | Hospital-based | Incident cases residing in six states (MN, WI, IA, IL, ND, SD) seen at the Mayo Clinic in Rochester, MN, from 2002–2005 | 22–89 | Olson et al (2007) |
| Melbourne Collaborative Cohort Study | Australia | Cohort | Incident cases diagnosed within the Melbourne Collaborative Cohort Study during the follow-up from baseline (1990–1994) to 2004 of the 2 4469 participating women | 30–82 | Giles and English (2002) |
| Nottingham Breast Cancer Case Series | UK | Hospital-based | Primary operable breast carcinoma patients presenting from 1986 to 1998, and entered into the Nottingham Tenovus Primary Breast Carcinoma Series | 26–93 | Rakha et al (2007) |
| Oulu Breast Cancer Study | Finland | Hospital-based | Consecutive incident cases diagnosed at the Oulu University Hospital, 2000–2004 | 28–92 | Erkko et al (2007) |
| NCI Polish Breast Cancer Study | Poland | Population-based | Incident cases from 2000–2003 identified through a rapid identification system in participating hospitals covering ∼90% of all eligible cases; periodic check against the cancer registries in Warsaw and Łódź to assure complete identification of cases | 27–75 | Garcia-Closas et al (2006) |
| Sheffield Breast Cancer Study | UK | Hospital-based | Women with pathologically confirmed breast cancer, recruited from surgical outpatient clinics at the Royal Hallamshire Hospital, Sheffield, 1998–2002; cases are a mixture of prevalent and incident disease | 29–93 | Rafii et al (2002); MacPherson et al (2004) |
| Study of Epidemiology and Risk factors in Cancer Heredity | UK | Population-based | Two groups of cases identified through East Anglian Cancer Registry: (1) prevalent cases diagnosed age <55 years from 1991–1996 and alive when study started in 1996; (2) incident cases diagnosed age <70 years diagnosed after 1996 | 23–69 | Callagy et al (2006) |
| University of British Columbia Breast Cancer Trials | Canada | Hospital-based | Women with stage I to III breast cancer, who participated in four different British Columbia Cancer Agency clinical trials between 1970 and 1990, and all received chemotherapy | 22–90 | Ragaz et al (1997); Ragaz et al (2005) |
| Vancouver General Hospital | Canada | Hospital-based | Women with primary breast cancer, who underwent surgery at Vancouver General Hospital 1975–1995 | 28–91 | Tischkowitz et al (2007) |
Table 2. Number of cases by study, ER status and HER2 status.
|
ER-positive
|
ER-negative
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study | HER2− (%) | HER2+ (%) | Total | HER2− (%) | HER2+ (%) | Total | ||||
| ABCS | 512 | (87) | 74 | (13) | 586 | 231 | (77) | 69 | (23) | 300 |
| BBCC | 585 | (88) | 77 | (12) | 662 | 164 | (73) | 62 | (27) | 226 |
| HEBCS | 173 | (88) | 23 | (12) | 196 | 37 | (77) | 11 | (23) | 48 |
| JGH | 166 | (94) | 11 | (6) | 177 | 77 | (79) | 21 | (21) | 98 |
| KBCS | 281 | (93) | 20 | (7) | 301 | 59 | (67) | 29 | (33) | 88 |
| MCBCS | 386 | (84) | 71 | (16) | 457 | 67 | (77) | 20 | (23) | 87 |
| MCCS | 307 | (93) | 24 | (7) | 331 | 99 | (76) | 31 | (24) | 130 |
| NOBCS | 1006 | (96) | 40 | (4) | 1046 | 346 | (82) | 76 | (18) | 422 |
| OBCS | 390 | (91) | 39 | (9) | 429 | 72 | (68) | 34 | (32) | 106 |
| PBCS | 656 | (95) | 34 | (5) | 690 | 289 | (80) | 71 | (20) | 360 |
| SBCS | 201 | (93) | 15 | (7) | 216 | 70 | (84) | 13 | (16) | 83 |
| SEARCH | 1447 | (91) | 135 | (9) | 1582 | 334 | (75) | 113 | (25) | 447 |
| UBC | 299 | (76) | 93 | (24) | 392 | 296 | (66) | 152 | (34) | 448 |
| VGH | 201 | (94) | 12 | (6) | 213 | 42 | (72) | 16 | (28) | 58 |
| Total | 6610 | (91) | 668 | (9) | 7278 | 2183 | (75) | 718 | (25) | 2901 |
Abbreviations: ABCS=Amsterdam Breast Cancer Study; BBCC=Bavarian Breast Cancer Case–Control Study; ER=oestrogen receptor; HEBCS=Helsinki Breast Cancer Study; HER2=human epidermal growth factor receptor-2; JGH=Jewish General Hospital; KBCS=Kuopio Breast Cancer Study; MCBCS=Mayo Clinic Breast Cancer Study; MCCS=Melbourne Collaborative Cohort Study; NOBCS=Nottingham Breast Cancer Case Series; OBCS=Oulu Breast Cancer Study; PBCS=NCI Polish Breast Cancer Study; SBCS=Sheffield Breast Cancer Study; SEARCH=Study of Epidemiology and Risk factors in Cancer Heredity; UBCBCT=University of British Columbia Breast Cancer Trials; VGH=Vancouver General Hospital.
