Abstract
It has been hypothesized that reproductive isolation should facilitate evolution under domestication. However, a systematic comparison of reproductive barrier strength between crops and their progenitors has not been conducted to test this hypothesis. Here, we present a systematic survey of reproductive barriers between 32 economically important crop species and their progenitors to better understand the role of reproductive isolation during the domestication process. We took a conservative approach, avoiding those types of reproductive isolation that are poorly known for these taxa (e.g., differences in flowering time). We show that the majority of crops surveyed are isolated from their progenitors by one or more reproductive barriers, despite the fact that the most important reproductive barrier in natural systems, geographical isolation, was absent, at least in the initial stages of domestication for most species. Thus, barriers to reproduction between crops and wild relatives are closely associated with domestication and may facilitate it, thereby raising the question whether reproductive isolation could be viewed as a long-overlooked “domestication trait.” Some of the reproductive barriers observed (e.g., polyploidy and uniparental reproduction), however, may have been favored for reasons other than, or in addition to, their effects on gene flow.
INTRODUCTION
The process of domestication was an inspiration to Charles Darwin when he developed the theory of evolution by natural selection (Darwin, 1859, 1868). Since then, studies of crop evolution have enhanced our understanding of evolutionary processes and speciation (Gepts, 2004; Ross-Ibarra et al., 2007). Surprisingly, speciation theory rarely has been employed for understanding the process of domestication. It has been recognized that reproductive isolation arising under domestication likely reduces gene flow between nascent crops and their nearby progenitors. Such barriers would have facilitated domestication via directional artificial selection imposed by protofarmers, as gene flow from wild relatives would not erode selective gains (Ladizinsky, 1985; Ellstrand, 2003).
There is widespread agreement that barriers to gene flow facilitate the accumulation of genetic differences between populations (Haldane, 1930; Bulmer, 1971; Felsenstein, 1976; Slatkin, 1985; Lenormand, 2002; Hendry and Taylor, 2004). Generally, strong barriers enable populations to diverge through either selection or drift, whereas weaker barriers typically permit divergence through selection, since even low levels of gene flow (effective number of migrants per generation ≥ 1) homogenize variation at neutral or nearly neutral loci (Hartl and Clark, 1997). Although strong divergent selection can allow population differentiation despite gene flow, such migration loads prevent populations from reaching their local optima and can generate weak indirect selection for traits that reduce gene flow between habitats (reviewed in Lenormand, 2002).
In plants, barriers that reduce gene flow can be divided into several categories (Rieseberg and Willis, 2007): (1) prepollination barriers: geographic, habitat, mechanical, and temporal isolation; (2) postpollination, prezygotic barriers: conspecific pollen precedence or gametic incompatibilities; (3) intrinsic postzygotic barriers: hybrid sterility, inviability, or breakdown; and (4) extrinsic postzygotic barriers: reductions in hybrid fitness due to the external environment. Prezygotic barriers are thought to make a greater contribution to speciation than postzygotic barriers, as they are often stronger in recently formed species and act before postzygotic barriers (Ramsey et al., 2003; Coyne and Orr, 2004; Lowry et al., 2008). Given that domesticated plants have arisen very recently, barriers between crops and their progenitors are expected to be mainly prezygotic, but as far as we are aware, this prediction has not previously been tested. Domestication via polyploidy may be an exception to this prediction, as whole-genome duplication results in substantial postzygotic isolation (Rieseberg and Willis, 2007) but may not necessarily impact prezygotic barriers.
Geographical isolation is reproductive isolation that arises because of limited contact among taxa due to geological and climatic processes that fragment populations, long-distance dispersal of founding populations, and/or the ecological range limits (ecogeographic isolation) of those taxa (Schemske, 2000; Lowry et al., 2008). Substantial geographic isolation is recognized as providing the most effective barrier to gene flow, and the vast majority of speciation events are believed to involve complete (allopatry) or partial (parapatry) geographic isolation (Coyne and Orr, 2004). Presumably, geographic isolation would facilitate domestication as well, but, to our knowledge, whether domestication generally occurs at the center, periphery, or well outside the geographic range of its progenitor has not been quantified.
Most discussions of reproductive isolation in natural populations focus on the means by which sexual, outcrossing species arise. The reproductive barriers discussed above reflect this bias. Additional barriers to gene flow found in natural populations involve certain reproductive modes that foster isolation and include high levels of self-fertilization (autogamy) as well as various forms of asexual reproduction (Levin, 1978). Like other kinds of reproductive barriers, autogamy and asexual reproduction offer a means for preserving adaptive gene combinations, but they differ from other barriers in that descendent lineages are as strongly isolated from each other as they are from their ancestors. Some authors have questioned whether autogamy and asexual reproduction should be viewed as reproductive barriers at all (Coyne and Orr, 2004), although other researchers disagree with this view, at least with respect to autogamy (Grant, 1981; Blackman and Rieseberg, 2004). However, both mechanisms offer a straightforward means for preserving desired genotypes and thus may have been intentionally or unintentionally exploited by early farmers, playing an important role in the domestication of some crops.
We are not aware of any systematic survey of reproductive barrier strength between crops and their progenitors. Here, we conduct such a survey of 32 economically important crop species (Table 1), exploring the hypothesis that reproductive isolation facilitates the process of domestication. We ask the following questions. (1) How frequently is reproductive isolation associated with plant domestication? (2) Does domestication, like speciation, typically occur in parapatry or allopatry with progenitor populations? (3) Are intrinsic postzygotic barriers rare relative to geographic isolation or other prezygotic barriers? (4) Are ploidy shifts more frequent during domestication than predicted by polyploid speciation rates in natural plant populations? (5) Does a transition toward higher rates of selfing or asexuality occur during domestication, or does the mating system influence the propensity for a species to be domesticated (Rick, 1988)? Our survey’s results not only shed light on the role of reproductive isolation in plant domestication but also are of practical relevance for the use of wild germplasm in crop improvement (Tanksley and McCouch, 1997) and for predicting the likelihood that engineered genes may “escape” from cultivated fields through crop–wild hybridization (Ellstrand, 2001; Snow et al., 2003). We show that the evolution of barriers to gene flow has accompanied the domestication of many crops, suggesting that reproductive isolation plays an important, if not crucial, role in the domestication histories for many crops (Hancock, 2004).
