This study provides insight into the evolution of flowering plants by demonstrating that a mechanism that modulates carpel margin development in Arabidopsis was recruited from light-regulated processes.
Abstract
A key innovation of flowering plants is the female reproductive organ, the carpel. Here, we show that a mechanism that regulates carpel margin development in the model flowering plant Arabidopsis thaliana was recruited from light-regulated processes. This recruitment followed the loss from the basic helix-loop-helix transcription factor SPATULA (SPT) of a domain previously responsible for its negative regulation by phytochrome. We propose that the loss of this domain was a prerequisite for the light-independent expression in female reproductive tissues of a genetic module that also promotes shade avoidance responses in vegetative organs. Striking evidence for this proposition is provided by the restoration of wild-type carpel development to spt mutants by low red/far-red light ratios, simulating vegetation shade, which we show to occur via phytochrome B, PHYTOCHROME INTERACTING FACTOR4 (PIF4), and PIF5. Our data illustrate the potential of modular evolutionary events to generate rapid morphological change and thereby provide a molecular basis for neo-Darwinian theories that describe this nongradualist phenomenon. Furthermore, the effects shown here of light quality perception on carpel development lead us to speculate on the potential role of light-regulated mechanisms in plant organs that, like the carpel, form within the shade of surrounding tissues.
INTRODUCTION
The flowering plants, or angiosperms, arose suddenly from an unknown ancestor in the early Cretaceous and rapidly became the dominant form of terrestrial vegetation. Angiosperms reproduce by means of the flower, whose principal defining feature is the carpel. This novel female reproductive organ encloses the ovules, providing numerous benefits in reproductive efficiency over the naked ovules typically present in the remaining seed plants or gymnosperms (Scutt et al., 2006). A number of genes are known to regulate the development of the two fused carpels that make up the gynoecium, or female floral whorl, in the model angiosperm Arabidopsis thaliana. Among these, the basic helix-loop-helix (bHLH) transcription factor SPATULA (SPT) (Heisler et al., 2001) plays a key role in regulating the development of the stigma, style, and septum, which emerge from the carpel margins to close the gynoecium and provide a route for pollen tube growth (Alvarez and Smyth, 1999). In spt mutants, these tissues show reduced cell elongation and lack extracellular matrix-secreting cells, which make up the pollen transmitting tissue of the style and septum (Alvarez and Smyth, 2002). Expression of SPT in the gynoecium is limited to carpel margin tissues (Heisler et al., 2001; Groszmann et al., 2010), where its effects are thus cell autonomous. However, misexpression of SPT produces no marked developmental effects in the gynoecium, which might be explained by the hypothesis that SPT functions as part of a heterodimer with the largely redundant bHLH factors HECATE1-3 (HEC1-3), whose expression in the gynoecium is also limited to the carpel margins (Gremski et al., 2007). In addition to its role in gynoecium development, SPT regulates seed dormancy (Penfield et al., 2005), leaf and cotyledon expansion (Ichihashi et al., 2010; Josse et al., 2011), and vegetative growth in response to temperature (Sidaway-Lee et al., 2010). SPT also plays a minor role (Girin et al., 2011; Groszmann et al., 2011), partially redundantly with its putative paralog ALCATRAZ (ALC) (Rajani and Sundaresan, 2001), in the development of fruit dehiscence zones. SPT and ALC occur within regions of the Arabidopsis genome that are derived from a large-scale duplication event that occurred relatively recently, after the separation of the Brassicaceae and Caricaceae lineages (Franzke et al., 2011; Groszmann et al., 2011).
SPT and ALC fall phylogenetically within Group 15 of the bHLH family of transcription factors (Toledo-Ortiz et al., 2003), most members of which function in light-regulated processes through interactions with the red/far-red (R/FR) reversible photoreceptor, phytochrome. These light-regulated factors, termed PHYTOCHROME INTERACTING FACTORs (PIFs) and/or PHYTOCHROME INTERACTING FACTOR 3-LIKE proteins (PILs), possess an active phytochrome binding (APB) domain that is capable of interacting specifically with the active Pfr form of phytochrome that predominates at high R/FR ratios (Khanna et al., 2004). This interaction leads to the phosphorylation and targeting of several PIFs for destruction in the proteasome (Bauer et al., 2004; Shen et al., 2005; Lorrain et al., 2008).
