Loss of function analysis reveals that the replication licensing factors CDT1a and CDT1b act redundantly during gametophyte development. In addition, reduced expression of these genes causes endogenous DNA stress in plants and results in spontaneous mutations, demonstrating that these two proteins are crucial to the maintenance of genome integrity both in vegetative and in reproductive cells.
Abstract
Meristems retain the ability to divide throughout the life cycle of plants, which can last for over 1000 years in some species. Furthermore, the germline is not laid down early during embryogenesis but originates from the meristematic cells relatively late during development. Thus, accurate cell cycle regulation is of utmost importance to avoid the accumulation of mutations during vegetative growth and reproduction. The Arabidopsis thaliana genome encodes two homologs of the replication licensing factor CDC10 Target1 (CDT1), and overexpression of CDT1a stimulates DNA replication. Here, we have investigated the respective functions of Arabidopsis CDT1a and CDT1b. We show that CDT1 proteins have partially redundant functions during gametophyte development and are required for the maintenance of genome integrity. Furthermore, CDT1-RNAi plants show endogenous DNA stress, are more tolerant than the wild type to DNA-damaging agents, and show constitutive induction of genes involved in DNA repair. This DNA stress response may be a direct consequence of reduced CDT1 accumulation on DNA repair or may relate to the ability of CDT1 proteins to form complexes with DNA polymerase ε, which functions in DNA replication and in DNA stress checkpoint activation. Taken together, our results provide evidence for a crucial role of Arabidopsis CDT1 proteins in genome stability.
INTRODUCTION
In contrast to animal development, plant development is mostly a postembryonic process, achieved by the activity of meristems in which cells divide throughout the plant’s life. In addition, the germline differentiates only late in development, implying that replication errors occurring in the shoot meristem would be transmitted to the next generation. Because of these specific features, accurate genome duplication and hence correct cell cycle regulation is of particular importance in plants. Although plants have evolved new regulators of cell cycle progression, the basic regulatory mechanisms are shared with other eukaryotes (reviewed in De Veylder et al., 2007; Costas et al., 2011). Cyclin-dependent kinase (CDK)-cyclin complexes are the core cell cycle regulators that allow the transition from one phase to another. Notably, entry into the S phase of the cell cycle requires phosphorylation of the RetinoBlastoma Retated (RBR) protein by CDK-cyclin D complexes. Upon phosphorylation, the inhibitory effect of RBR on E2F transcription factors is released, allowing the expression of downstream targets such as subunits of the prereplication complex (pre-RC) (reviewed in Inzé and De Veylder, 2006). Among these subunits, CDC10 Target1 (CDT1) and Cell Division Cycle6 (CDC6) are essential factors for DNA replication licensing in all eukaryotes because they recruit the DNA helicases called Mini-Chromosome Maintenance (MCM) proteins that open the replication forks (DePamphilis, 2003). Maintenance of genome integrity requires each part of the DNA to be replicated once and only once per cell cycle; therefore, the firing of replication origins must be tightly regulated. CDT1 is considered as the key factor that determines DNA replication licensing in all eukaryotes because its overexpression is sufficient to induce rereplication (Truong and Wu, 2011). Accordingly, this protein is the target of a wealth of regulatory mechanisms, including transcriptional control by E2F transcription factors, proteolysis, and interaction with the geminin, which functions as an inhibitor of CDT1 activity (reviewed in Truong and Wu, 2011). In animals, the dynamic formation of CDT1-geminin complexes, and the modulation of the stoichiometry between the two proteins, have recently been suggested to play a major role in controlling CDT1 activity during S-phase (Lutzmann et al., 2006; De Marco et al., 2009; Kisielewska and Blow, 2012).
In Arabidopsis thaliana, the pre-RC consists of six Origin Recognition Complex (ORC) proteins, each of which is encoded by a single gene, except ORC1, CDT1, and CDC6, which are encoded by duplicated genes, and six canonical MCM proteins (MCM2 to MCM7) (Masuda et al., 2004), but the function of only a few of these genes has been investigated. Loss-of-function approaches revealed that MCM7 is required for gametophyte development and is maternally required for embryo development (Springer et al., 2000), whereas MCM2 appears to be dispensable for gametogenesis but indispensable to embryo development (Ni et al., 2009). By contrast, overexpression of CDC6 or CDT1a did not affect overall plant development but stimulated endoreduplication (Castellano et al., 2004, 2001), an atypical cell cycle during which S-phase is not followed by mitosis (De Veylder et al., 2011), resulting in increased DNA content, suggesting that these two proteins accumulate in limiting amounts for DNA replication licensing. There is evidence for proteolytic regulation of CDT1a in Arabidopsis (Castellano et al., 2004), but bona fide geminin homologs appear to be absent from plant genomes, although the isolation of a CDT1-interacting protein that may function analogously to geminin, at least in some cell types, has been reported (Caro et al., 2007). Therefore, it is not clear whether other posttranslational regulation mechanisms affect CDT1 activity. Another open question is the respective role of the two CDT1 homologs CDT1a and CDT1b: only the function of CDT1a was investigated via overexpression in plants (Castellano et al., 2004). Interestingly, a loss-of-function approach using an RNA interference (RNAi) construct targeting both CDT1a and CDT1b allowed us to provide evidence for a role of CDT1 proteins in the coordination of plastid division and cell cycle progression, but again, we were not able to precisely assign distinctive roles to the two proteins (Raynaud et al., 2005).
Another crucial aspect of genome stability is the ability of cells to stop cell cycle progression upon DNA damage until repair completion. In this respect, one could expect these mechanisms to be of outstanding importance in plants. Indeed, plants are sessile organisms and therefore cannot escape from suboptimal growth conditions. Furthermore, they require light for their photosynthetic activity and thus are continuously exposed to DNA-damaging agents such as UV light or reactive oxygen species produced by chloroplasts. In all eukaryotes, including plants, genome integrity is under the surveillance of ataxia telangiectasia mutated and Rad3-related (ATR) and ataxia telangiectasia mutated (ATM) kinases: they are activated by DNA damage and simultaneously arrest cell cycle progression and promote the expression of genes encoding DNA repair proteins (Culligan et al., 2006; Bensimon et al., 2011; Nam and Cortez, 2011). As observed for the control of S-phase, regulating CDT1 activity is instrumental to the response to DNA damage in eukaryotes. First, CDT1 degradation is required to stop or delay S-phase progression until DNA is repaired, and this process involves the recruitment of CDT1 to the site of DNA damage (Roukos et al., 2011). Second, CDT1 activity is required after DNA repair is completed to reassemble pre-RC on unfired origins (Truong and Wu, 2011). Finally, CDT1 is required for break-induced DNA repair together with MCMs and pre-RC–activating proteins, whereas ORC and CDC6 are not (Lydeard et al., 2010). How plants respond to DNA-damaging agents and how this response affects cell cycle progression are beginning to be unraveled. One key player in this process is the Suppressor Of Gamma1 (SOG1) transcription factor, which functions downstream of ATR and ATM and is required both for cell cycle arrest and the induction of DNA repair genes (Yoshiyama et al., 2009). In addition, the WEE1 protein kinase has been shown to slow down S-phase progression upon DNA stress (De Schutter et al., 2007; Cools et al., 2011) and may act downstream of SOG1 (Yoshiyama et al., 2009). Interestingly, in plants, DNA stress not only induces an arrest of proliferating cells but also cell death in stem cell niches (Fulcher and Sablowski, 2009) and endoreduplication in other cell types (Cools and De Veylder, 2009; Adachi et al., 2011). Although there is no evidence to date for a role of plant CDT1 proteins in the control of genome integrity or DNA stress response, the observation that reduced expression of CDT1 genes both delays cell cycle progression and induces endoreduplication (Raynaud et al., 2005) suggests that a decrease of CDT1 accumulation may cause DNA stress or altered DNA stress sensing.
Based on our current knowledge of plant CDT1 proteins, the goal of this work was (1) to further investigate the respective functions of Arabidopsis CDT1a and CDT1b and (2) to determine whether these proteins are as important in plants as in other eukaryotes for the maintenance of genome integrity.