Table 1. Crop Species and Proposed Progenitors.
| Common Name | Family | Crop Species | Proposed Progenitor |
|---|---|---|---|
| 1. Banana | Musaceae | Musa acuminata (AAA Group) cv Dwarf Cavendish | Several Musa acuminata subspecies |
| 2. Barley | Poaceae | Hordeum vulgare | Hordeum vulgare subsp spontaneum (synonym of Hordeum spontaneum) |
| 3. Cassava | Euphrobiacea | Manihot esculenta | Manihot esculenta subsp flabellifolia (synonym of Manihot esculenta) |
| 4. Chickpea | Leguminosae | Cicer arietinum | Cicer reticulatum |
| 5. Cocoa | Malvaceae | Theobroma cacao | Theobroma cacao |
| 6. Coconut | Arecaceae | Cocos nucifera | Cocos nucifera |
| 7. Coffee (Arabica) | Rubiaceae | Coffea arabica | Coffea arabica |
| 8. Coffee (Robusta) | Rubiaceae | Coffea canephora | Coffea canephora |
| 9. Common bean | Leguminosae | Phaseolus vulgaris | Phaseolus vulgaris |
| 10. Upland cotton | Malvaceae | Gossypium hirsutum | Gossypium hirsutum |
| 11. Cowpea | Leguminosae | Vigna unguiculata subsp unguiculata | Vigna unguiculata subsp unguiculata var spontanea |
| 12. Grape | Vitaceae | Vitis vinifera | Vitis vinifera subsp sylvestris (synonym of Vitis vinifera) |
| 13. Maize | Poaceae | Zea mays | Zea mays subsp parviglumis (synonym of Zea mays) |
| 14. Finger millet | Poaceae | Eleusine coracana | Eleusine coracana subsp africana (synonym of Eleusine coracana) |
| 15. Pearl millet | Poaceae | Pennisetum glaucum | Pennisetum americanum subsp monodii (synonym of Pennisetum violaceum) |
| 16. Oat | Poaceae | Avena sativa | Avena sterilis |
| 17. Olive | Oleaceae | Olea europaea (also Olea europaea subsp europaea) | Olea europaea subsp oleaster (synonym of Olea europaea subsp europaea) |
| 18. Oil palm (African) | Arecaceae | Elaeis guineensis | Elaeis guineensis |
| 19. Pea | Leguminosae | Pisum sativum | Pisum sativum subsp humile (synonym of Pisum sativum subsp elatius) |
| 20. Peanut | Leguminosae | Arachis hypogaea | Arachis monticola |
| 21. Rapeseed | Brassicaceae | Brassica napus | Brassica rapa and Brassica oleracea |
| 22. Rice | Poaceae | Oryza sativa | Oryza rufipogon |
| 23. Rye | Poaceae | Secale cereale | Secale cereale |
| 24. Rubber tree | Euphorbiaceae | Hevea brasiliensis | Hevea brasiliensis |
| 25. Sesame | Pedaliaceae | Sesamum indicum | Sesamum indicum var malabaricum |
| 26. Sorghum | Poaceae | Sorghum bicolor | Sorghum bicolor subsp verticilliflorum (synonym of Sorghum arundinaceum) |
| 27. Soybean | Leguminosae | Glycine max | Glycine soja (synonym of Glycine max subsp soja) |
| 28. Sugarcane | Poaceae | Saccharum officinarum | Saccharum robustum |
| 29. Sunflower | Asteraceae | Helianthus annuus | Helianthus annuus |
| 30. Sweet potato | Convolvulaceae | Ipomoea batatas | Ipomoea trifida |
| 31. Common wheat | Poaceae | Triticum aestivum | Triticum turgidum and Aegilops tauschii |
| 32. Durum wheat | Poaceae | Triticum turgidum | Triticum turgidum subsp dicoccoides (synonym of Triticum dicoccoides) |
LITERATURE SURVEY
We started with the 30 economically most important “crop commodity items,” as measured in terms of area under cultivation and recorded in the FAOStat database of the Food and Agricultural Organization of the United Nations (http://faostat.fao.org/). This sample was chosen because a cursory review of a wider set of literature revealed that for most of the “economically less important crops,” insufficient information exists on the progenitor. Furthermore, our focus is on the more strongly domesticated crops, which we feel are represented well on this list. Only the major species that contribute to a crop’s total acreage are listed, so, for example, in the case of “plantain and banana,” only Musa acuminata (AAA Group) cv Dwarf Cavendish is listed. The crop species included in this analysis are diverse with respect to phylogeny and life history, and most have sufficient information available in terms of their domestication history and the identity of the progenitor for inclusion in our survey. Potato (Solanum tuberosum), for which there is much uncertainty regarding the identity of the progenitor, was removed from our list because of insufficient information (Spooner et al., 2005).
For banana, the precise identity of the progenitor is uncertain, and it is thought that several subspecies of M. acuminata have been involved in its domestication (Perrier et al., 2011). Therefore, we included a reference to “several subspecies of Musa acuminata” as the progenitor of banana in our analysis but treated it as a single crop–progenitor comparison. In the case of rapeseed, the two putative progenitors thought to have contributed to the origin of the crop, Brassica rapa and Brassica oleracea, were both considered (Smartt and Simmonds, 1995). “Millets” are listed as a single crop commodity item in the FAOStat database, but the two major millets are clearly distinct crop species, finger millet (Eleusine coracana) and pearl millet (Pennisetum glaucum); therefore, we treat them as separate crop species (http://faostat.fao.org/). The same holds true for wheat, where we treated durum wheat (Triticum turgidum) and common wheat (Triticum aestivum) as separate crop species as well, which results in a total of 32 crop species being considered here (Table 1). Where there is strong evidence of hybridization in the domestication history of the crop, such as bread wheat, information on all proposed progenitors is listed. Therefore, although we have included information on 32 economically important crop species, we are considering here a total of 34 crop–progenitor pair comparisons. The taxonomy of each crop and its proposed progenitor was verified using the standard reference The Plant List (http://www.theplantlist.org/). We recognize that some crop evolutionists and taxonomists might disagree with the taxonomy provided by these references.