Several lines of evidence indicate close links between SPT and light-regulated processes operating through phytochrome B (phyB) and PIFs. For example, phyB loss-of-function mutations suppress the spt phenotype in the gynoecium (Foreman et al., 2011), while SPT overexpression disrupts light signaling in seedlings, generating a long hypocotyl response identical to that observed in phyB mutants (Penfield et al., 2005). In addition, both SPT and PIF1 (PIL5) repress seed germination by regulating GA3 oxidase transcription (Penfield et al., 2005). Importantly, the promotion of cell elongation by SPT in the gynoecium (Alvarez and Smyth, 2002) parallels the effect of PIF4 and PIF5 (PIL6), which control shade avoidance responses in vegetative tissues (Lorrain et al., 2008). These growth responses are triggered by the low R/FR light ratios, perceived mainly by phyB, that characterize vegetation shade (Martinez-Garcia et al., 2010). Both SPT and shade avoidance mechanisms are closely linked to hormone signaling. For example, SPT has been proposed to promote and/or respond to an auxin peak that forms at the gynoecium apex (Nemhauser et al., 2000; Ståldal and Sundberg, 2009), while many auxin responsive genes are also known to be rapidly upregulated during shade avoidance responses (Martinez-Garcia et al., 2010). In addition, both SPT (Josse et al., 2011) and PIF4 (de Lucas et al., 2008) are negatively regulated by direct interactions with DELLA proteins that are themselves negatively regulated by gibberellin.
In this work, we investigate the mechanistic and evolutionary link between SPT-regulated carpel development and shade avoidance. We begin using phylogenetic analyses to demonstrate that SPT evolved from a light-regulated transcription factor, similar to present-day PIFs, by the loss of an APB domain. We then show that SPT shares a common set of target genes with light-regulated transcription factors and that several of these targets regulate carpel development as they do shade avoidance. We furthermore provide striking evidence for the link between SPT and shade avoidance by the restoration of wild-type gynoecium development to spt mutants grown under low R/FR ratios that simulate vegetation shade. We use a series of multiple mutants to demonstrate that this phenotypic restoration, which involves the upregulation of SPT targets, occurs via PHYB, PIF4, and PIF5. We discuss our observations in the context of the rapid evolutionary change that generated the flowering plants and also speculate on the potential role of light-regulated mechanisms in plant organs that, like the carpel, form within the shade of surrounding tissues.
RESULTS
SPT Evolved from a PIF by the Loss of an APB-Like Domain
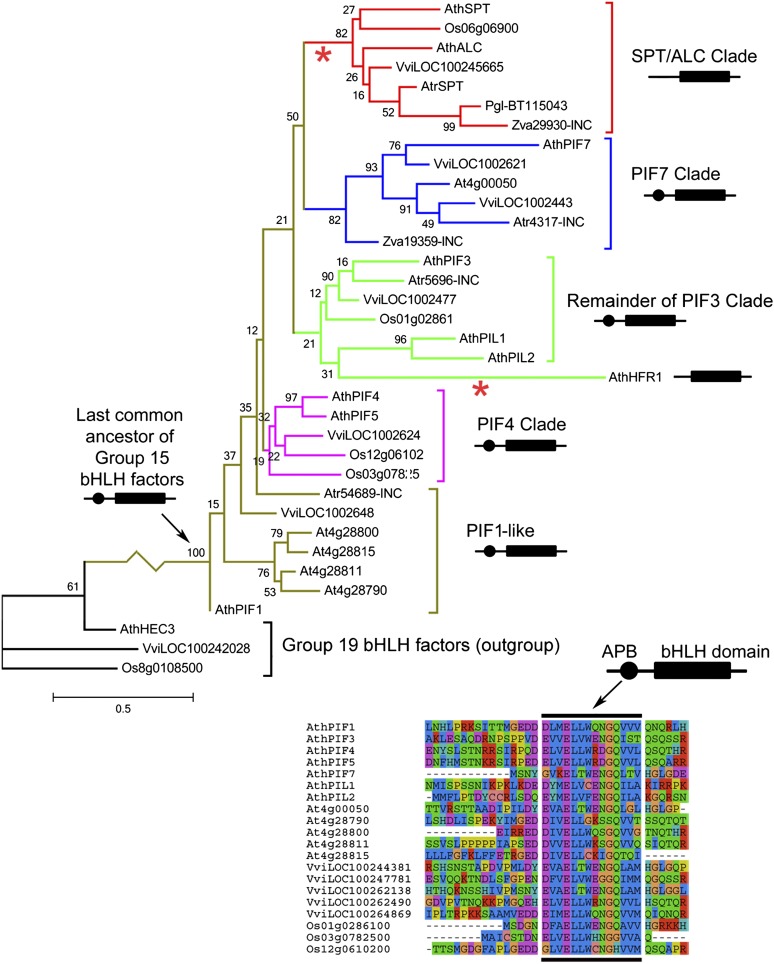
To investigate the evolution of SPT, we compiled a comprehensive set of Group 15 bHLH genes from the sequenced genomes of Arabidopsis (rosid eudocots), grape (Vitis vinifera) (asterid eudicots), and rice (Oryza sativa) (monocots). We also identified homologous ESTs from the ANA-grade (ANA for Amborellales-Nymphaeales-Austrobaileyales) angiosperm Amborella trichopoda, which is the probable sister to all other extant flowering plants, and conducted a comprehensive search for Group 15 bHLH sequences from gymnosperm sequence databases. This data set was completed by the addition of three HEC-like, Group 19 bHLH genes from Arabidopsis, grape, and rice to form an outgroup.