RESULTS
CDT1a and CDT1b Play Partially Redundant Roles during Gametophyte Development
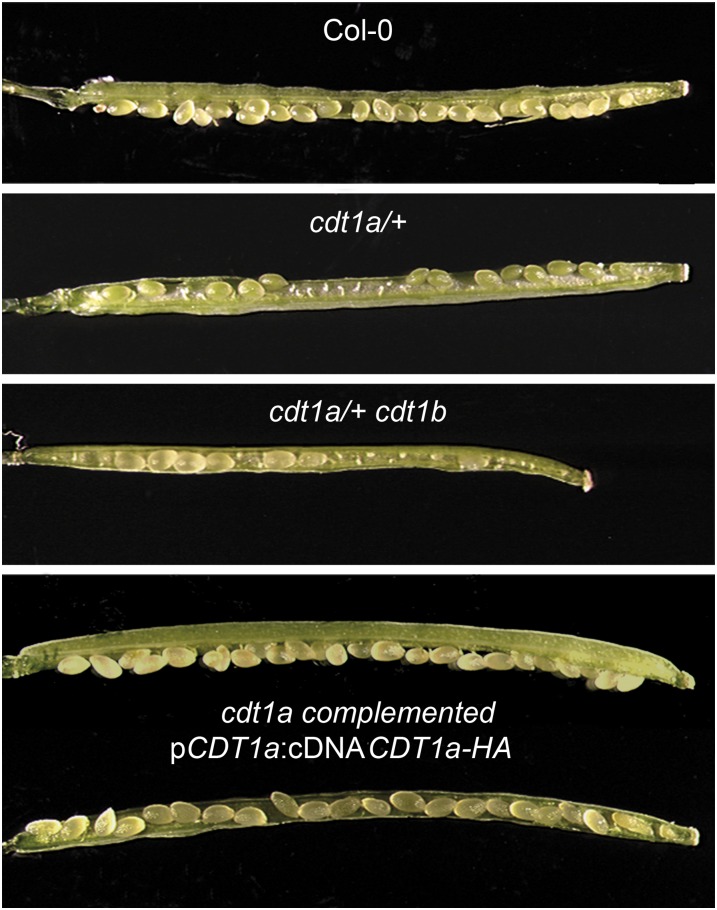
We reported previously that simultaneous silencing of CDT1a and CDT1b resulted in severe developmental defects and that cdt1b null mutants are phenotypically identical to the wild type (Raynaud et al., 2005). CDT1a and CDT1b have very similar expression patterns, suggesting that they could be functionally redundant (Castellano et al., 2004). To elucidate the respective functions of the two genes, we searched for cdt1a mutants in publicly available mutant collections. We found an insertion in CDT1a in the GABI-Kat collection (GABI_025G08). This mutant line harbors two insertions, one in the fourth exon of CDT1a (see Supplemental Figure 1 online) and the second 577 bp upstream of the start codon of HexoKinase Like3 (HKL3) (At4g37840), which encodes a putative hexokinase. Using a segregating population, the T-DNA insertion in CDT1a was separated from the second T-DNA (see Methods). For three lines, no plant harboring the HKL3 mutation was found, but all contained the CDT1a insertion, indicating that the parent line was mutated only for CDT1a. The presence of a sulfadiazine resistance maker in the GABI-Kat line allowed us to perform a segregation analysis of the T-DNA. Interestingly, the proportion of sulfadiazine-resistant plants in these lines was lower than expected (between one in four and one in three), and all cdt1a mutants identified in this first generation were hemizygous for the mutation (n = 12; hereafter referred to as cdt1a/+). These plants developed normally but formed short siliques containing aborted ovules (Figure 1). We counted normal and aborted seeds in siliques of seven plants (four siliques per plant) and found 49.8% aborted seeds. To confirm that this phenotype was due to the insertion in the CDT1a gene, we performed backcrosses. In the progeny, all sulfadiazine-resistant plants carried an insertion in CDT1a, and all showed a reduction in seed production. Furthermore, the mutant could be rescued by a construct encompassing a hemagglutinin (HA)-tagged version of CDT1a driven by its own promoter: cdt1a/+ plants were transformed with the pCDT1a:CDT1a-HA construct, and phenotypically wild-type plants homozygous for the cdt1a mutation expressing the HA-tagged protein could be recovered (Figure 1).
Figure 1.
cdt1 Mutants Are Partially Sterile.
Fully developed siliques from the wild type (Col-0), cdt1a/+, cdt1a/+cdt1b, and complemented cdt1a mutants were opened and observed using a binocular microscope.
[See online article for color version of this figure.]
To determine whether the cdt1a mutation was lethal during gametophyte development or during embryogenesis, we analyzed the transmission of the cdt1a mutation after self-pollination or reciprocal crosses. To follow the cdt1a mutation, we analyzed sulfadiazine resistance. Twelve plants were used for this analysis, seeds from three siliques obtained by each cross or self-fertilization were pooled, and ∼40 seeds were sown on selective medium (Table 1). When cdt1a/+ mutants were used as female, no sulfadiazine-resistant plants could be obtained, indicating that the mutation is not transmitted through the female side. When cdt1a/+ mutants were allowed to self-fertilize or used as male, ∼31 and 25% of their progeny were resistant to sulfadiazine. The difference between these two segregation values is not statistically significant (Student’s t test; P > 0.05). However, these segregation values are significantly lower than the expected 50% if the mutation was transmitted normally via pollen grains (χ2 = 6.48). Hence, segregation analyses indicated that disruption of CDT1a affects female gametophyte development and, to a lesser extent, pollen grain development.
Table 1. Result of T-DNA Transmission in Reciprocal Crosses of cdt1a/+.
| Cross | SulfR | SulfS | Percentage of SulfR plantlets |
|---|---|---|---|
| cdt1a/+ × cdt1a/+ | 164 | 343 | 31% |
| cdt1a/+ × Col-0 | 0 | 340 | 0% |
| Col-0 × cdt1a/+ | 140 | 398 | 25% |
Seeds obtained by each cross were sown on 0.5× MS medium containing sulfadiazine, and the sulfadiazine-sensitive (SulfS) and sulfadiazine-resistant (SulfR) plantlets were counted.
To confirm these observations and to determine which step of gametophyte development was affected by the cdt1a mutation, we tested pollen grain viability by Alexander staining and observed developing ovules from cleared siliques of cdt1a/+ mutants. As shown in Figure 2A, cdt1a/+ mutants show a proportion of green pollen grains after Alexander staining, indicating that they are not viable. We estimated that ∼15% (n = 200) of pollen grains were aborted in the mutant. 4′,6-diamidino-2-phenylindole (DAPI) staining on flower buds at various developmental stages revealed that meiospore formation was normal, but pollen grains stopped their development either before the first or second mitosis (Figures 2B and 2C). In developing siliques, we found that about half of the ovules contained normally developing embryos, whereas the other half appeared to have aborted. Closer examination revealed that half of the embryo sacs never reached maturity and stopped their development after one or two mitoses, giving rise to embryo sacs containing one to three nuclei (Figure 3). Consistently, CDT1-RNAi lines displayed similar defects during gametophyte development (data not shown). Taken together, these results suggest that CDT1a is strictly required for female gametophyte development and plays a role during male gametophyte development, although a majority of pollen grains harboring the cdt1a mutation develop normally.
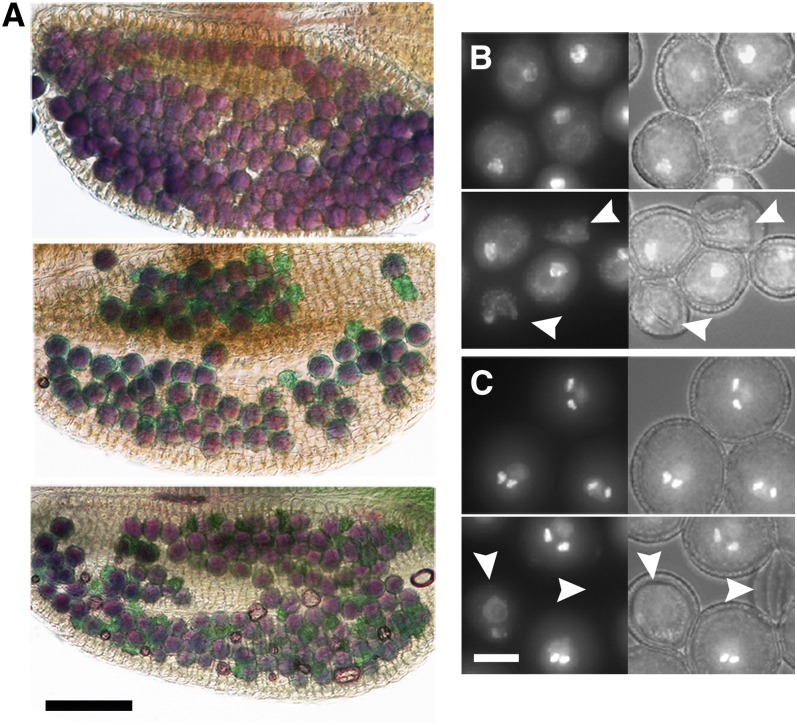
Figure 2.
Pollen Development Is Affected in cdt1a/+ Mutants.
(A) Alexander staining of mature anthers from the wild type (Col-0; top), cdt1a/+ (middle), and cdt1a/+cdt1b (bottom) mutants. Viable pollen grains stain purple. Nonviable pollen grains stain blue-green and can be observed in cdt1a/+ mutants, and their proportion is increased in the cdt1b background.
(B) DAPI staining of pollen grains from wild-type (top) and cdt1a/+ (bottom) plants at the binuclear stage. Aborting pollen grains can be detected in cdt1a/+ plants (white arrowheads).
(C) DAPI staining of pollen grains from wild-type (top) and cdt1a/+ (bottom) plants at the trinuclear stage. Aborting pollen grains can be detected in cdt1a/+ plants (white arrowheads), and some degenerating pollen grains contain two nuclei, indicating that they aborted after the first pollen mitosis.
In (B) and (C), left images are DAPI fluorescence and right images are overlays with bright-field images.