The hazard ratios estimated from the BCAC data set were then used to modify Predict, which is also based on a Cox proportional hazards models. However, Predict was developed using a case cohort of breast cancer cases of unknown HER2 status, and so the underlying baseline hazard is representative of cases of average HER2 status. The HER2 hazard ratio estimates based on the BCAC data are for HER2-positive cases compared with HER2-negative cases, and so these were rescaled to give an average hazard ratio of unity using an estimated prevalence of HER2 of 9% in ER-positive cases and 25% in ER-negative cases. The applied hazard ratios for ER-positive cases are shown in Table 3. A fixed hazard ratio of 0.93 for HER2–negative cases and 1.27 for HER2–positive cases compared with HER2 unknown cases was applied to the baseline hazard of the ER-negative model.
Table 3. Hazard ratio by HER2 status by time since diagnosis in ER-positive breast cancer.
|
HER2 status
|
|||
|---|---|---|---|
| Year after diagnosis | Unknown | Positive | Negative |
| 1 | 1 | 1.84 | 0.95 |
| 2 | 1 | 1.70 | 0.95 |
| 3 | 1 | 1.58 | 0.96 |
| 4 | 1 | 1.47 | 0.97 |
| 5 | 1 | 1.36 | 0.97 |
| 6 | 1 | 1.26 | 0.98 |
| 7 | 1 | 1.17 | 0.99 |
| 8 | 1 | 1.08 | 0.99 |
| 9 | 1 | 1.01 | 1.00 |
| 10 | 1 | 0.93 | 1.01 |
Abbreviations: ER=oestrogen receptor; HER2=human epidermal growth factor receptor-2.
Weighted average hazard ratio for HER2+ and HER2− =1.
Validation study population
We used the same case cohort from British Columbia, Canada, which we used to validate the original version of Predict. The data set has previously been described (Olivotto et al, 2005), but, in brief, includes data from 1653 patients with information on HER2 status out of a total of 3140 patients with stage I or II invasive breast cancer diagnosed in British Columbia, Canada, from 1989 to 1993, who were identified from the Breast Cancer Outcomes Unit (BCOU) of the BCCA. The BCOU prospectively records demographical information, pathological information, staging, initial treatment and outcome information including first loco-regional and distant relapse, as well as date and cause of death. Outcome data were reported annually by the treating oncologist, family physician or by monthly death certificate flagging through the British Columbia Cancer Registry and Department of Vital Statistics for British Columbia.
Information obtained from the BCOU database included age at diagnosis, sex, menopausal status, year of diagnosis, histology (ductal, lobular, other), histological grade, tumour size, number of lymph nodes sampled and number of lymph nodes positive, lymphovascular invasion status, ER status, HER2 status, type of local therapy (wide local excision, mastectomy, radiotherapy) and type of adjuvant systemic therapy (none, chemotherapy, endocrine therapy, both). The HER2 status was evaluated using TMAs as previously described (Chia et al, 2008). Chemotherapy regimens were categorised as four cycles of doxorubicin plus cyclophosphamide; 6 months of cyclophosphamide, methotrexate and fluorouracil, or other chemotherapy during this time period. None of these patients received trastuzumab.
Study endpoints were overall survival (OS) and BCSS. Of 1653 cases used in this analysis, cause of death was unknown in 5, and so 1648 cases were used for the analysis of BCSS. Ten-year-predicted OS and BCSS were calculated for each patient using Predict+, Predict and Adjuvant! (standard version 8) by investigators blinded to the actual outcome data for each patient after entry of patient age, tumour size, number of positive nodes, tumour grade, ER status, HER2 status (Predict+ only) and adjuvant systemic therapy. The default comorbidity setting of ‘minor symptoms’ and the chemotherapy option chosen was ‘anthracycline-containing; ⩾4 cycles’ were used in Adjuvant!. The ‘second-generation’ chemotherapy option (anthracycline-containing regimens) was used in Predict+ and Predict. The mode of detection input was not used in Predict or Predict+, as this information was not available in the British Columbia data set. Ten-year-predicted OS and BCSS from Predict+, Predict and Adjuvant were compared with observed 10-year OS and BCSS.
Model calibration is a comparison of the predicted mortality estimates from each model with the observed mortality. In addition to comparing calibration in the complete data set, we evaluated calibration within strata of other prognostic variables. We also evaluated calibration within quartiles of predicted mortality. A goodness-of-fit test was carried out by using a χ2-test based on the observed and predicted number of events (4 d.f.). Model discrimination was evaluated by calculating the AUC calculated for 10-year breast cancer-specific and overall mortality. This is a measure of how well the models identify those patients with worse survival. The AUC is the probability that the predicted mortality from a randomly selected patient who died will be higher than the predicted mortality from a randomly selected survivor.