In maximum likelihood phylogenies (Figure 1; see Supplemental Figure 1 online) performed using both nucleic acid and amino acid alignments (see Supplemental Data Set 1 online), SPT/ALC-like sequences form a well-supported clade in a sister position to a clade of PIF7-like sequences. The SPT/ALC clade is thus rooted within an overall clade of Group 15 bHLH sequences, though the six individual nodes separating the SPT/ALC clade from the base of the Group 15 clade were not supported by high bootstrap values (which vary from 12 to 50% in Figure 1 and from 13 to 41% in Supplemental Figure 1 online). To confirm the position of the SPT/ALC clade as emerging from within the remaining Group 15 bHLH sequences, we examined the topologies of the first 200 individual bootstrap trees used to support the phylogeny shown in Figure 1. Of these, 158 trees (79%) contained an intact SPT/ALC clade, in close agreement with the bootstrap value for this clade of 82% (Figure 1), which was based on all 1000 bootstrap replicates performed. In 152 of these 158 trees, the SPT/ALC clade was rooted within an overall clade of Group 15 bHLH genes, rather than as sister to a remaining clade of Group 15 genes or as part of a basally diverging polytomy of Group 15 bHLH genes. Our analysis thus gives a high level (152/200 = 76%) of support for the hypothesis that the SPT/ALC clade shown in Figure 1 arose from within an overall clade of Group 15 bHLH genes rather than the hypothesis that this clade forms a sister group to one or more remaining clades of Group 15 bHLH genes (6/200 = 3% support) or that it does not form a monophyletic clade (42/200 = 21% support). This conclusion is in general agreement with previous phylogenies that focused on bHLH sequences from Arabidopsis (Toledo-Ortiz et al., 2003; Khanna et al., 2004).
Figure 1.
Phylogenetic Analysis of Group 15 bHLH Sequences Showing Loss of the APB Domain in the SPT/ALC Lineage.
A maximum likelihood phylogeny from the nucleic acid alignment in Supplemental Data Set 1A online. The percentage bootstrap support values are shown at the nodes. N-terminal APB-like domains are indicated by black circles in schematic diagrams of PIF proteins, and an alignment of these domains, where present, is shown beneath the phylogeny. The inferred positions of two losses of APB-like domains are indicated on the phylogeny by asterisks. Three HEC-like genes from Group 19 of the bHLH family are included in the analysis as an outgroup. Species of origin are indicated by the prefixes: At or Ath for Arabidopsis thaliana, Atr for Amborella trichopoda, Os for Oryza sativa, Pgl for Picea glauca, Vvi for Vitis vinifera, and Zva for Zamia vasquezii. ESTs that are incomplete at their 5′-terminus, and for which the presence of an N-terminal ABP-like domain is therefore unknown, are indicated by the suffix “INC.” The branch lengths and scale bar indicate the mean number of nucleic acid substitutions per site.