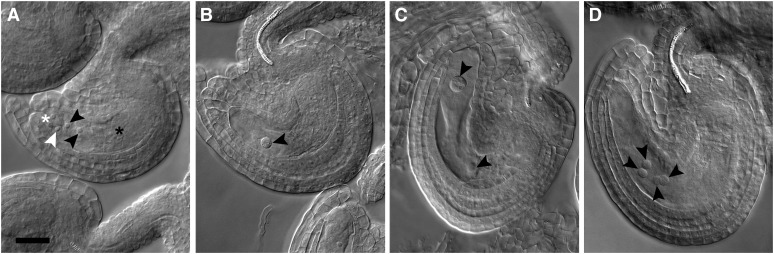
Figure 3.
Embryo Sac Development Is Compromised in cdt1a/+ Mutants.
(A) Mature embryo sac from a wild-type plant (Col-0). The positions of synergids (white star), egg cell (white arrowhead), central cell nuclei (black arrowheads), and antipodal cells (black stars) are indicated.
(B) to (D) Abnormal embryo sacs in cdt1a/+ mutants. Embryo sacs containing one (B), two (C), or four (D) nuclei were observed in 50% of ovules in cdt1a/+ mutants.
Normal development of pollen grains lacking CDT1a may be allowed by the presence of CDT1b. To test this hypothesis, we obtained cdt1a/+cdt1b−/− sesquimutants and analyzed the transmission of the cdt1a mutation in their progeny. We found that ∼5% of plants inherited the cdt1a mutation (n = 1857) versus 30% in the cdt1a/+ mutant (n = 1344). Thus, the cdt1b mutation significantly reduced the transmission of the cdt1a mutation (χ2 = 27.15). Observation of developing pollen grains revealed a higher proportion of aborted pollen in the sesquimutant. Flow cytometry analysis of the pollen DNA content in the wild type and mutants did not allow us to detect a difference between cdt1a/+ and wild-type plants: pollen isolated from flowers before dehiscence distributed in a single narrow peak corresponding to three cellular pollen grains, with a minor peak toward the lower DNA contents corresponding to cell debris and empty pollen grains. However, the distribution of pollen grains from cdt1a/+cdt1b plants displayed a shoulder on the peak toward lower DNA content, suggesting that developing pollen grains failed to complete S-phase (see Supplemental Figures 2 and 3 online). However, we cannot rule out the possibility that the lower fluorescence observed could be due to the degeneration of nonviable pollen grains. This result indicates that CDT1a and CDT1b play partially redundant functions during pollen grain development but that pollen grains can be viable in the absence of both genes. By contrast, CDT1a is strictly required for female gametophyte development, suggesting that some of its functions cannot be taken over by CDT1b.
Downregulation of CDT1 Proteins Affects Genome Integrity
The observation that a proportion of pollen grains have a reduced DNA content in cdt1a/+cdt1b mutants suggests that genome integrity is compromised in the absence of CDT1. Indeed, upon closer examination, we noticed that ∼10% (n = 270) of the sulfadiazine-resistant plantlets in the progeny of cdt1a/+cdt1b plants were smaller than their siblings (Figure 4A). When these plants were transferred to soil and allowed to grow, they displayed a variety of phenotypic alterations (summarized in Table 2; Figures 4B to 4E). Out of 36 plants, 21 grew and developed like their cdt1a/+cdt1b siblings, but 15 displayed a variety of developmental defects: in the most extreme cases, plants had a drastically reduced stature and deformed leaves, as shown in Figures 4B to 4E. Such a frequency of abnormal plants was never observed in the progeny of ecotype Columbia (Col-0), cdt1a/+, or cdt1b plants, indicating that these anomalies were a consequence of the loss of both CDT1 genes in pollen grains. Most of these abnormal plants were almost completely sterile, but some produced a few seeds and gave rise to both aberrant and wild-type–looking plants in their progeny. Flow cytometry analysis of the DNA content in the second generation of aberrant plants (n = 40) allowed us to identify four aneuploid plants with a genome size either 10% larger or 10% smaller than the wild type. This technique did not reveal any modification in the genome size of other plants but would not allow the detection of small variations. These results indicate that the loss of both CDT1 genes results in severe modifications of the genome during pollen grain formation.
Figure 4.
Development of Some cdt1a/+cdt1b Mutants Is Affected.
(A) The progeny of a cdt1a/+cdt1b mutant were sown on 0.5× MS medium supplemented with sulfadiazine. After 10 d, the development of sulfadiazine-sensitive plants is arrested and they appear yellow. Some resistant plantlets show delayed growth (white arrowheads).
(B) to (E) Examples of developmental alterations observed in these smaller plantlets after transfer to soil and growth in the greenhouse. Magenta arrowheads point at abnormal plants.
Table 2. Summary of the Phenotypic Alterations Observed in the Progeny of cdt1a/+cdt1b Plants.
| Plant No. | Plant Height | Leaf Phenotype | Fertility |
|---|---|---|---|
| 21 | Normal | Normal | Reduced as shown in Figure 1 |
| 7 | Reduced | Pale green | Sterile |
| 2 | Normal | Slightly pale | Sterile |
| 1 | Drastically reduced | Small, pale green, and twisted leaves (Figure 7B, right) | Entered senescence and died before flowering |
| 1 | Reduced | Round and small leaves | Fasciated stem, sterile |
| 2 | Dramatically reduced | Pale green and curly leaves (Figures 7C and 7E) | Sterile |
| 2 | Normal/slightly reduced | Slightly pale (Figures 7B, left, and 7D) | Delayed flowering, abnormal flower morphology, altered phyllotaxy, sterile |
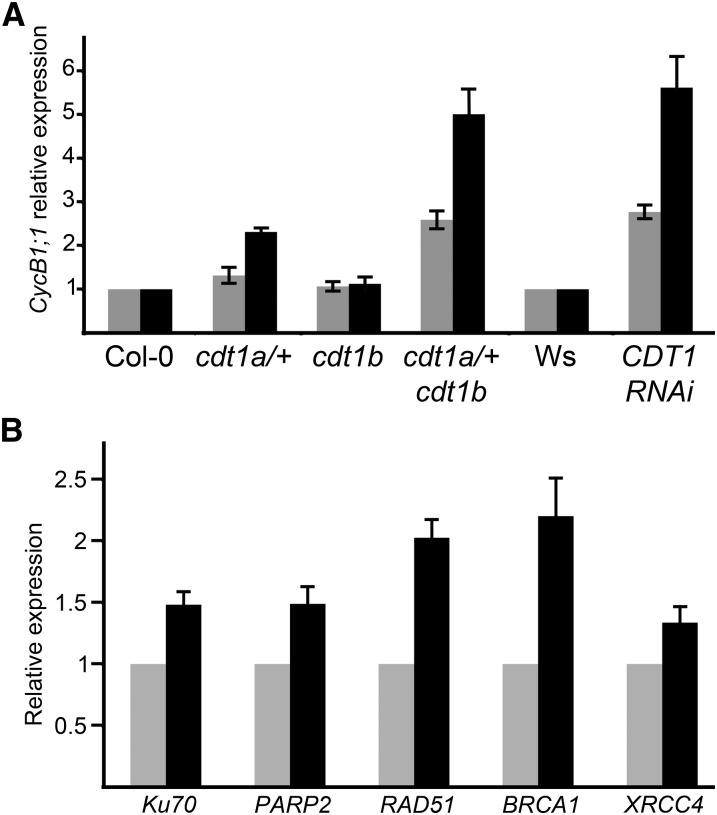
To gain further insight into the function of CDT1 proteins in the maintenance of genome integrity in sporophytic tissues, we investigated the consequences of reduced CDT1 accumulation in plantlets, taking advantage of the CDT1-RNAi lines because homozygous cdt1a mutants cannot be obtained. We reported previously that CDT1-RNAi plants incorporate less bromodeoxyuridine (an analog of thymidine that incorporates into replicated DNA during S-phase) than the wild type and have smaller leaves due to a reduction of both cell expansion and cell proliferation (Raynaud et al., 2005). Furthermore, the root tip mitotic index is reduced in these plants (see Supplemental Figure 4C online). In addition to this reduction of proliferative activity, CDT1-RNAi lines display enhanced endoreduplication; these two features have been reported in several mutants subjected to endogenous DNA stress (reviewed in Cools and De Veylder, 2009). Many of these mutants exhibit enhanced expression of Cyclin B1;1 (CYCB1;1); therefore, we monitored the expression of CYCB1;1 in CDT1-RNAi plantlets. Expression of CYCB1;1 was found to be increased CDT1-RNAi plantlets and in cdt1a/+cdt1b but not cdt1b mutants (Figure 5A). This observation suggests that reduced expression of CDT1 genes results in DNA stress. To confirm this, the expression of several genes involved in various DNA repair pathways was monitored in wild-type and CDT1-RNAi plants. As shown in Figure 5B, the expression of Ku70, PARP2, RAD51, BRCA1, and XRCC4 was found to be increased in CDT1-RNAi lines. This increase was low (between 1.3- and 2.3-fold depending on the gene) but reproducible, providing evidence for endogenous DNA stress induced by reduced CDT1 accumulation.