CHARACTERIZATION OF REPRODUCTIVE BARRIERS
Several different approaches can be taken to assess reproductive barrier strength. A popular method is to identify and quantity the individual barriers that exist between a given pair of taxa and then combine them to estimate total isolation strength (Ramsey et al., 2003; Lowry et al., 2008). However, the necessary information for such an approach does not exist for the species pairs considered here. A second approach is to use molecular markers to examine ongoing gene flow or to estimate gene flow indirectly from overall genetic divergence (Klinger et al., 1992; Arias and Rieseberg, 1994; Morjan and Rieseberg, 2004). This approach suffers because it is difficult to estimate range-wide realized gene flow from experimental studies, and demographic effects likely invalidate indirect estimates of gene flow between crops and their wild relatives.
Given these difficulties, we have not attempted to assess the identity and strength of the entire suite of reproductive barriers for each crop. Instead, we focused our efforts on what we suspected were the important reproductive barriers, especially those for which information could be found in the literature. These include (1) geographical isolation; (2) mating system isolation, especially transitions from outcrossing to selfing; (3) isolation through asexual propagation; (4) ploidy changes, because whole-genome duplications typically generate substantial reproductive isolation (Coyne and Orr, 2004; Linder and Rieseberg, 2004; Mallet, 2007); and (5) the fitness of hybrids between the crop and its proposed progenitor. Estimates of outcrossing rates using genetic markers for both the progenitor and crop were limited, and we relied on reports in the literature by experts in the field if no quantitative mating system estimates were available. If mating system data were available, we considered a taxon outcrossing if its mean outcrossing rate estimate was >80% and selfing if it was <20%. Also, a taxon was classified as outcrossing if it had a sexual system that enforced outcrossing. Similarly, as comparable quantitative estimates of hybrid fitness were lacking for the majority of the crops and their wild progenitors considered here, we classified hybrid fitness according to the following four categories: (1) generally no reduction in hybrid fitness; (2) reduced fitness in post-F1 generations (wherever such data were available); (3) reduction of fitness in some F1 plants; (4) no or few fertile hybrids formed. Furthermore, we also recorded information on the predominant mode of propagation for each crop.
In cases where hybridization between two or more species is thought to have played a key role during domestication, we considered each case separately for the purpose of our assessment of taxonomic differences and hybrid fitness. However, in our comparisons of transition in mating systems and the assessment of breakdown of self-incompatibility systems and of differences in ploidy level, data on all progenitors were treated as a single case for each crop in order to avoid pseudoreplication. This was the case for common wheat and rapeseed in our data set.
REPRODUCTIVE BARRIER STRENGTH IN DOMESTICATED SPECIES
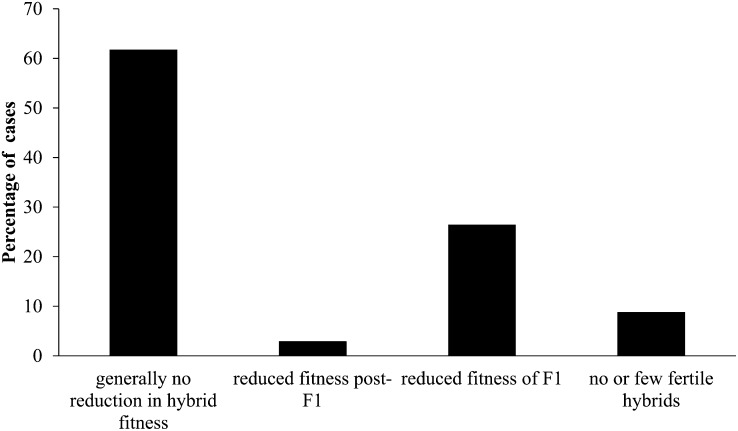
Our literature survey revealed that three-fourths of the world’s most important crops are isolated from their wild progenitors by a minimum of one reproductive barrier, at least if these barriers are defined broadly to include autogamy and asexual reproduction (see Supplemental Appendix 1 online). If we restrict our consideration to traditional barriers, such as ploidy differences and reduced hybrid fitness, then more than one-third of the cases considered here (38%) show evidence of reproductive isolation (Figure 1; see Supplemental Appendix 1 online). These estimates are conservative, because some reproductive barriers were not included (e.g., flowering time, phenology, conspecific pollen precedence, and gametic incompatibilities). In addition, components of isolation that operate mainly in agricultural environments rather than the greenhouse are likely to have been missed. We conclude that reproductive isolation frequently exists between crops and their progenitors, despite the short time span of domestication (<12,000 years) (Hancock, 2004). Nearly complete barriers to gene flow are observed in ∼10% of cases: banana, common wheat (progenitor: Aegilops tauschii), and rapeseed (progenitor: B. oleracea), all of which involve ploidy barriers.
Figure 1.
Hybrid Fitness of Crop–Progenitor Crosses.
The percentage of all cases considered is shown.