Results
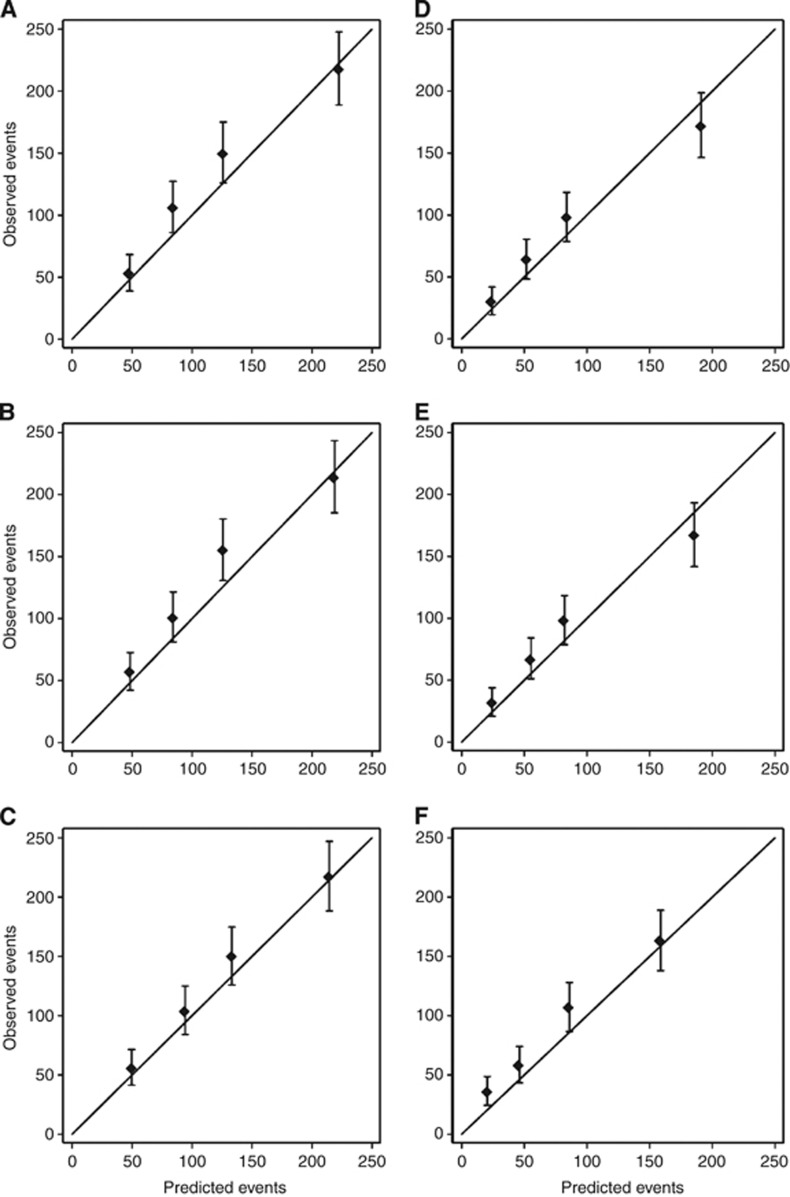
The calibration of Predict for both all-cause mortality and for breast cancer-specific mortality was improved by the incorporation of HER2 status. Adjuvant! performed slightly better than Predict and Predict+ for all-cause mortality, but both Predict and Predict+ outperformed Adjuvant! for breast cancer-specific mortality. All three models slightly underestimated the observed number of deaths. The total number of deaths predicted by Adjuvant was within 6.1% of that observed (492 vs 524, P=0.16) compared with 8.8% for Predict (478 vs 524, P=0.04) and 8.4% for Predict+ (480 vs 524, P=0.05). The total number of breast cancer-specific deaths predicted by Adjuvant was within 14% of that observed (311 vs 360, P=0.01) compared with 3.6% for Predict (347 vs 360, P=0.49) and 2.5% for Predict+ (351 vs 360, P=0.60). Table 4 shows the observed and predicted all-cause 10-year OS by various clinico-pathological sub-groups. All models performed well in most subgroups. Notable exceptions were the performance in women aged 20–35 years, in which all three models underpredicted the actual number of deaths by 32%, and in HER2-positive cases, in which Adjuvant and Predict underestimated the number of deaths by close to 20% compared with just 9% for Predict+. None of these differences were statistically significant (P>0.05). We also compared the predicted mortality with that observed for women in each quartile of predicted risk. Calibration was good for Adjuvant! across all risk categories (goodness-of-fit, P=0.51), and reasonable for Predict+ (goodness-of-fit, P=0.042) and Predict (goodness-of-fit, P=0.032) (Figures 1A–C). Table 5 shows the observed 10-year breast cancer-specific mortality compared with that predicted by Predict+, Predict and Adjuvant. Again, each model performed well across most subgroups, except in women aged 20–35 years wherein they all underpredicted the actual number of deaths by 32% (P>0.05). In HER2-positive patients, the total number of breast cancer-specific deaths predicted by Predict+ was within 5% of observed (71 vs 75, P=0.64) compared with 20% for Predict (n=60 vs 75, P=0.08) and 29% for Adjuvant (53 vs 75, P=0.01). Calibration across the quartiles of predicted breast cancer risk was good for Predict+ (goodness-of-fit, P=0.11) and Predict (goodness-of-fit, P=0.068), and reasonable for Adjuvant! (goodness-of-fit, P=0.001) (Figures 1D–F). Model discrimination was similar for all three models with AUCs for breast cancer-specific mortality of 0.665, 0.661 and 0.649 for Predict+, Predict and Adjuvant!, respectively.
Table 4. Observed and predicted 10-year all-cause mortality by demographical, pathological and treatment characteristics.