Figure 5.
Downregulation of CDT1 Genes Results in Endogenous DNA Stress.
(A) Expression of CycB1;1 was monitored by quantitative RT-PCR. Total RNA was extracted from plantlets. Data presented here are averages of two technical replicates obtained from two biological replicates (black and gray bars). Expression of CycB1;1 was reproducibly found to be increased in CDT1-RNAi and cdt1a/+cdt1b mutants. The increase observed in the second experiment in cdt1a/+ mutants was not reproducible, suggesting that expression of CycB1;1 varies a little in this background.
(B) Expression of genes involved in various DNA repair pathways was monitored by quantitative RT-PCR. All genes were found to be induced in CDT1-RNAi lines (black bars) compared with the wild type (Wassilewskija [Ws]; gray bars). Data presented here represent two biological replicate experiments. Each quantitative RT-PCR was repeated at least twice on each biological replicate.
For all panels, bars are averages ± sd.
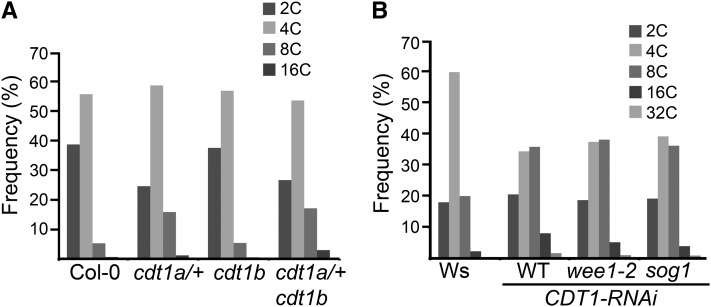
As an independent test for the presence of DNA stress, the DNA ploidy distribution was measured in the cdt1a/+ and cdt1a/+cdt1b mutants by flow cytometry. Twelve-day-old cdt1a/+ and cdt1a/+cdt1b plantlets showed a clear increase in endoreduplication in the first leaf compared with the wild-type (Figure 6A; see Supplemental Figure 5 online). This increase was also observed in whole plantlets for cdt1a/+cdt1b mutants but not for cdt1a/+ mutants (data not shown) and was not observed in cauline leaves of mature plants, indicating that increased endoreduplication is transient in cdt1a/+ and cdt1a/+cdt1b mutants and occurs only at early stages of development. Indeed, all viable cdt1 mutants still possess one wild-type copy of CDT1a. CDT1-RNAi lines, therefore, are likely to accumulate lower levels of CDT1 proteins than the mutant throughout their vegetative development. Taken together, these results support the view that reduced CDT1 accumulation results in DNA stress in Arabidopsis.
Figure 6.
Endoreduplication Is Increased in Plants with Reduced CDT1 Accumulation in a WEE1- and SOG1-Independent Manner.
(A) Endoreduplication in cdt1 mutants. Analyses were performed on the first leaf of Col-0, cdt1a/+, cdt1b, and cdt1a/+cdt1b plants harvested 14 d after stratification. Ten leaves were used for each sample. Representative cytometry profiles are shown in Supplemental Figure 4 online.
(B) Endoreduplication in CDT1-RNAi plants is independent of WEE1 and SOG1. The first cauline leaf of Wassilewskija (Ws), CDT1-RNAi, wee1-2 CDT1-RNAi, and sog1 CDT1-RNAi was used for this analysis. Increased levels of endoreduplication were observed in CDT1-RNAi in the wild-type (WT), wee1-2, and sog1 backgrounds. Representative cytometry profiles are shown in Supplemental Figure 5 online.
Increased Endoreduplication in CDT1-RNAi Plants Is Not Dependent on WEE1 or SOG1 Activation
Genome integrity is under the control of several checkpoints during the cell cycle. In Arabidopsis, WEE1 activation has been proposed to govern DNA stress–dependent cell cycle arrest (De Schutter et al., 2007), and double-strand breaks have been reported to activate CYCB1;1 expression and endoreduplication in a SOG1-dependent but WEE1-independent manner (Adachi et al., 2011). We tried to determine which of these checkpoints was activated in CDT1-RNAi plants. We first introduced the CDT1-RNAi construct in the wee1 background. Endoreduplication in plants displaying the CDT1-RNAi phenotype was measured in eight wee1-2 homozygous mutant lines and compared with wee1-2/+ hemizygous siblings as well as wild-type plants and wee1-2 mutants. As described by De Schutter et al. (2007), the wee1 mutation did not affect endoreduplication under normal growth conditions. As shown in Figure 6B and Supplemental Figure 6 online, the endoreduplication effect of the CDT1-RNAi construct was similar in the wild-type and the wee1-2 mutant backgrounds, indicating that the increase in endoreduplication was not WEE1 dependent. Similarly, CDT1-RNAi lines still displayed increased endoreduplication in the sog1 background, indicating that neither WEE1 nor SOG1 is responsible for the endoreduplication phenotype observed in CDT1-RNAi lines. Also, transmission of the cdt1a mutation remained unchanged in the wee1-2 and sog1 backgrounds, indicating that the observed developmental arrest in gametophytes does not require WEE1 or SOG1 activity. This may either suggest that the observed increase in endoreduplication is not related to the response to DNA stress or that downregulation of CDT1 occurs downstream of WEE1 and/or SOG1 activation to promote endoreduplication upon DNA damage. To gain further insight into the role of CDT1 proteins in the DNA damage response, we next investigated the sensitivity of the different lines to DNA stress.
CDT1-RNAi Plants Display Enhanced Tolerance to DNA Stress
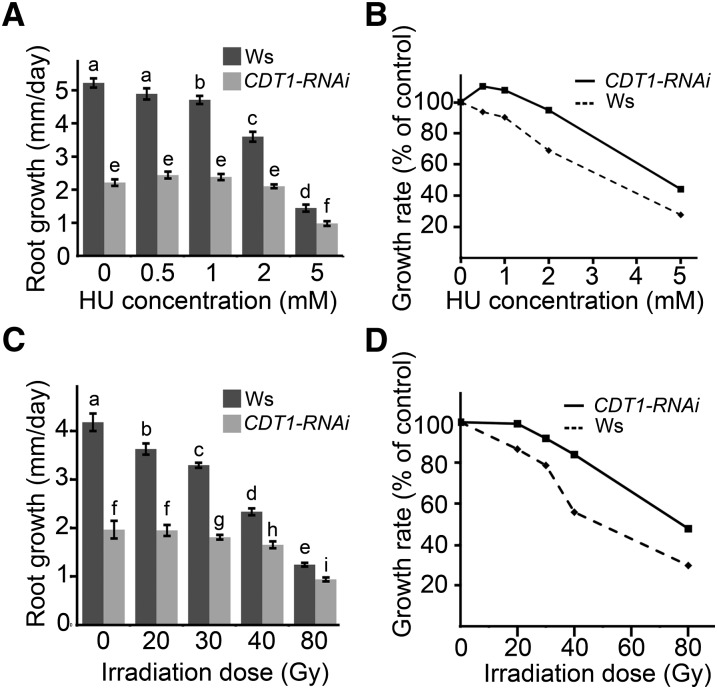
The appearance of mutations in the progeny of cdt1a/+cdt1b mutants could either be due to an increase in DNA damage or to a defect in DNA stress checkpoints. To discriminate between these two hypotheses, we tested the resistance of cdt1 mutants and CDT1-RNAi lines to DNA stress. We first investigated the role of CDT1 proteins in the response to replication fork stalling. To this end, plants were grown for 10 d on Murashige and Skoog (MS) medium and transferred to plates containing increasing concentrations of hydroxyurea (HU), which is an inhibitor of ribonucleoside reductase. HU treatment depletes the cellular content of deoxyribonucleotide and thereby induces stalling of the replication fork. Root growth was measured 2 d after transfer on HU-containing medium (Figures 7A and 7B). On MS plates, root growth of CDT1-RNAi plantlets was much lower than that of the wild type, and this difference was still observed on HU-containing medium. However, root length was significantly reduced by 1 mM HU treatment in the wild type but remained unchanged for CDT1-RNAi plants on medium containing 1 or 2 mM HU, suggesting that they show increased tolerance to stalling of the replication fork. To further confirm that growth of CDT1-RNAi plantlets is not as strongly inhibited as the wild type by DNA stress, we tested their tolerance to γ-irradiation. Root growth was measured 3 d after irradiation at various doses. Root growth was reduced in the wild type at a dose of 20 Gray but remained unchanged in CDT1-RNAi lines up to 30 Gray (Figure 7B). Furthermore, relative growth was lower in the wild type than for CDT1-RNAi plants in all conditions tested (Figure 7D).
Figure 7.
CDT1-RNAi Plants Are Tolerant to DNA Stress.
(A) Root growth of wild-type (Wassilewskija [Ws]) and CDT1-RNAi (RNAi) plants after HU treatment. Plantlets were germinated and grown on 0.5× MS medium for 10 d and transferred either to 0.5× MS medium or 0.5× MS medium supplemented with the indicated amount of HU. Root elongation was measured after 2 d. Bars are averages ± se. Letters indicate significantly different values (Student’s t test; for a and b, P < 0.05; for all other letters, P < 0.001).