It is also possible that certain isolating barriers between crops and their proposed progenitors arise in wild populations well after the beginning of the domestication process, once the crop is common and the wild relative has become rare. Under such a scenario, certain crop alleles may become detrimental in wild populations and the selection pressure for reproductive isolation in the wild may increase (Ladizinsky, 1985), but there is as yet no empirical evidence for this. Furthermore, the association between domestication and reproductive isolation does not necessarily mean that reproductive isolation was a cause of domestication or even that the ease with which reproductive isolation arose affected the domestication process. Some of the reproductive barriers observed likely were favored for reasons in addition to or other than their effects on gene flow. For example, selfing and asexual reproduction may have been selected by early farmers because of reproductive assurance (Rick, 1988; Allard, 1999; Gepts, 2004). Likewise, changes in ploidy might have been favored because of effects on development or on fruit and seed size (Smartt and Simmonds, 1995; Villar and Veneklaas, 1998; Well and Fossey, 1998; Otto and Whitton, 2000).
We also found differences in how reproductive isolation has evolved in domesticated plants versus wild species. For example, unlike the origin of wild plant species, geographical isolation was not associated with the origin of most of the domesticated plants studied here (see Supplemental Appendix 1 online; Stebbins, 1950; Rieseberg and Willis, 2007). In some instances, the presumed geographic location of domestication was deduced partly from the current distribution of the progenitor (e.g., upland cotton [Gossypium hirsutum] in Yucatan [Brubaker and Wendel, 1994]) rather than exclusively from archaeological records or the center of crop diversity. With this caveat in mind, almost all examples of domestication are thought to have occurred in sympatry (located in the same geographic area) with the wild progenitor species. The only exception in our data appears to be sunflower (Helianthus annuus) domestication, which occurred on the periphery of the range of the common sunflower, so it is best classified as parapatric.
Thus, it appears that sympatry was not a major impediment to domestication. This could be because (1) other barriers to gene flow arose quickly, such as polyploidy, thereby mitigating the effects of sympatry (Rieseberg and Willis, 2007); (2) microgeographic variation in habitat provided local geographic isolation; or (3) both conscious and unconscious selection were strong enough to overcome the homogenizing effects of gene flow between the incipient crop and its wild progenitor (Papa and Gepts, 2003; Papa et al., 2005, 2007). The high level of sympatry compared with speciation in the wild suggests that reproductive barriers other than geographical isolation are likely of more importance, particularly during the initial stages of domestication.
There are a number of crops for which some researchers have suggested that certain domestication traits might have evolved after the incipient crop was transferred to areas where the wild relatives were absent. Such a pattern, if widespread, would support the importance of reproductive isolation in domestication. One example is finger millet, which has been suggested to have been domesticated initially in the East African Highlands, yet the earliest known remains date to 2000 BC in India, and it only shows up later in the paleorecord of East Africa (Fuller, 2006). Similar patterns have also been suggested for pearl millet and sorghum (Sorghum bicolor) domestication (Haaland, 1995, 1999; Fuller, 2007), although this remains controversial. However, there are too few such crops in our list to allow quantification of this potential “allopatry effect.”
Another surprise was the high frequency of intrinsic postzygotic isolation (38% of cases assayed) given the fairly short time scale of domestication (typically <12,000 years [Hancock, 2004]) and the lack of geographic isolation. Specifically, 21 crop–wild relative pairs out of the 34 pairs displayed little or no reduction in hybrid fitness (category 1, 61%; Figure 1); one pair, barley (Hordeum vulgare), exhibited reduced fertility in later generation hybrids (category 2, 3%); nine pairs showed evidence of reduced F1 fitness (category 3, 26%); and three pairs have been shown to produce few or no fertile hybrids (category 4, 9%). Only 50 (13%) of 397 wild plant species surveyed by Rieseberg et al. (2006) showed significant intraspecific variation for intrinsic postzygotic reproductive barriers. This is a conservative estimate, because the wild population crosses typically were assayed for a smaller number of reproductive barriers than the crop–wild hybrids. Also, the crop–wild complexes are enriched for selfers, which tend to exhibit higher levels of intraspecific reproductive isolation than outcrossers (Grundt et al., 2006). Nonetheless, it appears that highly domesticated crops are more strongly isolated from their progenitors than is expected for a typical intraspecific cross in plants.
We are not sure why postzygotic barriers have arisen so quickly’ in some crops. Presumably, a substantial fraction of these barriers result from the sorting of hybrid incompatibilities that already existed in progenitor populations. A recent review of the attributes of genes underlying reproductive isolation in plants, many of which were characterized in crop plants, revealed strong roles for diversifying selection and genetic drift in the evolution of intrinsic hybrid incompatibilities (Rieseberg and Blackman, 2010). Both forces are likely to be strong in crop evolution, perhaps contributing to the patterns observed here. Also, all six cases where the history of domestication includes polyploidy show varying degrees of postzygotic isolation. This is not surprising given that interploidy hybrids often exhibit difficulties in chromosome pairing during meiosis, which pose a significant challenge to successful reproduction (Rieseberg and Willis, 2007).
In our survey, four cases exhibited ploidy differences between the crop and the wild progenitor: rapeseed, sweet potato (Ipomoea batatas), banana, and common wheat (see Supplemental Appendix 1 online). The case of sugarcane (Saccharum officinarum) is unclear, since lineages with a wide range of chromosome numbers exist in both the putative progenitor and the crop. The frequency of ploidy changes in our data (13%) is similar to the estimates of polyploid speciation in angiosperms (∼15%) (Wood et al., 2009), and in all cases it is associated with uniparental reproduction in the crop (selfing or asexual reproduction), a feature that could aid in overcoming the minority cytotype disadvantage experienced by neopolyploids (Otto and Whitton, 2000).
Interestingly, reproductive barrier strength between crops and their wild relatives does not correlate closely with taxonomy. Based on the recently published Plant List (http://www.theplantlist.org/), we found that 14 crop–progenitor pairs are differentiated at the species level, three pairs are differentiated at the subspecies level, two at the variety level, and 16 pairs are recognized as the same taxon. Of the 14 cases in which taxonomists consider the crop and the progenitor to belong to different taxa, in seven instances (oat [Avena sativa], chickpea [Cicer arietinum], pearl millet, sorghum, durum wheat, sugarcane, and cowpea [Vigna unguiculata]) it appears from the literature that the crop and the progenitor are fully compatible reproductively. Overdifferentiation by taxonomists has been found for wild plant species (Rieseberg et al., 2006), and the same appears to be true for the designation of species formed during domestication.