Sequences from the ANA-grade angiosperm Amborella and from two gymnosperms, Picea and Zamia, occur within the SPT/ALC clade, strongly supporting the view that its root lineage separated through gene duplication (probably from that of the PIF7 clade) before the last common ancestor of all seed plants. The Arabidopsis genes LONG HYPOCOTYL IN FAR RED1 (HFR1), PIL1, and PIL2, for which probable orthologs in the other species analyzed could not be conclusively identified, showed unstable positions in our phylogenetic analyses (cf. Figure 1 and Supplemental Figure 1 online), possibly due to recent and rapid evolution of these sequences. Inclusion in phylogenies of species close to Arabidopsis could be used to better investigate the phylogenetic placement of these three genes.
Alignment of Group 15 bHLH sequences indicates the presence of an APB-like domain near the N terminus of all full-length proteins analyzed in this study (Figure 1), except those of the SPT/ALC clade and HFR1, which is a negative regulator of shade avoidance responses. We therefore conclude that an APB-like domain was present in the last common ancestor of Group 15 bHLH genes from angiosperms and gymnosperms but was lost in the SPT/ALC and HFR1 lineages. The absence of an APB domain in SPT/ALC pro-orthologs (Sharman, 1999) throughout the flowering plants, including the basally diverging A. trichopoda, provides strong evidence for the loss of this domain before the last common ancestor of the flowering plants. Furthermore, the absence of this domain in an SPT/ALC pro-ortholog of the gymnosperm Picea glauca suggests that it was probably lost before the last common ancestor of all seed plants, as indicated by asterisks in Figure 1 and Supplemental Figure 1 online. If the APB domain was lost before the radiation of the extant seed plants, then this loss could not have made an immediate contribution to the origin carpel in the angiosperm lineage. However, such an early loss of the APB domain could have been a prerequisite for the subsequent origin of the developmental roles of SPT in the carpel and in other angiosperm tissues.
SPT Regulates Gynoecium Development by Activating Genes Involved in Shade Avoidance
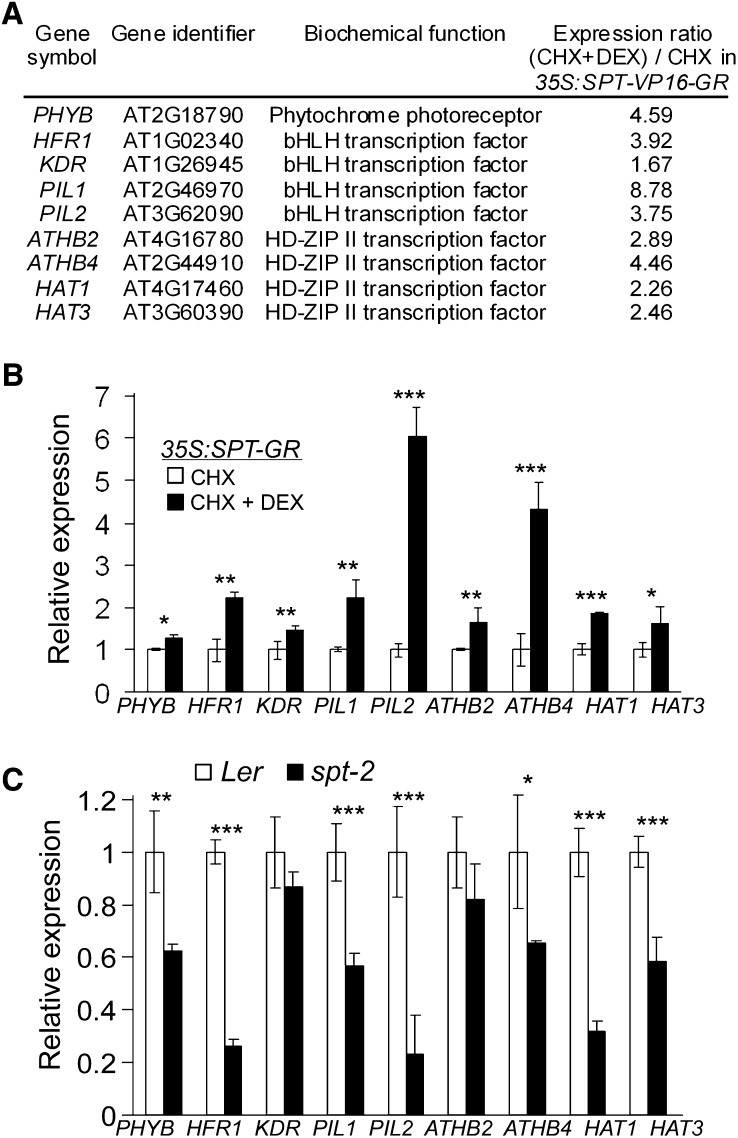