(B) Relative growth of roots from Ws (dashed line) and CDT1-RNAi (solid line) as a function of HU concentration in the growth medium. The reduction of root growth in CDT1-RNAi is less severe than for Ws plantlets at all concentrations tested.
(C) Root growth of wild-type (Ws) and CDT1-RNAi (RNAi) plants after γ-irradiation. Plantlets were germinated on 0.5× MS medium and irradiated at the indicated doses. Root growth was measured after 3 d. Letters indicate significantly different values (Student’s t test; for a, b, f, and g, P < 0.05; for all other letters, P < 0.001 ). ns, Nonsignificantly different values (P > 0.05).
(D) Relative growth of roots from Ws (dashed line) and CDT1-RNAi (solid line) as a function of γ-ray dose (Gy). The reduction of root growth in CDT1-RNAi is less severe than for Ws plantlets at all doses tested.
Several hypotheses may account for the reduced sensitivity of CDT1-RNAi plantlets to DNA-damaging agents. One possibility would be that sensing of DNA damage is impaired in these plants, resulting in the absence of cell cycle arrest and DNA repair, as observed in the sog1 mutant. Alternatively, the apparent resistance could be due to basal activation of the DNA stress checkpoint, resulting in a less severe growth inhibition after DNA damage. Because growth of CDT1-RNAi plantlets is further reduced after γ-irradiation compared with untreated CDT1-RNAi plantlets, cell cycle arrest induced by DNA damage seemed to be functional in this background. The second hypothesis, therefore, appeared the most likely. To confirm this, we tested whether the response to γ irradiation was normal in CDT1-RNAi plants by assessing recognition of double-strand breaks (DSB), induction of DNA repair genes, and cell cycle arrest after γ-irradiation. All steps of the DSB response were found to be functional in CDT1-RNAi lines (for details, see Supplemental Figure 4 and Supplemental Methods 1 online). Response to DNA damage was actually enhanced in CDT1-RNAi plants in terms of γ-H2AX incorporation at the site of DNA damage and RAD51 expression, demonstrating that sensing and repair of DSB are functional in these plants and suggesting that they are “primed” by endogenous stress, allowing a faster or stronger response, at least at early steps of the pathway.
CDT1a and CDT1b Interact with DNA Polymerase ε
To further investigate the role of CDT1a and CDT1b in the maintenance of genome integrity, we searched for interacting partners by tandem affinity purification (TAP). We reasoned that this method may allow us to identify new protein complexes containing CDT1a and/or CDT1b together with proteins involved in DNA repair or DNA damage sensing. Protein complexes could be purified when the tag was positioned at the N-terminal end of CDT1 proteins. Few proteins were identified by this method, but the results were very reproducible (Table 3; see Supplemental Table 1 online). CDT1a and CDT1b were found to copurify with the two subunits of DNA polymerase ε (DPB2 and POL2A). Additionally, we found a transcription factor that appeared to copurify specifically with CDT1b.
Table 3. Proteins Identified by Tandem Affinity Purification Using CDT1a or CDT1b as Bait.
| Bait Protein | Identified Proteins |
||||||
|---|---|---|---|---|---|---|---|
| Accession No. | Protein Name | No. Found/No. Expected | Protein Score | Expected | Best Ion Score | Expected | |
| CDT1a | AT1G08260 | EMB2284, POL2A, TIL1, EMB529 | 2/2 | 781 | 2.6E-74 | 65 | 4.20E-06 |
| CDT1a | AT5G22110 | DBP2, CYL2 | 2/2 | 452 | 2.10E-41 | 115 | 4.60E-11 |
| CDT1a | AT2G31270 | CDT1a | 2/2 | 88 | 5.70E-05 | – | – |
| CDT1b | AT1G08260 | EMB2284, POL2A, TIL1, EMB529 | 2/2 | 911 | 2.60E-87 | 78 | 2.80E-07 |
| CDT1b | AT5G22110 | DBP2, CYL2 | 2/2 | 777 | 6.50E-74 | 133 | 7.10E-13 |
| CDT1b | AT2G27470 | CCAAT box binding transcription factor subunit HAP3 related | 2/2 | – | – | 36 | 5.60E-04 |
| CDT1b | AT3G54710 | CDT1b | 2/2 | 910 | 3.30E-87 | 115 | 6.60E-11 |
All identified proteins were found in both experiments. Values for protein score and best ion score are for two independent experiments. –, values below the threshold score. Detailed MS data can be found in Supplemental Table 1 online.
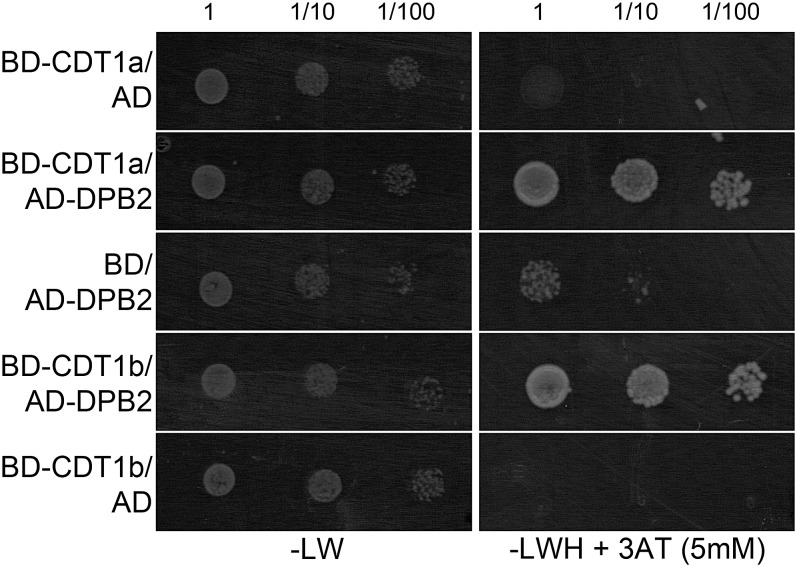
To confirm that CDT1 proteins can interact with DNA polymerase ε, we used the yeast two-hybrid system. Both CDT1a and CDT1b were found to interact with the regulatory subunit DPB2 (Figure 8). The catalytic subunit is very large, and two clones encompassing ∼500 amino acids at the C terminus of the protein (POL3 and POL5), corresponding to the region involved in DPB2 binding, were tested (Ronceret et al., 2005). We could not find an interaction between CDT1 proteins and these fragments, although the POL3 fragment could interact with DPB2 as reported (Ronceret et al., 2005) (data not shown). These results suggest that CDT1 proteins form a complex with DNA polymerase ε through their association with DPB2.
Figure 8.
CDT1a and CDT1b Interact with the DPB2 Subunit of DNA Polymerase ε in the Yeast Two-Hybrid System.
Yeast cells were transformed with constructs encompassing fusions between CDT1a or CDT1b and the GAL4 DNA binding domain (BD-CDT1a and BD-CDT1b, respectively), between DPB2 and the GAL4 activation domain (AD-DPB2), or empty vectors (AD and BD). Transformants were resuspended in water to a final absorbance (600 nm) of 1, and serial dilutions were spotted on nonselective medium lacking only Leu and Trp (−LW) or on selective medium lacking Leu, Trp, and His and supplemented with 3-amino-triazole to a final concentration of 5mM (−LWH + 3AT). Restoration of His prototrophy indicates that both CDT1a and CDT1b can interact with DPB2.
DISCUSSION
Although many reports have highlighted the importance of cell cycle regulation during plant development, reports regarding the relationships between cell cycle regulation and genome stability are scarce. However, it is well known in other eukaryotes and more specifically in human cells that altered regulation of DNA replication licensing due to changes in CDT1 activity results in aberrant replication, activates DNA damage checkpoints and predisposes for malignant transformation (Petropoulou et al., 2008). Therefore, regulation of pre-RC activity is likely to be crucial for the maintenance of genome integrity also in plants. Overexpression analysis suggests that accumulation of CDC6 and CDT1 is rate limiting for the initiation of S-phase, both in proliferating and in endoreduplicating cells, but that misexpression of these genes is not sufficient to alter the size of meristems (Castellano et al., 2001, 2004). We previously reported that downregulation of Arabidopsis CDT1a and CDTb affects cell cycle progression, plastid division, and chloroplast biogenesis, but we were not able to investigate the consequences of a complete loss of function of these genes (Raynaud et al., 2005). Our new results reveal that the two CDT1 genes are crucial to gametophyte development and to the maintenance of genome integrity in Arabidopsis both in gametophytic and sporophytic tissues.