UNIPARENTAL REPRODUCTION AND DOMESTICATION
The evolution of uniparental reproduction is one factor that can contribute to reproductive isolation, as it allows individuals to produce fertile offspring without the incorporation of genes from a different lineage. Selfing as well as asexual reproduction can be particularly advantageous in crops, because it enables farmers to more easily select combinations of traits, because a selfing plant will “breed true to type” (Rick, 1988; Zohary and Hopf, 2000; Gepts, 2004; Glémin and Bataillon, 2009). In addition to their potential role in preserving desirable combinations of traits and genes (Baker, 1959; Dickinson and Antonovics, 1973; Epinat and Lenormand, 2009), uniparental reproduction may facilitate domestication through reproductive assurance (Darwin, 1876; Baker, 1955; Kalisz et al., 2004), allowing populations to survive demographic bottlenecks (mate limitation) and pollinator limitation (reviewed in Holsinger, 1996). Avoidance of pollinator limitation might have been especially critical to domestication, because it would allow farmers to successfully grow the species in a greater number of environments. Consequently, the advantages of these breeding systems suggest that they should evolve frequently during domestication.
On the other hand, selfing can be associated with significant short-term fitness costs through inbreeding depression (Darwin, 1876; Charlesworth and Charlesworth, 1987), and both selfing and asexual breeding systems can slow adaptation (in the long term) and inhibit the purging of deleterious recessive alleles (Williams, 1975; Kondrashov, 1984; Schoen and Brown, 1991; Schultz and Lynch, 1997; Agrawal, 2006; Glémin et al., 2006; Morran et al., 2009). Given the brief time scale of domestication, as well as reduced competition within agricultural fields, which could ameliorate some of the short-term costs of selfing (Armbruster and Reed, 2005), it might be that the benefits of these breeding systems outweigh their long-term liabilities, particularly when combined with occasional episodes of outcrossing.
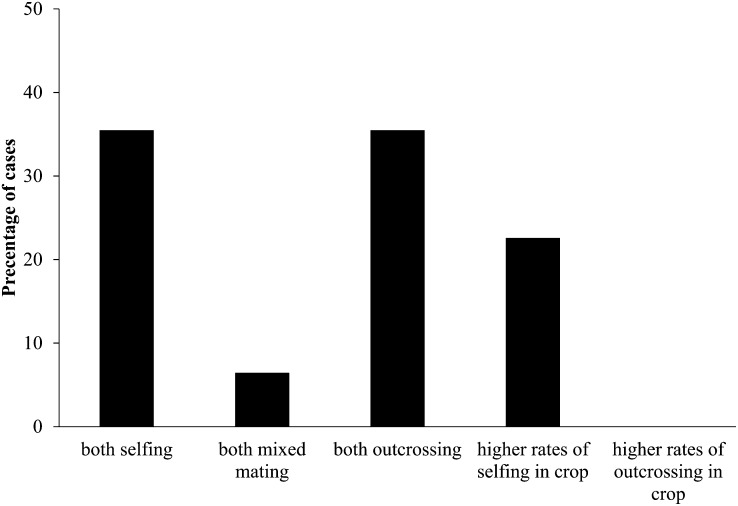
Data on mating system transitions were used to place each of the crop–wild relative pairs into one of five categories; the case of sesame (Sesamum indicum) was excluded because of insufficient information on the mating system of the progenitor. In 11 out of 31 cases (35%), the crop and progenitor are predominantly selfing (category 1); coconut (Cocos nucifera) as well as cotton and its wild progenitor have mixed mating systems with no evidence for differences in the selfing rates (category 2, 6%); both the crop and its progenitor are predominantly outcrossing in 11 more cases (category 3, 35%); seven pairs show higher rates of selfing in the crop (category 4, 23%); whereas there are no pairs that show higher rates of outcrossing in the crop (category 5) (Figure 2). Therefore, of the crop–progenitor pairs analyzed for transitions in mating systems during domestication, almost one-quarter (Figure 2) show higher rates of selfing in the crop, but none show higher rates of selfing in the progenitor. Empirical studies have shown that transitions in mating systems from selfing to outcrossing rarely, if ever, occur in nature, whereas transitions from outcrossing to selfing are more frequent (Stebbins, 1957; Rick, 1988; Takebayashi and Morrell, 2001, Goldberg et al., 2010), a pattern also repeated in our data.
Figure 2.
Transition in Mating System during Domestication.
The percentage of all cases considered is shown.
The majority of crop–progenitor pairs do not appear to show a transition in mating systems. It is worth noting that the majority of outcrossing pairs, with the exception of maize (Zea mays), rye (Secale cereale), and pearl millet, are either perennial crops, such as oil palm (Elaeis guineensis), rubber tree (Hevea brasiliensis), olive (Olea europaea), sugarcane, banana, and robusta coffee (Coffea canephora), and/or are propagated predominantly asexually through vegetative means, such as sweet potato, olive, cassava (Manihot esculenta), banana, and rubber tree. The lack of mating system transitions in the latter is not surprising, given that the sexual mode of reproduction is much less relevant for such species in cultivated environments. One could argue that these species have undergone a transition to asexuality via cultural coevolution with humans rather than through intrinsic mechanisms. Likewise, perennial species are thought to become domesticated at a much slower pace (and to exhibit domestication syndromes to a much lower degree) than annuals because of longer generation times.