To identify the targets of SPT, we generated transgenic plants in which the translocation to the nucleus of constitutively produced recombinant SPT protein, fused to the VP16 transcriptional activation domain and the hormone binding domain of the rat glucocorticoid receptor (GR; 35S:SPT-VP16-GR plants), could be induced by application of the hormone analog dexamethasone (DEX). The VP16 domain was used in constructs to generate changes to SPT target gene expression, even if the expression of those targets in wild-type plants was limited by some other factor than SPT. A similar strategy had previously proved necessary for the investigation of LEAFY targets (Busch et al., 1999). To limit the genes identified to be probable immediate targets of SPT, both DEX and mock treatments were performed in the presence of cycloheximide (CHX) to block protein synthesis. Gene expression changes were monitored 2 h after SPT induction in three independent experiments using Complete Arabidopsis Transcriptome Microarrays, and the results of these were compared with a list of 24 putative immediate SPT targets (see Supplemental Data Set 2 online, derived from the complete dataset available at GEOmnibus, accession number GSE12913). Interestingly, nine of the targets identified (Figure 2A) are known to be involved in shade avoidance, including PHYB and four transcription factors from each of the bHLH (Salter et al., 2003; Hyun and Lee, 2006; Roig-Villanova et al., 2006) and Homeodomain-Leu Zipper Class II (HD-ZIP II) families (Steindler et al., 1999; Roig-Villanova et al., 2006; Ciarbelli et al., 2008; Sorin et al., 2009). Of these factors, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2 (ATHB2) is a positive effector of shade avoidance (Steindler et al., 1999), while HFR1 acts to prevent exaggerated shade avoidance responses (Sessa et al., 2005) by forming inactive complexes with PIF4 and PIF5 (Hornitschek et al., 2009). KIDARI may act as a positive effector of shade avoidance through a specific negative interaction with HFR1 (Hyun and Lee, 2006). A further six of the SPT targets identified here (listed as “hormone-related genes” in Supplemental Data Set 2 online) are directly connected with auxin or brassinosteroid signaling, which are known to be involved in shade avoidance and may thus represent further mechanistic links between this process and SPT (Carabelli et al., 2007; Roig-Villanova et al., 2007; Garcia et al., 2008; Tao et al., 2008).
Figure 2.
Induction by SPT of Light-Regulated Genes Associated with Shade Avoidance.
(A) Shade avoidance genes identified as SPT targets in microarray experiments and their levels of induction in inflorescence tissues of 35S:SPT-VP16-GR transformants following treatment with CHX+DEX, compared with CHX-treated controls.