The cdt1a mutation prevents female gametophyte development and results in partially penetrant pollen grain development defects: ∼30% of cdt1a pollen grains did not reach the three-nuclei stage. Although complete knockout of the CDT1b gene does not affect development or fertility, the penetrance of the cdt1a mutation was increased in this background, and only 10% of cdt1a/+cdt1b pollen grains were viable and allowed fertilization. These results indicate that CDT1a and CDT1b have partially redundant functions during male gametophyte development and that CDT1a, but not CDT1b, is required for female gametophyte development. Both male and female gametophyte development require accurate cell cycle regulation. After meiosis, the microspore undergoes an asymmetric division, giving birth to the vegetative cell that exits the cell cycle in the G1-phase and a generative cell. This cell divides again to produce the twin sperm cells required for double fertilization that are arrested in the S-phase of the cell cycle (Berger and Twell, 2011); hence, S-phase is initiated three times during pollen grain formation. Likewise, embryo sac development requires three mitotic events, and there is compelling evidence that inhibition of cell cycle progression severely affects this process (Yang et al., 2010). By contrast, failure to arrest cell division because of the loss of the RBR1 gene results in a gametophyte-lethal phenotype due to overproliferation of the embryo sac cells (Ebel et al., 2004). Intriguingly, all mutants deficient for one pre-RC subunit described so far do not display a gametophyte-lethal phenotype: mcm2 mutants arrest development at the globular stage (Ni et al., 2009), and the MCM7 gene is maternally required for embryo development (Springer et al., 2000) and results in leaky gametophyte lethality (Springer et al., 1995). Considering the essential function of all these proteins at the onset of S-phase, one would rather expect mutant microspores and megaspores to arrest their development before the first gametophytic mitosis. This discrepancy could result either from some functional redundancy between MCM proteins or between CDT1a and CDT1b, respectively, or from the accumulation of maternally inherited pools of the proteins, as hypothesized for the incomplete penetrance of the cdka1;1 mutation in pollen grain development (Nowack et al., 2006). Such a hypothesis would imply that the whole pool of CDT1 proteins is not degraded after the initiation of DNA replication but inactivated in a reversible way, which hints at the existence of a yet unidentified functional homolog of geminin. The fact that CDT1b cannot compensate for the absence of CDT1a during embryo sac development and can only partially do so during male gametophyte development could mean that CDT1a accumulates at higher levels than CDT1b in these tissues or could reflect a specific function of CDT1a. Indeed, we found that CDT1a, but not CDT1b, harbored a plastid-targeting sequence and that this protein could indeed accumulate in plastids (Raynaud et al., 2005). Recently, the plastid-localized PPR2 has been reported to be essential for both male and female gametophyte development in Arabidopsis (Lu et al., 2011); therefore, the inability of CDT1b to fully compensate for the absence of CDT1a may relate to the function of CDT1a in plastids.
Another interesting consequence of the cdt1a mutation is that pollen grains arrested at the two-cell stage apparently degenerate. This is at variance with the phenotypes reported in several mutants in which CDKA;1 activity is reduced and the last pollen division is delayed (reviewed in Berger and Twell, 2011): in this case, the two-celled pollen grains are able to fertilize ovules, resulting in the fertilization of only one cell in the embryo sac (Nowack et al., 2006). The degenerative phenotype observed in cdt1a and cdt1a/+cdt1b pollen grains could be due to incomplete genome replication during S-phase, as evidenced by flow cytometry analysis and the induction of programmed cell death. Indeed, programmed cell death has been proposed to act as a cellular surveillance mechanism to ensure the successful progression of male gametogenesis (Zhang et al., 2011). Incomplete genome replication or failure to repair some DNA damage may account for the abnormal frequency of mutant plants in the progeny of cdt1a/+cdt1b mutants. Indeed, plants with a modified genome size compared with the wild type were found in the progeny of cdt1a/+cdt1b plants. The technique used in this study would not have allowed us to identify changes in nuclear DNA content representing less than 10% of the whole genome, which corresponds to an entire chromosome. Aberrant plants with apparently normal DNA content, therefore, are likely to present genomic rearrangements, deletions, or other types of mutations that are more difficult to identify.
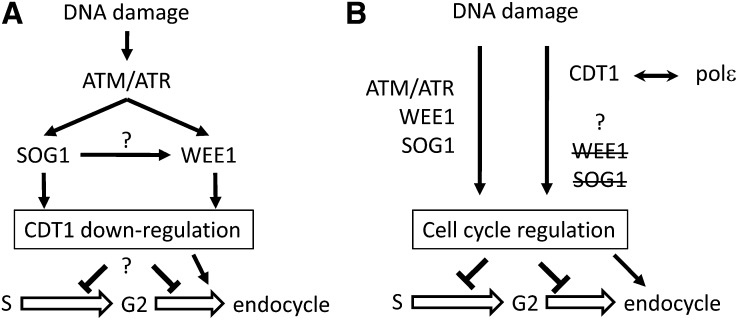
In addition to their implication in the onset of S-phase evidenced by the above-described gametophytic defects, our results point to a role for the CDT1 proteins in the maintenance of genome integrity. Indeed, elevated levels of CYCB1;1 expression in CDT1-RNAi plants and in cdt1a/+cdt1b mutants together with increased endoreduplication, and constitutive induction of genes involved in DNA repair in CDT1-RNAi lines, also point to endogenous DNA stress when the expression of CDT1 is low (Cools and De Veylder, 2009). Two possible models that could account for the observed DNA damage responses observed in the CDT1-RNAi plants are depictured in Figure 9. In both models, the reduction of CDT1 accumulation results in DNA damage, causing delayed cell cycle progression and increased expression of DNA repair–related genes, whereby the basal activation of the DNA stress response could have a priming effect on the plants, allowing them to better tolerate exogenously applied stress. Indeed, treatment with low doses of DNA-damaging agents has been shown to improve repair efficiency after exposure to higher doses of the same drug in plants (Baranczewski et al., 1997). The accompanying increase in endoreduplication observed in CDT1-RNAi plants does not appear to require WEE1 and SOG1 activity. In this context, two alternative hypotheses may account for the data reported here. Downregulation of CDT1 may be a component of the known pathways leading from DNA damage to endoreduplication situated downstream of WEE1 and SOG1 (Figure 9A). This is in agreement with several reports demonstrating that CDT1 proteins are degraded upon DNA stress in animal cells (Roukos et al., 2011). Such a model would imply that reduced CDT1 levels promote endoreduplication. CDT1a overexpression also stimulates endoreduplication (Castellano et al., 2004), but the similar outcome of opposite variations in CDT1 accumulation probably reflects a dual role of CDT1 in the promotion of replication licensing and in DNA damage response. Alternatively, the ability of CDT1 proteins to form complexes with DNA polymerase ε suggests that they could function in a new signaling cascade regulating cell cycle progression upon DNA stress occurring during S-phase (Figure 9B). Indeed, we found that both proteins interact with the regulatory subunit of DNA polymerase ε, DPB2. The primary function of DNA polymerase ε is the synthesis of the leading strand during DNA replication (Pursell et al., 2007), but it is also involved in a variety of other cellular functions, including the S-phase checkpoint (Pursell and Kunkel, 2008). The ability of plant CDT1 proteins to bind this polymerase is unexpected, as CDT1 is thought to be released from chromatin upon pre-RC activation or to be maintained at replication origins (Xouri et al., 2007; Truong and Wu, 2011). However, DUP, the CDT1 homolog of Drosophila, has been shown to travel with the replication fork (Claycomb et al., 2002), and CDT1 has recently been proposed to participate in a surveillance mechanism that blocks nascent strand elongation upon illegitimate replication licensing in Xenopus (Tsuyama et al., 2009). Therefore, the polymerase ε/CDT1 complex may be involved in monitoring the proper progression of S-phase and in the activation of cellular responses upon events such as replication fork stalling or illegitimate licensing, which would result in the expression of DNA repair genes, inhibition of cell cycle progression, and induction of endoreduplication. This hypothesis implies that a WEE1- and SOG1-independent signaling cascade can be activated by some types of DNA stress in plant cells. Further genetic studies will be required to identify factors involved in this pathway.
Figure 9.
Two Possible Models Regarding the Role of CDT1 Proteins in Response to DNA Stress.
(A) Downregulation of CDT1 upon DNA stress may be promoted by WEE1 and/or SOG1. This would result in a delay in S-phase progression, but how this would induce endoreduplication remains unclear.
(B) CDT1 proteins, possibly via their ability to form complexes with DNA polymerase ε, may be at the top of a signaling pathway that could regulate cell cycle progression and endoreduplication in parallel with the WEE1- and SOG1-dependent pathway.
Although further work is required to fully elucidate the molecular mechanisms involving CDT1 proteins in the maintenance of genome integrity, our results highlight two contrasting situations in sporophytic tissues and in gametophytes. Reduction of CDT1 accumulation in somatic cells would slow cell cycle progression and induce the activation of DNA repair genes, whereas in gametophytes lacking CDT1 proteins completely, replication errors would not systematically lead to pollen abortion: aberrant plants observed in the progeny of cdt1a/+cdt1b mutants likely arise from aberrant pollen grains that failed to degenerate in spite of replication defects. Taken together, this work provides strong evidence for a role of CDT1 proteins in the maintenance of genome integrity throughout the life cycle of the plant.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana seeds were surface-sterilized by treatment with Bayrochlor (Bayrol) for 20 min and washed and imbibed in sterile water for 2 to 4 d at 4°C to obtain homogeneous germination. Seeds were sown on commercially available 0.5× MS medium (Basalt Salt Mixture M0221; Duchefa) with the suitable antibiotic if needed and solidified with 0.8% agar (Phyto-Agar HP696; Kalys) and grown in a long-day (16 h of light/8 h of dark, 21°C) growth chamber. After 2 weeks, the plants were transferred to soil in a glasshouse or in a growth chamber under short-day conditions (8 h of light, 20°C/16 h of dark, 18°C) for 2 weeks before being transferred to long-day conditions.