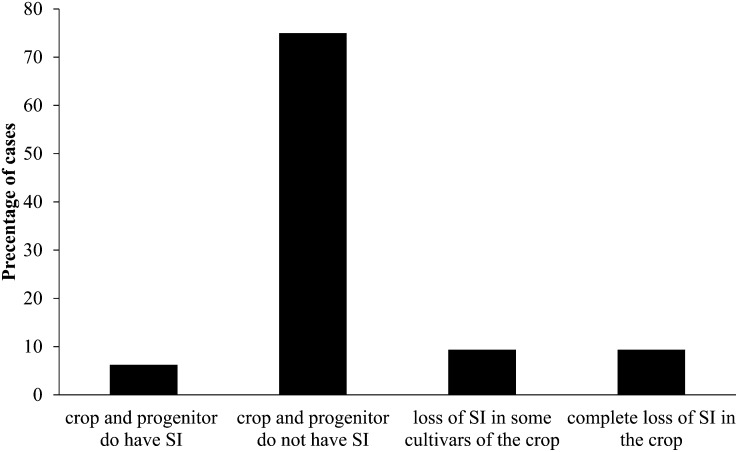
The transition from outcrossing to selfing can be facilitated by the breakdown of sexual systems that enforce outcrossing, such as self-incompatibility systems or dioecy (Igic and Kohn, 2006). A transition from separate sexes to combined sexes appears to be rare and is evident in our data set only in grape (Vitis vinifera) (Di Vecchi-Staraz et al., 2009). For cosexual taxa, most progenitors and crops (75%) do not exhibit a self-incompatibility system, and only in two cases, rye and robusta coffee, are the crops and the progenitors both self-incompatible (Figure 3). In three cases, rapeseed, sugarcane, and sunflower, there is a complete loss of the self-incompatibility system during domestication, and a partial breakdown of the self-incompatibility systems in some cultivars seems to have occurred in three more cases (cocoa [Theobroma cacao], olive, and sweet potato).
Figure 3.
Breakdown of Self-Incompatibility (SI) during Domestication.
The percentage of all cases considered is shown.
Estimates place the percentage of self-fertilizing angiosperms at ∼20%, while the percentage of obligate outcrossers (i.e., dioecious or self-incompatibility systems) is ∼50% (Vogler and Kalisz, 2001; Igic and Kohn, 2006). Therefore, the higher frequency of selfing progenitors (35% of all cases considered here) and the lower frequency of obligate outcrossers (35% of all cases considered here) could indicate that the ability to self-fertilize represents a preadaptation to domestication. However, as Barrett and Harder (1996), Barrett (1998), and Morgan et al. (1997) show, annual species have considerably higher rates of selfing than woody perennials, which are predominantly outcrossing. Interestingly, annuals tend to have high reproductive output (Primack, 1979; Wilson and Thompson, 1989; Karlsson and Méndez, 2005), an attractive feature for a farmer, and a short generation time, which would speed domestication. This is not a benefit of selfing per se, but it could partially explain the high incidence of selfers that are domesticated. In order to investigate whether selfing species are preadapted to become domesticated, larger sample sizes than included here will be required to disentangle information on mating systems from other correlated factors, such as an annual habit.
CONCLUSIONS
In summary, our review of 29 of the most economically important crops and their progenitors has shown that domestication is frequently associated with the existence or evolution of prezygotic and/or postzygotic reproductive barriers, despite the fact that the most important reproductive barrier in natural systems, geography, was absent, at least in the initial stages of domestication for most species. Strong barriers to reproduction between crops and wild relatives could increase the rate of domestication and/or facilitate the maintenance of gene combinations favored by early human farmers. However, our hypothesis that reproductive isolation may have facilitated domestication will require additional evidence. This could include, for example, comparison with levels of reproductive isolation in semidomesticated crops, quantification of the allopatry effect, if it exists, as well as selection studies that attempt to redomesticate crops under varying levels of gene flow. In addition, there is strong evidence that higher rates of selfing are frequently associated with domestication and that a breakdown of the self-incompatibility system is common. Whether selfing species are preadapted to becoming domesticated remains an open question.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Appendix 1. Crop Species and Proposed Progenitor Pairs Analyzed.
Supplementary Material
Acknowledgments
We acknowledge Sarah Otto, who provided many useful comments and suggestions for revisions during the preparation of the article. We are also thankful for helpful advice from Jeffrey Ross-Ibarra, Catherine de D’Andrea, Crispin Jordan, Paul Gepts, and Quentin Cronk. N.C.E. thanks Carl Schlichting for calling this important evolutionary issue to his attention well over a decade ago. Research on plant domestication in the Rieseberg laboratory has been supported by Genome Canada, the U.S. National Science Foundation (Grant DBI 0820451), and the Canadian International Development Agency. Additional support came from the John Simon Guggenheim Memorial Foundation and the U.S. National Science Foundation (Grant DEB 1020799 to N.C.E.).
AUTHOR CONTRIBUTIONS
H.D., L.H.R., and K.A.H. conceived the study. S.E.R. and H.D. did the literature survey. H.D., K.A.H., L.H.R., and N.C.E. wrote the article.