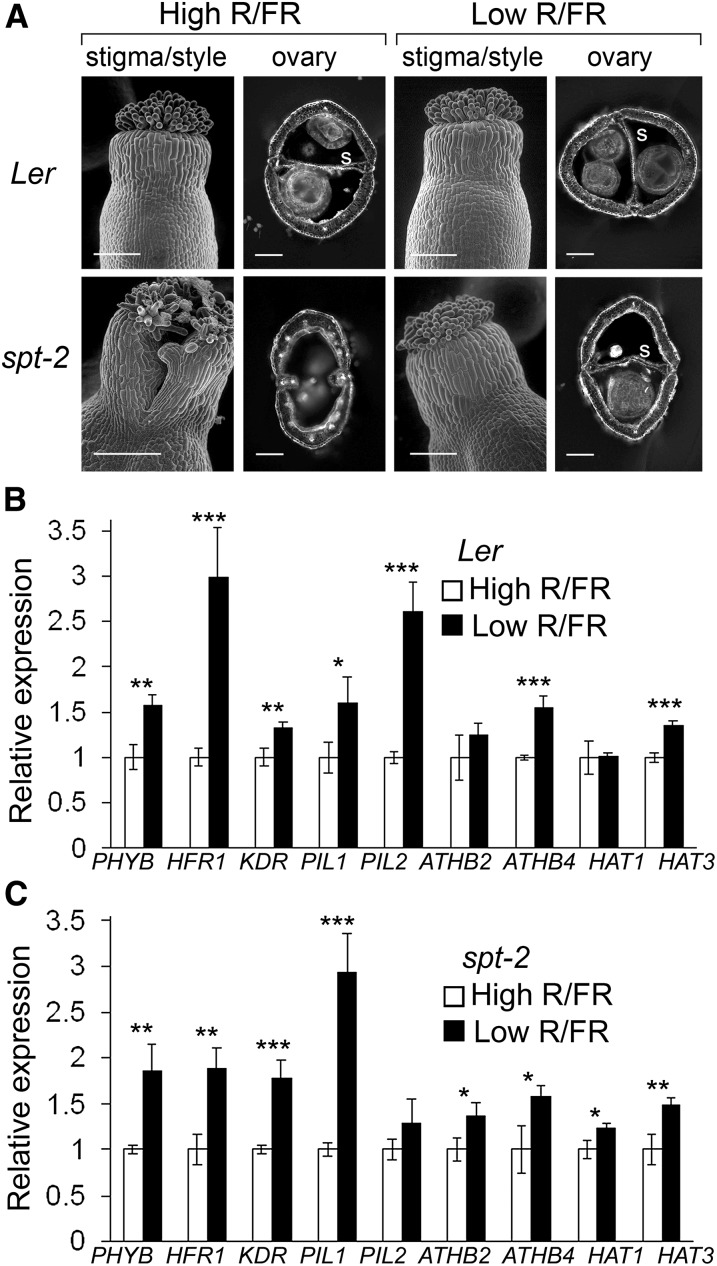
(B) qRT-PCR analysis showing induction of SPT targets in inflorescence tissues of 35S:SPT-GR transformants following treatment with CHX+DEX, compared with CHX-treated controls. Expression levels were normalized using GAPDH expression. Four technical replicates were performed, and sd error bars are shown. Asterisks indicate significant expression differences in t tests at *P < 0.05, **P < 0.01, and ***P < 0.001.
(C) qRT-PCR analyses showing upregulation of SPT targets in wild-type (Landsberg erecta [Ler]) compared with spt-2 inflorescences. Expression levels were normalized using GAPDH expression. Four technical replicates were performed, and sd error bars are shown. Asterisks indicate significant differences in t tests at *P < 0.05, **P < 0.01, and ***P < 0.001.
In this work, we focus on the nine SPT targets (Figure 2A) that are directly implicated in shade avoidance mechanisms. To determine whether SPT induction had a positive or negative effect on the transcription of these targets in the absence of the VP16 transcriptional activation domain, we performed quantitative RT-PCR (qRT-PCR) analysis on transgenic plants containing a 35S:SPT-GR construct. The functionality of this construct was verified in an independent experiment involving the restoration of wild-type gynoecium development to transgenic spt-2 mutants, which show a strong loss-of-function phenotype, following DEX application (see Supplemental Figure 2 online). Expression of the nine SPT targets analyzed in 35S:SPT-GR plants (in the Columbia-0 [Col-0] background) increased significantly following DEX treatment (Figure 2B), and seven of these genes were also significantly upregulated in the wild type compared spt-2 inflorescences (Figure 2C), confirming that SPT activates their transcription.