T-DNA insertion lines from the SALK collection (SALK_001298) and from GABI (GABI_025G08) were obtained from the Nottingham Arabidopsis Stock Centre.
The CDT1-RNAi lines and cdt1b mutants have been described previously (Raynaud et al., 2005). The cdt1a/+ mutant was obtained from the GABI-Kat collection (Scholl et al., 2000), and mutants were identified by PCR. The GABI T-DNA and CDT1a mut primers allowed us to screen for the mutated allele, and the CDT1a WT and CDT1a mut primers were used to amplify the wild-type allele. Plants harboring an insertion in the HKL3 gene were identified using the GABI-T-DNA and HLK3mut primers. Sequences of these primers are given in Supplemental Table 2 online.
Complementation of cdt1a/+ Mutants
For complementation of cdt1a/+ mutants, we generated a HA-tagged version of CDT1a driven by its own promoter. To this end, a triple HA tag was amplified by PCR using the HA-dir (containing a KpnI site) and HA-rev (containing a SacI site) primers. This fragment was sequenced and cloned between the KpnI and SacI sites of the pPZP111 vector (Hajdukiewicz et al.,1994) to generate the pPZP111-HA vector. The putative promoter sequence for CDT1a (815 bp upstream of the translation initiation codon) and the first 24 bp of the CDT1a coding sequence were amplified using the pCDT1-HindIII (containing a HindIII site) and pCDT1-XbaI (containing the XbaI site found in the CDT1a cDNA) primers, and this fragment was digested using HindIII and XbaI. The 3′ end of the CDT1a coding sequence was amplified using the CDT1-ClaI (situated upstream of the ClaI site found in the CDT1a cDNA) and CDT1a-KpnI (containing a KpnI site) primers; after sequencing, this fragment was digested using the ClaI and KpnI sites. These two fragments and the center part of the CDT1a cDNA cut at the XbaI and ClaI sites were ligated into the pPZP111-HA vector to generate the pCDT1a:CDT1a-HA construct. The sequences of primers are indicated in Supplemental Table 2 online. The pCDT1a:CDT1a-HA plasmid was introduced into Agrobacterium tumefaciens by electroporation, and Arabidopsis transgenic lines were generated by the floral dip method (Clough and Bent, 1998). T1 transformants were selected on kanamycin (50 µg/mL) and sulfadiazine (5 µg/mL). Fertile cdt1a/+ plants were identified, and their progeny were sown on selective medium. This allowed us to identify cdt1a homozygous mutants expressing the CDT1a-HA protein, and these plants were identical to the wild type.
Plant Treatments
For HU treatments, plants were transferred and aligned either to fresh control medium or HU-containing medium (Sigma-Aldrich) and grown vertically under long-day conditions for 2 d.
γ-Irradiation assays were performed as described previously (Domenichini et al., 2006). For comparison of sensitivity to γ-rays between the wild type and CDT1-RNAi, plantlets irradiated as above were transferred and grown on 0.5× MS medium in a vertical position under long-day conditions for 3 d.
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted from seedlings with the RNeasy MiniPrep kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 µg of total RNA using Improm-II reverse transcriptase (A3802; Promega) according to the manufacturer’s instructions. One-twenty-fifth of the synthesized cDNA was mixed with 100 nM of each primer and LightCycler 480 Sybr Green I master mix (Roche Applied Science) for quantitative PCR analysis. Products were amplified and fluorescent signals acquired with the LightCycler 480 detection system. The specificity of amplification products was determined by melting curves. PP2AA3 was used as an internal control for signal normalization. Exor4 relative quantification software (Roche Applied Science) automatically calculates the relative expression level of the selected genes with algorithms based on the ΔΔcycle threshold method. Data were from duplicates of at least two biological replicates. Quantitative RT-PCR analysis was performed at least twice for each biological replicate. The sequences of primers can be found in Supplemental Table 2 online.
Light and Fluorescence Microscopy
Fresh siliques were opened using a stereomicroscope (SVII; Zeiss), and images were captured with a color charge-coupled device camera (Power HAD; Sony). Anthers were stained in Alexander solution to stain pollen grains and observed by light microscopy (Alexander, 1969).
Differential interference contrast microscopy was used to observe female gametophytes that had been fixed in ethanol:acetic acid (3:1) and cleared using chloral hydrate solution (8 g of chloral hydrate, 1 mL of glycerol, and 2 mL of water). Images were captured on an Axioskop microscope (Zeiss) with a Spot RT slider camera (Diagnostic Instrument), and Z-stack projections (average of intensity) were performed using ImageJ software (rsbweb.nih.gov/ij/) and enhanced using Adobe Photoshop software.
For pollen mitosis, inflorescences were fixed as above, and for each flower, pollen grains were sorted from anthers on a polysine slide (Thermo Scientific) with a drop of Vectashield with DAPI (H-1200; Vector Laboratories). Images (Z-stacks) were captured on an epifluorescence videomicroscope (DMI6000B; Leica) with an ER-Hamamatsu camera, and Z-stack projections (maximum of intensity) were performed using ImageJ software.
Flow Cytometry
For flow cytometric analysis of nuclei, tissues were chopped with a razor blade in 1 mL of Galbraith buffer (Galbraith, 1983) supplemented with 1% polyvinylpyrrolidone 10,000, 5 mM metabisulfite, and 5 mg/mL RNase from a stock solution at 50 units/mg.
Propidium iodide was added to the filtered supernatants at 50 µg/mL. Endoreplication levels of 5,000 to 10,000 stained nuclei were determined using a Cyflow SL flow cytometer (Partec) with a 532-nm solid state laser (100 mW) for excitation, and emission data were collected after a 590-nm long-pass filter.
For quantification of genome size, a piece to tomato (Solanum lycopersicum) leaf was chopped with each sample to include an internal control.
TAP Experiments
CDT1a and CDT1b cDNAs were cloned between the KpnI and EcoRI sites of the pENTR3C vector. For C-terminal fusions to the TAP tag, we used the CDT1a KpnI dir and CDT1a EcoRI rev primers for CDT1a and CDT1b KpnI dir and CDT1b EcoRI rev primers for CDT1b. For N-terminal fusion to the TAP tag, the original stop codon of the cDNA was conserved using the CDT1a rev stop and CDT1b rev stop primers for CDT1a and CDT1b, respectively. Cloning of transgenes encoding tag fusions under the control of the constitutive cauliflower mosaic virus 35S promoter and transformation of Arabidopsis cell suspension cultures were then performed as described previously (Van Leene et al., 2007). Tandem affinity purification of protein complexes was done using the protein G and streptavidin binding peptide tag (Bürckstümmer et al., 2006) followed by protein precipitation and separation, according to Van Leene et al. (2008). For the protocols of proteolysis and peptide isolation, acquisition of mass spectra by a 4800 Proteomics Analyzer (Applied Biosystems), and MS-based protein homology identification based on The Arabidopsis Information Resource 8.0 genomic database, we refer to Van Leene et al. (2010). Experimental background proteins were subtracted based on ∼40 TAP experiments on wild-type cultures and cultures expressing the TAP-tagged mock proteins β‑glucuronidase, red fluorescent protein, and green fluorescent protein (Van Leene et al., 2010).
Yeast Two-Hybrid Experiments
Yeast two-hybrid constructs encompassing CDT1a and CDT1b were obtained by recombination using Gateway technology. Gateway-compatible yeast two-hybrid vectors were derived from the pGADT7 and pGBKT7 vectors and were a gift from Pascale Rossignol (John Innes Centre, Norwich, UK). The DPB2 cDNA was amplified by PCR using the Pol ε EcoRI dir and Pol ε XhoI stop primers and cloned between the EcoRI and XhoI sites of pENTR1A. Yeast two-hybrid constructs were obtained by recombination. Yeast transformation and interaction assays were performed as described previously (Raynaud et al., 2005).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CDT1a, At2g31270; CDT1b, At3g54710; DPB2, At5g22110; POL2A, At1g08260; CYCB1;1, At4g37490; PP2AA3, At1g13320; Ku70, At1g48050; XRCC4, At3g23100; PARP2, At2g31320; RAD51, At5g20850; BRCA1, At4g21070.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Position of the T-DNA Insertion in the GABI_025G08 Line.
Supplemental Figure 2. DNA Content in Pollen Grains Isolated from a Whole Inflorescence of a Col-0 Plant.
Supplemental Figure 3. Altered DNA Content in Pollen Grains Isolated from cdt1a/+cdt1b Mutants.
Supplemental Figure 4. Endoreduplication Is Increased in cdt1a/+ and cdt1a/+cdt1b Mutants.
Supplemental Figure 5. Endoreduplication Is Enhanced in CDT1-RNAi Plants in a WEE1- and SOG1-Independent Manner.
Supplemental Figure 6. Cellular Response to DNA Damage Is Normal in CDT1-RNAi Plants.