References
- Agrawal A.F. (2006). Evolution of sex: Why do organisms shuffle their genotypes? Curr. Biol. 16: R696–R704 [DOI] [PubMed] [Google Scholar]
- Allard R.W. (1999). History of plant population genetics. Annu. Rev. Genet. 33: 1–27 [DOI] [PubMed] [Google Scholar]
- Arias D.M., Rieseberg L.H. (1994). Gene flow between cultivated and wild sunflowers. Theor. Appl. Genet. 89: 655–660 [DOI] [PubMed] [Google Scholar]
- Armbruster P., Reed D.H. (2005). Inbreeding depression in benign and stressful environments. Heredity (Edinb) 95: 235–242 [DOI] [PubMed] [Google Scholar]
- Baker H. (1959). Reproductive methods as factors in speciation in flowering plants. In Cold Spring Harbor Symposia on Quantitative Biology. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), p. 177. [DOI] [PubMed]
- Baker H.G. (1955). Self-compatibility and establishment after ‘long-distance’ dispersal. Evolution 9: 347–349 [Google Scholar]
- Barrett S.C.H. (1998). The evolution of mating strategies in flowering plants. Science 3: 335–341 [Google Scholar]
- Barrett S.C.H., Harder L.D. (1996). Ecology and evolution of plant mating. Trends Ecol. Evol. (Amst.) 11: 73–79 [DOI] [PubMed] [Google Scholar]
- Blackman B.K., Rieseberg L.H. (2004). How species arise? Science 305: 612–613 [Google Scholar]
- Brubaker C.L., Wendel J.F. (1994). Reevaluating the origin of domesticated cotton (Gossypium hirsutum; Malvaceae) using nuclear restriction fragment length polymorphisms (RFLPs). Am. J. Bot. 81: 1309–1326 [Google Scholar]
- Bulmer M. (1971). The effect of selection on genetic variability. Am. Nat. 105: 201–211 [Google Scholar]
- Charlesworth D., Charlesworth B. (1987). Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 18: 237–268 [Google Scholar]
- Coyne J.A., Orr H.A. (2004). Speciation. (Sunderland, MA: Sinauer Associates)
- Darwin C. (1859). On the Origin of Species by Means of Natural Selection. (London: J. Murray)
- Darwin C. (1868). The Variation of Plants and Animals under Domestication. (London: J. Murray)
- Darwin C. (1876). The Effects of Cross and Self Fertilization in the Vegetable Kingdom. (London: J. Murray)
- Dickinson H., Antonovics J. (1973). Theoretical considerations of sympatric divergence. Am. Nat. 107: 256–274 [Google Scholar]
- Di Vecchi-Staraz M., Laucou V., Bruno G., Lacombe T., Gerber S., Bourse T., Boselli M., This P. (2009). Low level of pollen-mediated gene flow from cultivated to wild grapevine: Consequences for the evolution of the endangered subspecies Vitis vinifera L. subsp. silvestris. J. Hered. 100: 66–75 [DOI] [PubMed] [Google Scholar]
- Ellstrand N.C. (2001). When transgenes wander, should we worry? Plant Physiol. 125: 1543–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand N.C. (2003). Dangerous Liaisons? When Cultivated Plants Mate with Their Wild Relatives (Baltimore, MD: Johns Hopkins University Press) [Google Scholar]
- Epinat G., Lenormand T. (2009). The evolution of assortative mating and selfing with in- and outbreeding depression. Evolution 63: 2047–2060 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1976). The theoretical population genetics of variable selection and migration. Annu. Rev. Genet. 10: 253–280 [DOI] [PubMed] [Google Scholar]
- Fuller D.Q. (2006). Agricultural origins and frontiers in South Asia: A working synthesis. J. World Prehist. 20: 1–86 [Google Scholar]
- Fuller D.Q. (2007). Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann. Bot. (Lond.) 100: 903–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P. (2004). Crop domestication as a long-term selection experiment. Plant Breed. Rev. 24: 1–44 [Google Scholar]
- Glémin S., Bataillon T. (2009). A comparative view of the evolution of grasses under domestication. New Phytol. 183: 273–290 [DOI] [PubMed] [Google Scholar]
- Glémin S., Bazin E., Charlesworth D. (2006). Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc. Biol. Sci. 273: 3011–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg E.E., Kohn J.R., Lande R., Robertson K.A., Smith S.A., Igić B. (2010). Species selection maintains self-incompatibility. Science 330: 493–495 [DOI] [PubMed] [Google Scholar]
- Grant V. (1981). Plant Speciation. (New York: Columbia University Press)
- Grundt H.H., Kjølner S., Borgen L., Rieseberg L.H., Brochmann C. (2006). High biological species diversity in the arctic flora. Proc. Natl. Acad. Sci. USA 103: 972–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaland R. (1995). Sedentism, cultivation, and plant domestication in the Holocene Middle Nile region. J. Field Archaeol. 22: 157–174 [Google Scholar]
- Haaland R. (1999). The puzzle of the late emergence of domesticated sorghum in the Nile valley. In The Prehistory of Food: Appetites for Change, C. Gosden and J. Hather, eds (London and New York: Routledge), pp. 397–418.
- Haldane J. (1930). A mathematical theory of natural and artificial selection. Part IV. Isolation. Transactions of the Cambridge Philosophical Society 26: 220–230 [Google Scholar]
- Hancock J.F. (2004). Plant Evolution and the Origin of Crop Species, 2nd ed. (Wallingford, UK: CABI Publishing)
- Hartl D.L. Clark A.G. (1997). Principles of Population Genetics, 4th ed. (Sunderland, MA: Sinauer Associates)
- Hendry A.P., Taylor E.B. (2004). How much of the variation in adaptive divergence can be explained by gene flow? An evaluation using lake-stream stickleback pairs. Evolution 58: 2319–2331 [DOI] [PubMed] [Google Scholar]
- Holsinger K.E. (1996). Pollination biology and the evolution of mating systems in flowering plants. Evol. Biol. 29: 107–149 [Google Scholar]
- Igic B., Kohn J.R. (2006). The distribution of plant mating systems: Study bias against obligately outcrossing species. Evolution 60: 1098–1103 [PubMed] [Google Scholar]
- Kalisz S., Vogler D.W., Hanley K.M. (2004). Context-dependent autonomous self-fertilization yields reproductive assurance and mixed mating. Nature 430: 884–887 [DOI] [PubMed] [Google Scholar]
- Karlsson P.S., Méndez M. (2005). The resource economy of plant reproduction. In Reproductive Allocation in Plants, E.G. Reekie and F.A. Bazzaz, eds (Burlington, UK: Elsevier Academic Press), pp. 1–49.