|
Deaths
|
|||||
|---|---|---|---|---|---|
| Number | Observed | Adjuvant | Predict | Predict+ | |
| Age group (years) | |||||
| 20–35 | 55 | 22 | 15 | 15 | 15 |
| 36–50 | 492 | 120 | 112 | 119 | 119 |
| 51–65 | 523 | 152 | 133 | 132 | 133 |
| 66–75 | 433 | 153 | 153 | 141 | 142 |
| 76+ | 150 | 76 | 79 | 71 | 71 |
| Menopausal status | |||||
| Pre- | 520 | 135 | 121 | 128 | 128 |
| Post | 1089 | 380 | 360 | 339 | 341 |
| Peri-/unknown | 44 | 8 | 10 | 11 | 11 |
| Morphology | |||||
| Ductal | 1489 | 479 | 440 | 428 | 432 |
| Lobular | 132 | 38 | 43 | 40 | 40 |
| Other | 32 | 5 | 8 | 8 | 8 |
| Grade | |||||
| 1 | 104 | 22 | 19 | 15 | 15 |
| 2 | 778 | 208 | 203 | 176 | 177 |
| 3 | 694 | 269 | 247 | 265 | 268 |
| Unknown | 77 | 24 | 23 | 20 | 20 |
| LV invasion | |||||
| Negative | 868 | 233 | 230 | 218 | 218 |
| Positive | 715 | 262 | 239 | 238 | 241 |
| Unknown | 70 | 26 | 22 | 20 | 20 |
| Node status | |||||
| Negative | 976 | 235 | 230 | 215 | 216 |
| Positive | 677 | 288 | 262 | 262 | 265 |
| Tumour size | |||||
| 1–10 | 210 | 40 | 39 | 40 | 40 |
| 11–20 | 745 | 196 | 189 | 186 | 187 |
| 21–50 | 698 | 287 | 264 | 251 | 253 |
| ER status | |||||
| ER− | 350 | 123 | 124 | 129 | 130 |
| ER+ | 1303 | 400 | 367 | 348 | 350 |
| Local Rx | |||||
| BCS+RT | 814 | 200 | 205 | 200 | 200 |
| Mast+RT | 169 | 77 | 74 | 77 | 78 |
| Mast | 670 | 246 | 213 | 201 | 202 |
| Systemic Rx | |||||
| None | 660 | 175 | 170 | 161 | 162 |
| Hormone | 526 | 201 | 188 | 175 | 176 |
| Chemo | 317 | 96 | 93 | 97 | 98 |
| Both | 150 | 51 | 41 | 43 | 44 |
| HER2 | |||||
| Negative | 1450 | 434 | 420 | 405 | 398 |
| Positive | 203 | 90 | 72 | 73 | 82 |
| Total | 1653 | 524 | 492 | 478 | 480 |
Abbreviations: BCS=breast conserving surgery; ER=oestrogen receptor; HER2=human epidermal growth factor receptor-2; LV=lymphovascular; RT=radiotherapy.
The sum of the predicted events within categories may differ from the sum of the observed events because of rounding errors in the predicted events.
Figure 1.
Calibration plots of observed outcomes with 95% confidence intervals against predicted outcomes by quartiles of the predicted value. Overall survival predicted by (A) Predict+ (B) Predict and (C) Adjuvant!, and breast cancer-specific survival predicted by (D) Predict+ (E) Predict and (F) Adjuvant!.
Table 5. Observed and predicted 10-year breast cancer-specific mortality by demographical, pathological and treatment characteristics.
|
Deaths from breast cancer
|
|||||
|---|---|---|---|---|---|
| Number | Observed | Adjuvant | Predict | Predict+ | |
| Age group | |||||
| 20–35 | 55 | 22 | 15 | 15 | 15 |
| 36–50 | 492 | 118 | 102 | 110 | 110 |
| 51–65 | 522 | 102 | 96 | 106 | 108 |
| 66–75 | 430 | 88 | 76 | 86 | 87 |
| 76+ | 149 | 30 | 23 | 31 | 31 |
| Menopausal status | |||||
| Pre− | 520 | 133 | 110 | 118 | 119 |
| Peri-/Unknown | 1085 | 221 | 192 | 220 | 222 |
| Post− | 43 | 6 | 9 | 9 | 9 |
| Morphology | |||||
| Ductal | 1485 | 340 | 281 | 314 | 318 |
| Lobular | 131 | 18 | 24 | 26 | 26 |
| Other | 32 | 2 | 7 | 7 | 7 |
| Grade | |||||
| 1 | 104 | 4 | 5 | 5 | 5 |
| 2 | 775 | 133 | 108 | 104 | 104 |
| 3 | 693 | 206 | 183 | 225 | 228 |
| Unknown | 76 | 17 | 15 | 14 | 14 |
| LV invasion | |||||
| Negative | 864 | 144 | 129 | 142 | 142 |
| Positive | 714 | 196 | 168 | 190 | 193 |
| Unknown | 70 | 18 | 13 | 14 | 14 |
| Node status | |||||
| Negative | 972 | 132 | 117 | 128 | 129 |
| Positive | 676 | 228 | 194 | 219 | 221 |
| Tumour size | |||||
| 1–10 | 210 | 22 | 14 | 21 | 21 |
| 11–20 | 741 | 118 | 101 | 121 | 122 |
| 21–50 | 697 | 220 | 196 | 205 | 208 |
| ER status | |||||
| ER− | 350 | 104 | 102 | 113 | 114 |
| ER+ | 1298 | 256 | 209 | 234 | 236 |
| Local Rx | |||||
| BCS+RT | 810 | 140 | 128 | 142 | 142 |
| Mast+RT | 169 | 59 | 59 | 69 | 70 |
| Mast | 669 | 161 | 124 | 137 | 139 |
| Systemic Rx | |||||
| None | 658 | 100 | 88 | 99 | 99 |
| Hormone | 523 | 121 | 102 | 117 | 118 |
| Chemo | 317 | 94 | 86 | 92 | 93 |
| Both | 150 | 45 | 36 | 40 | 40 |
| HER2 | |||||
| Negative | 1445 | 285 | 258 | 287 | 280 |
| Positive | 203 | 75 | 53 | 60 | 71 |
| Total | 1648 | 360 | 311 | 347 | 351 |
Abbreviations: BCS=breast conserving surgery; ER=oestrogen receptor; HER2=human epidermal growth factor receptor-2; LV=lymphovascular; RT=radiotherapy.