Supplemental Table 1. Detailed Results for TAP Analysis.
Supplemental Table 2. Primer Sequences.
Supplemental Methods 1. Immunofluorescence.
Supplementary Material
Acknowledgments
We thank Martine Devic (Institut de Recherche pour le Développement, Montpellier, France) for kindly providing us yeast two-hybrid constructs encompassing fragments of POL2A and Pascale Rossignol (John Innes Centre, Norwich, UK) for kindly providing us Gateway-compatible yeast two-hybrid vectors. We are especially grateful to Daniel Vezon (Institut National de Recherche Agronomique, Versailles, France) for helpful advice on gametophyte development analysis and to Spencer Brown (Institut des Sciences du Végétal, Gif-sur-Yvette cedex, France) for assistance with flow cytometry experiments. We thank two anonymous reviewers for constructive remarks on the manuscript. This work was supported by the Agence Nationale de la Recherche of France (Grant ANR 2010 JCJC1207 01).
AUTHOR CONTRIBUTIONS
S.D. and M.B. designed and performed the research, G.D.J. and E.V.D.S. performed the research and analyzed the data, S.B. and M.B. performed the research, L.D.V. and C.B. wrote the article, and C.R. designed and performed the research and wrote the article.
Glossary
- HA
hemagglutinin
- DAPI
4′,6-diamidino-2-phenylindole
- Col-0
ecotype Columbia
- HU
hydroxyurea
- MS
Murashige and Skoog
- TAP
tandem affinity purification
References
- Adachi S., et al. (2011). Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Baranczewski P., Nehls P., Rieger R., Rajewsky M.F., Schubert I. (1997). Removal of O6-methylguanine from plant DNA in vivo is accelerated under conditions of clastogenic adaptation. Environ. Mol. Mutagen. 29: 400–405 [DOI] [PubMed] [Google Scholar]
- Bensimon A., Aebersold R., Shiloh Y. (2011). Beyond ATM: The protein kinase landscape of the DNA damage response. FEBS Lett. 585: 1625–1639 [DOI] [PubMed] [Google Scholar]
- Berger F., Twell D. (2011). Germline specification and function in plants. Annu. Rev. Plant Biol. 62: 461–484 [DOI] [PubMed] [Google Scholar]
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006). An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Caro E., Castellano M.M., Gutierrez C. (2007). A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447: 213–217 [DOI] [PubMed] [Google Scholar]
- Castellano M.d.M., Boniotti M.B., Caro E., Schnittger A., Gutierrez C. (2004). DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16: 2380–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano M.M., del Pozo J.C., Ramirez-Parra E., Brown S., Gutierrez C. (2001). Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13: 2671–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J.M., MacAlpine D.M., Evans J.G., Bell S.P., Orr-Weaver T.L. (2002). Visualization of replication initiation and elongation in Drosophila. J. Cell Biol. 159: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cools T., De Veylder L. (2009). DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 12: 23–28 [DOI] [PubMed] [Google Scholar]
- Cools T., Iantcheva A., Weimer A.K., Boens S., Takahashi N., Maes S., Van den Daele H., Van Isterdael G., Schnittger A., De Veylder L. (2011). The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas C., Sanchez M.d.L., Sequeira-Mendes J., Gutierrez C. (2011). Progress in understanding DNA replication control. Plant Sci. 181: 203–209 [DOI] [PubMed] [Google Scholar]
- Culligan K.M., Robertson C.E., Foreman J., Doerner P., Britt A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48: 947–961 [DOI] [PubMed] [Google Scholar]
- De Marco V., et al. (2009). Quaternary structure of the human Cdt1-geminin complex regulates DNA replication licensing. Proc. Natl. Acad. Sci. USA 106: 19807–19812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M.L. (2003). The ‘ORC cycle’: A novel pathway for regulating eukaryotic DNA replication. Gene 310: 1–15 [DOI] [PubMed] [Google Scholar]
- De Schutter K., Joubès J., Cools T., Verkest A., Corellou F., Babiychuk E., Van Der Schueren E., Beeckman T., Kushnir S., Inzé D., De Veylder L. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Inzé D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665 [DOI] [PubMed] [Google Scholar]
- De Veylder L., Larkin J.C., Schnittger A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Domenichini S., Raynaud C., Ni D.A., Henry Y., Bergounioux C. (2006). Atmnd1-delta1 is sensitive to gamma-irradiation and defective in meiotic DNA repair. DNA Repair (Amst.) 5: 455–464 [DOI] [PubMed] [Google Scholar]
- Ebel C., Mariconti L., Gruissem W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Fulcher N., Sablowski R. (2009). Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl. Acad. Sci. USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Maddox J.M., Ayres N.M., Sharma D.P., Firoozabady E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 512–527 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Kisielewska J., Blow J.J. (2012). Dynamic interactions of high Cdt1 and geminin levels regulate S phase in early Xenopus embryos. Development 139: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Li C., Wang H., Chen H., Berg H., Xia Y. (2011). AtPPR2, an Arabidopsis pentatricopeptide repeat protein, binds to plastid 23S rRNA and plays an important role in the first mitotic division during gametogenesis and in cell proliferation during embryogenesis. Plant J. 67: 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Maiorano D., Méchali M. (2006). A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus. EMBO J. 25: 5764–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard J.R., Lipkin-Moore Z., Sheu Y.J., Stillman B., Burgers P.M., Haber J.E. (2010). Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 24: 1133–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H.P., Ramos G.B., de Almeida-Engler J., Cabral L.M., Coqueiro V.M., Macrini C.M., Ferreira P.C., Hemerly A.S. (2004). Genome based identification and analysis of the pre-replicative complex of Arabidopsis thaliana. FEBS Lett. 574: 192–202 [DOI] [PubMed] [Google Scholar]
- Nam E.A., Cortez D. (2011). ATR signalling: More than meeting at the fork. Biochem. J. 436: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni D.A., Sozzani R., Blanchet S., Domenichini S., Reuzeau C., Cella R., Bergounioux C., Raynaud C. (2009). The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytol. 184: 311–322 [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C., Schnittger A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67 [DOI] [PubMed] [Google Scholar]
- Petropoulou C., Kotantaki P., Karamitros D., Taraviras S. (2008). Cdt1 and geminin in cancer: Markers or triggers of malignant transformation? Front. Biosci. 13: 4485–4494 [DOI] [PubMed] [Google Scholar]
- Pursell Z.F., Isoz I., Lundström E.B., Johansson E., Kunkel T.A. (2007). Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell Z.F., Kunkel T.A. (2008). DNA polymerase epsilon: A polymerase of unusual size (and complexity). Prog. Nucleic Acid Res. Mol. Biol. 82: 101–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Perennes C., Reuzeau C., Catrice O., Brown S., Bergounioux C. (2005). Cell and plastid division are coordinated through the prereplication factor AtCDT1. Proc. Natl. Acad. Sci. USA 102: 8216–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A., Guilleminot J., Lincker F., Gadea-Vacas J., Delorme V., Bechtold N., Pelletier G., Delseny M., Chabouté M.E., Devic M. (2005). Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J. 44: 223–236 [DOI] [PubMed] [Google Scholar]
- Roukos V., Kinkhabwala A., Colombelli J., Kotsantis P., Taraviras S., Nishitani H., Stelzer E., Bastiaens P., Lygerou Z. (2011). Dynamic recruitment of licensing factor Cdt1 to sites of DNA damage. J. Cell Sci. 124: 422–434 [DOI] [PubMed] [Google Scholar]
- Scholl R.L., May S.T., Ware D.H. (2000). Seed and molecular resources for Arabidopsis. Plant Physiol. 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer P.S., Holding D.R., Groover A., Yordan C., Martienssen R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G(1) phase and is required maternally for early Arabidopsis development. Development 127: 1815–1822 [DOI] [PubMed] [Google Scholar]
- Springer P.S., McCombie W.R., Sundaresan V., Martienssen R.A. (1995). Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268: 877–880 [DOI] [PubMed] [Google Scholar]
- Truong L.N., Wu X. (2011). Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol 3: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyama T., Watanabe S., Aoki A., Cho Y., Seki M., Enomoto T., Tada S. (2009). Repression of nascent strand elongation by deregulated Cdt1 during DNA replication in Xenopus egg extracts. Mol. Biol. Cell 20: 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Van Leene J., Witters E., Inzé D., De Jaeger G. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Xouri G., Dimaki M., Bastiaens P.I., Lygerou Z. (2007). Cdt1 interactions in the licensing process: A model for dynamic spatiotemporal control of licensing. Cell Cycle 6: 1549–1552 [DOI] [PubMed] [Google Scholar]
- Yang W.C., Shi D.Q., Chen Y.H. (2010). Female gametophyte development in flowering plants. Annu. Rev. Plant Biol. 61: 89–108 [DOI] [PubMed] [Google Scholar]
- Yoshiyama K., Conklin P.A., Huefner N.D., Britt A.B. (2009). Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 106: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Teng C., Liang Y. (2011). Programmed cell death may act as a surveillance mechanism to safeguard male gametophyte development in Arabidopsis. Protein Cell 2: 837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.