- Klinger T., et al. (1992). Crop-weed hybridization in radish (Raphanus sativus): Effects of distance and population size. Am. J. Bot. 79: 1431–1435 [Google Scholar]
- Kondrashov A.S. (1984). Deleterious mutations as an evolutionary factor. 1. The advantage of recombination. Genet. Res. 44: 199–217 [DOI] [PubMed] [Google Scholar]
- Ladizinsky G. (1985). Founder effect in crop-plant evolution. Econ. Bot. 39: 191–199 [Google Scholar]
- Lenormand T. (2002). Gene flow and the limits to natural selection. Trends Ecol. Evol. 17: 183–189 [Google Scholar]
- Levin D.A. (1978). The origin of isolation mechanisms in plants. Evol. Biol. 11: 185–317 [Google Scholar]
- Linder C.R., Rieseberg L.H. (2004). Reconstructing patterns of reticulate evolution in plants. Am. J. Bot. 91: 1700–1708 [PMC free article] [PubMed] [Google Scholar]
- Lowry D.B., Modliszewski J.L., Wright K.M., Wu C.A., Willis J.H. (2008). The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363: 3009–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. (2007). Hybrid speciation. Nature 446: 279–283 [DOI] [PubMed] [Google Scholar]
- Morgan M.T., Schoen D.J., Bataillon T.M. (1997). The evolution of self-fertilization in perennials. Am. Nat. 150: 618–638 [DOI] [PubMed] [Google Scholar]
- Morjan C.L., Rieseberg L.H. (2004). How species evolve collectively: Implications of gene flow and selection for the spread of advantageous alleles. Mol. Ecol. 13: 1341–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L.T., Parmenter M.D., Phillips P.C. (2009). Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature 462: 350–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.P., Whitton J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34: 401–437 [DOI] [PubMed] [Google Scholar]
- Papa R., Acosta J., Delgado-Salinas A., Gepts P. (2005). A genome-wide analysis of differentiation between wild and domesticated Phaseolus vulgaris from Mesoamerica. Theor. Appl. Genet. 111: 1147–1158 [DOI] [PubMed] [Google Scholar]
- Papa R., Bellucci E., Rossi M., Leonardi S., Rau D., Gepts P., Nanni L., Attene G. (2007). Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples. Ann. Bot. (Lond.) 100: 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa R., Gepts P. (2003). Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theor. Appl. Genet. 106: 239–250 [DOI] [PubMed] [Google Scholar]
- Perrier X., et al. (2011). Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA 108: 11311–11318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primack R.B. (1979). Reproductive effort in annual and perennial species of Plantago (Plantaginaceae). Am. Nat. 114: 51–62 [Google Scholar]
- Ramsey J., Bradshaw H.D., Jr., Schemske D.W. (2003). Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution 57: 1520–1534 [DOI] [PubMed] [Google Scholar]
- Rick C. (1988). Evolution of mating systems in cultivated plants. In Plant Evolutionary Biology, L. Gottlieb and S. Jain, eds (London: Chapman and Hall), pp. 133–147.
- Rieseberg L.H., Blackman B.K. (2010). Speciation genes in plants. Ann. Bot. (Lond.) 106: 439–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H., Willis J.H. (2007). Plant speciation. Science 317: 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H., Wood T.E., Baack E.J. (2006). The nature of plant species. Nature 440: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J., Morrell P.L., Gaut B.S. (2007). Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. USA 104 (suppl. 1): 8641–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemske D.W. (2000). Understanding the origin of species. Evolution 54: 1069–1073 [Google Scholar]
- Schoen D.J., Brown A.H. (1991). Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc. Natl. Acad. Sci. USA 88: 4494–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S.T., Lynch M. (1997). Mutation and extinction: The role of variable mutational effects, synergistic epistasis, beneficial mutations, and degree of outcrossing. Evolution 51: 1363–1371 [DOI] [PubMed] [Google Scholar]
- Slatkin M. (1985). Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16: 393–430 [Google Scholar]
- Smartt J., Simmonds N.W. (1995). Evolution of Crop Plants (Singapore: Longman Scientific & Technical) [Google Scholar]
- Snow A.A., et al. (2003). A Bt transgene reduces herbivory and enhances fecundity in wild sunflowers. Ecol. Appl. 13: 279–286 [Google Scholar]
- Spooner D.M., McLean K., Ramsay G., Waugh R., Bryan G.J. (2005). A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc. Natl. Acad. Sci. USA 102: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G.L. (1950). Variation and Evolution in Plants. (New York: Columbia University Press)
- Stebbins G.L. (1957). Self fertilization and population variability in higher plants. Am. Nat. 91: 337–354 [Google Scholar]
- Takebayashi N., Morrell P.L. (2001). Is self-fertilization an evolutionary dead end? Revisiting an old hypothesis with genetic theories and a macroevolutionary approach. Am. J. Bot. 88: 1143–1150 [PubMed] [Google Scholar]
- Tanksley S.D., McCouch S.R. (1997). Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277: 1063–1066 [DOI] [PubMed] [Google Scholar]
- Villar R., Veneklaas E. (1998). Relative growth rate and biomass allocation in 20 Aegilops (Poaceae) species. New Phytol. 140: 425–437 [DOI] [PubMed] [Google Scholar]
- Vogler D.W., Kalisz S. (2001). Sex among the flowers: The distribution of plant mating systems. Evolution 55: 202–204 [DOI] [PubMed] [Google Scholar]
- Well E.V., Fossey A. (1998). A comparative investigation of seed germination, metabolism and seedling growth between two polyploid Triticum species. Euphytica 101: 83–89 [Google Scholar]
- Williams G.C. (1975). Sex and Evolution (Princeton, NJ: Princeton University Press)
- Wilson A.M., Thompson K. (1989). A comparative study of reproductive allocation in 40 British grasses. Funct. Ecol. 3: 297–302 [Google Scholar]
- Wood T.E., Takebayashi N., Barker M.S., Mayrose I., Greenspoon P.B., Rieseberg L.H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 106: 13875–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D., Hopf M. (2000). Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia, Europe, and the Nile Valley, 3rd ed. (New York: Oxford University Press)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.