The sum of the predicted events within categories may differ from the sum of the observed events because of rounding errors in the predicted events.
Discussion
Predict was developed using a flexible, Cox proportional hazards model that enables the easy incorporation of additional prognostic factors using external estimates of the prognostic effect of such a factor. We have used this flexibility to incorporate the prognostic effect of HER2 derived from a large multi-centre study. A key feature of Predict, and one that differentiates it from Adjuvant!, is that it uses different underlying models for ER-positive and ER-negative disease. This is particularly important as the effect of other prognostic variables is often different according to ER status. This difference is particularly marked for HER2 status, where the effect of HER2 is strongly time-dependent in ER-positive cases but not for ER-negative cases. Our results confirm that the inclusion of HER2 status in the clinical prognostication tool Predict improves both model discrimination and calibration. The improvement in model fit was most pronounced in the HER2-positive subset of patients and was particularly marked for breast cancer-specific mortality. Given that it is breast cancer-specific mortality that is reduced by adjuvant therapy (Early Breast Cancer Trialists Collaborative Group, 1998a, 1998b), it is likely that improved model performance will lead to more accurate predictions of the absolute benefit of treatment. Adjuvant! performed better than either Predict or Predict+ for overall mortality. This might be expected as the background mortality data on which Adjuvant! is based are from North America, whereas Predict is based on background mortality data from the United Kingdom.
The weaknesses of our study also need to be considered in interpreting our results. There were some missing data for the BCAC cohorts used to derive the HER2-specific hazard ratios and for the BCOU cohort. Data were missing for multiple reasons, but the major reason was because of unavailability of archival pathology material. Although it is possible that there is some selection bias in the missing data – for example, pathology material is more likely to be unavailable for small tumours – any bias is unlikely to be large. This notion is supported by the results of the validation in the independent data from the BCOU cohort. As we have described previously, none of the models performed very well in the youngest age group. The reasons for this are unclear, but probably reflect the fact that the data on which the models were based included relatively small numbers of patients in this age group.
The online version of Predict includes survival estimates and treatment benefit at 5 and 10 years post diagnosis. Four clinical trials have now reported on the benefit of trastuzumab therapy – FinHER (Joensuu et al, 2009), HERA (Smith et al, 2007), B31/N9831 (Romond et al, 2005; Perez et al, 2010) and BCIRG006 (Slamon et al, 2011). The relative reduction in the all-cause mortality has ranged from 0.33 to 0.45. Two studies reported results stratified by tumour hormone-receptor status neither of which found evidence for heterogeneity (Romond et al, 2005; Smith et al, 2007; Perez et al, 2010). The new version of Predict+, including trastuzumab benefit at 5 years is available at www.predict.nhs.uk.
One of the key advantages of the Predict models is that they include mode of detection as one of the input parameters. As previously discussed, screen detection appears to confer an additional survival advantage over and above known prognostic factors. Unfortunately, as screening data was not available in the British Columbia data set, it was not possible to use this feature when running the Predict models, so the default setting was ‘unknown’ for all patients. Thus, we cannot estimate the performance of the Predict models if the mode of detection were known. However, it is very likely that model performance would improve with the addition of mode-of-detection data. In addition, a recent publication has called for the mode of detection to be taken into account when deciding optimal adjuvant therapy for an individual patient (Mook et al, 2011). Predict+ can now provide such a model with the added benefit of the inclusion of HER2 status as well as the absolute treatment benefit of trastuzumab at 5 years. Further validations of Predict+ in data sets where the mode of detection, HER2 status and trastuzumab treatment are recorded are planned in the near future.
This study reports the first successful inclusion of HER2 status in a prognostic model (Predict+) based on known clinical and pathological factors. The study has demonstrated a marked improvement in 10-year BCSS estimates using Predict+ compared with the original Predict model for HER2-positive patients. Both Predict models provide better BCSS estimates than Adjuvant! in HER2-positive patients in this data set. This improvement in BCSS prognostication for HER2-positive patients was not achieved at the expense of the HER2-negative cohort, wherein both Predict and Predict+ performed better than Adjuvant. Predict+ also provided better breast cancer-specific mortality estimates that Predict and Adjuvant. This is extremely important for optimal model function, as it is the breast cancer-specific mortality that is reduced by the relative risk reductions of adjuvant therapy, and this improvement in model performance by Predict+ should lead to more accurate absolute treatment benefit predictions for individual patients.
Acknowledgments
We thank all the patients who took part in the component BCAC studies and the many other individuals who have made these studies possible. In particular, we thank Hans Peterse, Rob Tollenaar, Vincent Smit, Renate de Groot, Renate Udo, Flora van Leeuwen (ABCS); Claudia Rauh, Julia Wessel (BBCC); the BC Cancer Registry and Breast Cancer Outcomes Unit (BCCA); Päivi Heikkilä, Kirsimari Aaltonen, Kristiina Aittomäki, Ari Ristimäki, Laura Hautala, Mira Heinonen, RN Hanna Jäntti, Irja Erkkilä, and the Finnish Cancer Registry (HEBCS); Lars A Akslen (JGH); Vicky Cafourek, Matthew Kosel and Zachary Fredericksen (MCBCS); John Hopper, Dallas English and Helen Kelsall (MCCS), (NOBCS); Arja Jukkola-Vuorinen, Taina Turpeenniemi-Hujanen, Mervi Grip, Saila Kauppila, Kari Mononen and Meeri Otsukka (OBCS); Louise Brinton, Jonine Figueroa, Kelly Bolton, Neonila, Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, and Michael Stagner (PBCS); Sabapathy, Balasubramanian, Malcolm WR Reed, Helen Cramp, and Dan Connley (SBCS); Sarah-Jane Dawson and the SEARCH team (SEARCH). The contributing studies are funded by grants from Breast Cancer Campaign (2004Nov49), Cancer Research UK (C490/A10119, C490/A10124); US National Institutes of Health (CA122340, CA122340Z, CA116201); special Government Funding (EVO) of Kuopio University Hospital; the Cancer Fund of North Savo; the Finnish Cancer Organisation; the Academy of Finland (132473); strategic funding of the University of Eastern Finland; the Helsinki University Central Hospital Research Fund; the Finnish Cancer Society; the Sigrid Juselius Foundation; Susan G Komen for the Cure, Yorkshire Cancer Research. GCW and CC received research funding from the Cambridge NIHR Biomedical Research Centre and the Cambridge Experimental Cancer Medicine Centre. CDB, TN and DH are partly supported through Career Awards from the Michael Smith Foundation For Health Research. MKS is funded by the Dutch Cancer Society (NKI DCS 2009-4363).
Author contributions
The Predict project was initiated by GCW, CC and PDPP. Study design: GCW, CDB and PDPP. Data analysis: ED, CDB and PDPP. Patient enrolment and data collection: DCG, MKS, LJvtV, AB, PAF, MWB, MC, TON, KG, DH, HN, TH, LRB, WDF, RW, KP, AM, GG, GS, IE, AG, MG-C, MS, AC, SSC, FMB, EP, CC and PDPP. Drafting of the manuscript: GCW and PDPP. All authors contributed to the editing of the final manuscript.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflicts of interest.
References
- Blamey RW, Ellis IO, Pinder SE, Lee AH, Macmillan RD, Morgan DA, Robertson JF, Mitchell MJ, Ball GR, Haybittle JL, Elston CW (2007a) Survival of invasive breast cancer according to the Nottingham Prognostic Index in cases diagnosed in 1990–1999. Eur J Cancer 43(10): 1548–1555 [DOI] [PubMed] [Google Scholar]
- Blamey RW, Pinder SE, Ball GR, Ellis IO, Elston CW, Mitchell MJ, Haybittle JL (2007b) Reading the prognosis of the individual with breast cancer. Eur J Cancer 43(10): 1545–1547 [DOI] [PubMed] [Google Scholar]
- Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, Heikkila P, Heikkinen T, Nevanlinna H, Akslen LA, Begin LR, Foulkes WD, Couch FJ, Wang X, Cafourek V, Olson JE, Baglietto L, Giles GG, Severi G, McLean CA, Southey MC, Rakha E, Green AR, Ellis IO, Sherman ME, Lissowska J, Anderson WF, Cox A, Cross SS, Reed MW, Provenzano E, Dawson SJ, Dunning AM, Humphreys M, Easton DF, Garcia-Closas M, Caldas C, Pharoah PD, Huntsman D (2010) Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med 7(5): e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callagy GM, Pharoah PD, Pinder SE, Hsu FD, Nielsen TO, Ragaz J, Ellis IO, Huntsman D, Caldas C (2006) Bcl-2 is a prognostic marker in breast cancer independently of the Nottingham Prognostic Index. Clin Cancer Res 12(8): 2468–2475 [DOI] [PubMed] [Google Scholar]
- Campbell HE, Taylor MA, Harris AL, Gray AM (2009) An investigation into the performance of the Adjuvant! Online prognostic programme in early breast cancer for a cohort of patients in the United Kingdom. Br J Cancer 101(7): 1074–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia S, Norris B, Speers C, Cheang M, Gilks B, Gown AM, Huntsman D, Olivotto IA, Nielsen TO, Gelmon K (2008) Human epidermal growth factor receptor 2 verexpression as a prognostic factor in a large tissue microarray series of node-negative breast cancers. J Clin Oncol 26(35): 5697–5704 [DOI] [PubMed] [Google Scholar]
- D'Eredita G, Giardina C, Martellotta M, Natale T, Ferrarese F (2001) Prognostic factors in breast cancer: the predictive value of the Nottingham Prognostic Index in patients with a long-term follow-up that were treated in a single institution. Eur J Cancer 37(5): 591–596 [DOI] [PubMed] [Google Scholar]
- Dawson SJ, Duffy SW, Blows FM, Driver KE, Provenzano E, LeQuesne J, Greenberg DC, Pharoah P, Caldas C, Wishart GC (2009) Molecular characteristics of screen-detected vs symptomatic breast cancers and their impact on survival. Br J Cancer 101(8): 1338–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists Collaborative Group (1998a) Polychemotherapy for early breast cancer: an overview of the randomised trials. Early Breast Cancer Trialists' Collaborative Group. Lancet 352(9132): 930–942 [PubMed] [Google Scholar]
- Early Breast Cancer Trialists Collaborative Group (1998b) Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet 351: 1451–1467 [PubMed] [Google Scholar]
- Erkko H, Xia B, Nikkila J, Schleutker J, Syrjakoski K, Mannermaa A, Kallioniemi A, Pylkas K, Karppinen SM, Rapakko K, Miron A, Sheng Q, Li G, Mattila H, Bell DW, Haber DA, Grip M, Reiman M, Jukkola-Vuorinen A, Mustonen A, Kere J, Aaltonen LA, Kosma VM, Kataja V, Soini Y, Drapkin RI, Livingston DM, Winqvist R (2007) A recurrent mutation in PALB2 in Finnish cancer families. Nature 446(7133): 316–319 [DOI] [PubMed] [Google Scholar]
- Fagerholm R, Hofstetter B, Tommiska J, Aaltonen K, Vrtel R, Syrjakoski K, Kallioniemi A, Kilpivaara O, Mannermaa A, Kosma VM, Uusitupa M, Eskelinen M, Kataja V, Aittomaki K, von Smitten K, Heikkila P, Lukas J, Holli K, Bartkova J, Blomqvist C, Bartek J, Nevanlinna H (2008) NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat Genet 40(7): 844–853 [DOI] [PubMed] [Google Scholar]
- Fasching PA, Loehberg CR, Strissel PL, Lux MP, Bani MR, Schrauder M, Geiler S, Ringleff K, Oeser S, Weihbrecht S, Schulz-Wendtland R, Hartmann A, Beckmann MW, Strick R (2008) Single nucleotide polymorphisms of the aromatase gene (CYP19A1), HER2/neu status, and prognosis in breast cancer patients. Breast Cancer Res Treat 112(1): 89–98 [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, Welch R, Lissowska J, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Bardin-Mikolajczak A, Struewing JP (2006) Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet 119(4): 376–388 [DOI] [PubMed] [Google Scholar]
- Giles GG, English DR (2002) The Melbourne Collaborative Cohort Study. IARC Sci Publ 156: 69–70 [PubMed] [Google Scholar]
- Hartikainen JM, Tuhkanen H, Kataja V, Dunning AM, Antoniou A, Smith P, Arffman A, Pirskanen M, Easton DF, Eskelinen M, Uusitupa M, Kosma VM, Mannermaa A (2005) An autosome-wide scan for linkage disequilibrium-based association in sporadic breast cancer cases in eastern Finland: three candidate regions found. Cancer Epidemiol Biomarkers Prev 14(1): 75–80 [PubMed] [Google Scholar]
- Haybittle JL, Blamey RW, Elston CW, Johnson J, Doyle PJ, Campbell FC, Nicholson RI, Griffiths K (1982) A prognostic index in primary breast cancer. Br J Cancer 45(3): 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu H, Bono P, Kataja V, Alanko T, Kokko R, Asola R, Utriainen T, Turpeenniemi-Hujanen T, Jyrkkio S, Moykkynen K, Helle L, Ingalsuo S, Pajunen M, Huusko M, Salminen T, Auvinen P, Leinonen H, Leinonen M, Isola J, Kellokumpu-Lehtinen PL (2009) Fluorouracil, epirubicin, and cyclophosphamide with either docetaxel or vinorelbine, with or without trastuzumab, as adjuvant treatments of breast cancer: final results of the FinHer Trial. J Clin Oncol 27(34): 5685–5692 [DOI] [PubMed] [Google Scholar]
- Joensuu H, Lehtimaki T, Holli K, Elomaa L, Turpeenniemi-Hujanen T, Kataja V, Anttila A, Lundin M, Isola J, Lundin J (2004) Risk for distant recurrence of breast cancer detected by mammography screening or other methods. JAMA 292(9): 1064–1073 [DOI] [PubMed] [Google Scholar]
- Kilpivaara O, Bartkova J, Eerola H, Syrjakoski K, Vahteristo P, Lukas J, Blomqvist C, Holli K, Heikkila P, Sauter G, Kallioniemi OP, Bartek J, Nevanlinna H (2005) Correlation of CHEK2 protein expression and c.1100delC mutation status with tumor characteristics among unselected breast cancer patients. Int J Cancer 113(4): 575–580 [DOI] [PubMed] [Google Scholar]
- MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MW, Pharoah PD, Ponder BA, Meuth M, Bhattacharyya NP, Cox A (2004) Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst 96(24): 1866–1869 [DOI] [PubMed] [Google Scholar]
- Mook S, Schmidt MK, Rutgers EJ, van de Velde AO, Visser O, Rutgers SM, Armstrong N, van't Veer LJ, Ravdin PM (2009) Calibration and discriminatory accuracy of prognosis calculation for breast cancer with the online Adjuvant! program: a hospital-based retrospective cohort study. Lancet Oncol 10(11): 1070–1076 [DOI] [PubMed] [Google Scholar]
- Mook S, Van ‘t Veer LJ, Rutgers EJ, Ravdin PM, van de Velde AO, van Leeuwen FE, Visser O, Schmidt MK (2011) Independent prognostic value of screen detection in invasive breast cancer. J Natl Cancer Inst 103(7): 585–597 [DOI] [PubMed] [Google Scholar]
- Olivotto IA, Bajdik CD, Ravdin PM, Speers CH, Coldman AJ, Norris BD, Davis GJ, Chia SK, Gelmon KA (2005) Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol 23(12): 2716–2725 [DOI] [PubMed] [Google Scholar]
- Olson JE, Ingle JN, Ma CX, Pelleymounter LL, Schaid DJ, Pankratz VS, Vierkant RA, Fredericksen ZS, Wu Y, Couch FJ, Vachon CM, Sellers TA, Weinshilboum RM (2007) A comprehensive examination of CYP19 variation and risk of breast cancer using two haplotype-tagging approaches. Breast Cancer Res Treat 102(2): 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EA, Reinholz MM, Hillman DW, Tenner KS, Schroeder MJ, Davidson NE, Martino S, Sledge GW, Harris LN, Gralow JR, Dueck AC, Ketterling RP, Ingle JN, Lingle WL, Kaufman PA, Visscher DW, Jenkins RB (2010) HER2 and chromosome 17 effect on patient outcome in the N9831 adjuvant trastuzumab trial. J Clin Oncol 28(28): 4307–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii S, O'Regan P, Xinarianos G, Azmy I, Stephenson T, Reed M, Meuth M, Thacker J, Cox A (2002) A potential role for the XRCC2 R188H polymorphic site in DNA-damage repair and breast cancer. Hum Mol Genet 11(12): 1433–1438 [DOI] [PubMed] [Google Scholar]
- Ragaz J, Jackson SM, Le N, Plenderleith IH, Spinelli JJ, Basco VE, Wilson KS, Knowling MA, Coppin CM, Paradis M, Coldman AJ, Olivotto IA (1997) Adjuvant radiotherapy and chemotherapy in node-positive premenopausal women with breast cancer. N Engl J Med 337(14): 956–962 [DOI] [PubMed] [Google Scholar]
- Ragaz J, Olivotto IA, Spinelli JJ, Phillips N, Jackson SM, Wilson KS, Knowling MA, Coppin CM, Weir L, Gelmon K, Le N, Durand R, Coldman AJ, Manji M (2005) Locoregional radiation therapy in patients with high-risk breast cancer receiving adjuvant chemotherapy: 20-year results of the British Columbia randomized trial. J Natl Cancer Inst 97(2): 116–126 [DOI] [PubMed] [Google Scholar]
- Rakha EA, El-Sayed ME, Green AR, Paish EC, Lee AH, Ellis IO (2007) Breast carcinoma with basal differentiation: a proposal for pathology definition based on basal cytokeratin expression. Histopathology 50(4): 434–438 [DOI] [PubMed] [Google Scholar]
- Ravdin PM, Siminoff LA, Davis GJ, Mercer MB, Hewlett J, Gerson N, Parker HL (2001) Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol 19(4): 980–991 [DOI] [PubMed] [Google Scholar]
- Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel VG, Visscher DW, Yothers G, Jenkins RB, Brown AM, Dakhil SR, Mamounas EP, Lingle WL, Klein PM, Ingle JN, Wolmark N (2005) Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 353(16): 1673–1684 [DOI] [PubMed] [Google Scholar]
- Schmidt MK, Tollenaar RA, de Kemp SR, Broeks A, Cornelisse CJ, Smit VT, Peterse JL, van Leeuwen FE, Van't Veer LJ (2007) Breast cancer survival and tumor characteristics in premenopausal women carrying the CHEK2*1100delC germline mutation. J Clin Oncol 25(1): 64–69 [DOI] [PubMed] [Google Scholar]
- Shen Y, Yang Y, Inoue LY, Munsell MF, Miller AB, Berry DA (2005) Role of detection method in predicting breast cancer survival: analysis of randomized screening trials. J Natl Cancer Inst 97(16): 1195–1203 [DOI] [PubMed] [Google Scholar]
- Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, Mackey J, Glaspy J, Chan A, Pawlicki M, Pinter T, Valero V, Liu MC, Sauter G, von Minckwitz G, Visco F, Bee V, Buyse M, Bendahmane B, Tabah-Fisch I, Lindsay MA, Riva A, Crown J (2011) Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med 365(14): 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I, Procter M, Gelber RD, Guillaume S, Feyereislova A, Dowsett M, Goldhirsch A, Untch M, Mariani G, Baselga J, Kaufmann M, Cameron D, Bell R, Bergh J, Coleman R, Wardley A, Harbeck N, Lopez RI, Mallmann P, Gelmon K, Wilcken N, Wist E, Sanchez Rovira P, Piccart-Gebhart MJ (2007) 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet 369(9555): 29–36 [DOI] [PubMed] [Google Scholar]
- Syrjakoski K, Vahteristo P, Eerola H, Tamminen A, Kivinummi K, Sarantaus L, Holli K, Blomqvist C, Kallioniemi OP, Kainu T, Nevanlinna H (2000) Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J Natl Cancer Inst 92(18): 1529–1531 [DOI] [PubMed] [Google Scholar]
- Tischkowitz M, Brunet JS, Begin LR, Huntsman DG, Cheang MC, Akslen LA, Nielsen TO, Foulkes WD (2007) Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer 7: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JH, Dowle C, Williams MR, Elston CW, Ellis IO, Hinton CP, Blamey RW, Haybittle JL (1987) Confirmation of a prognostic index in primary breast cancer. Br J Cancer 56(4): 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart GC, Azzato EM, Greenberg DC, Rashbass J, Kearins O, Lawrence G, Caldas C, Pharoah PD (2010) PREDICT: a new UK prognostic model that predicts survival following surgery for invasive breast cancer. Breast Cancer Res 12(1): R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart GC, Bajdik CD, Azzato EM, Dicks E, Greenberg DC, Rashbass J, Caldas C, Pharoah PD (2011) A population-based validation of the prognostic model PREDICT for early breast cancer. Eur J Surg Oncol 37(5): 411–417 [DOI] [PubMed] [Google Scholar]
- Wishart GC, Greenberg DC, Britton PD, Chou P, Brown CH, Purushotham AD, Duffy SW (2008) Screen-detected vs symptomatic breast cancer: is improved survival due to stage migration alone? Br J Cancer 98(11): 1741–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]