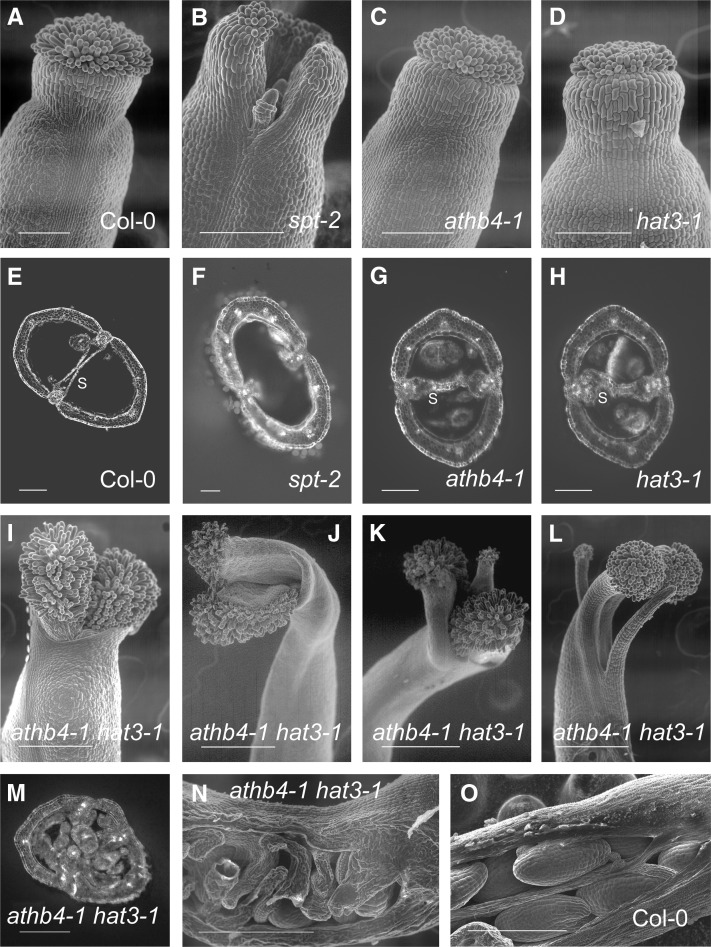
The identification of known mediators of shade avoidance as SPT targets suggested that these genes may also function in gynoecium development. We used a mutant-based approach to test this hypothesis for the HD-ZIP II genes ATHB4 and HAT3 (Figure 3), which redundantly regulate shade avoidance responses (Sorin et al., 2009). Though both athb4 (Figures 3C and 3G) and hat3 (Figures 3D and 3H) single mutants showed gynoecium development as for wild-type plants (Figures 3A and 3E), the athb4 hat3 double mutant showed incomplete apical fusion in the gynoecium (Figures 3I to 3L) and severely reduced development of the septum (Figures 3M and 3N), as do spt mutants (Figures 3B and 3F). Further effects in athb4 hat3 double mutants, compared with wild-type plants (Figures 3A, 3E, and 3O), included branching of the gynoecium apex into multiple processes (Figures 3K and 3L) and highly disorganized ovule development (Figures 3M and 3N). Thus, the athb4 hat3 phenotype is consistent with a role for these genes as SPT targets in the development of carpel margin tissues, though they clearly also function in other developmental processes in the gynoecium. Interestingly, a pronounced gynoecium phenotype has recently been described for jaiba mutants, which are affected in another of the SPT targets identified here, HAT1 (Zúñiga-Mayo et al., 2012).
Figure 3.
Similarity of Gynoecium Phenotypes between spt Single and athb4 hat3 Double Mutants.
Gynoecium phenotypes of wild-type Col-0 plants ([A], [E], and [O]); spt-2 ([B] and [F]), athb4-1 ([C] and [G]), and hat3-1 ([D] and [H]) single mutants; and athb4-1 hat3-1 double mutants ([I] to [N]). Images in (I) to (L) show an increasing range of severity in athb4-1 hat3-1 phenotypes. All ovary sections are transverse, except longitudinal in (N) and (O). s, septum. Bars = 100 µm in stigma/style scanning electron microscopy images ([A] to [D] and [I] to [L]) and 200 µm in ovary sections ([E] to [H] and [M] to [O]).
SPT Regulates Its Targets by Binding to G-Box Motifs
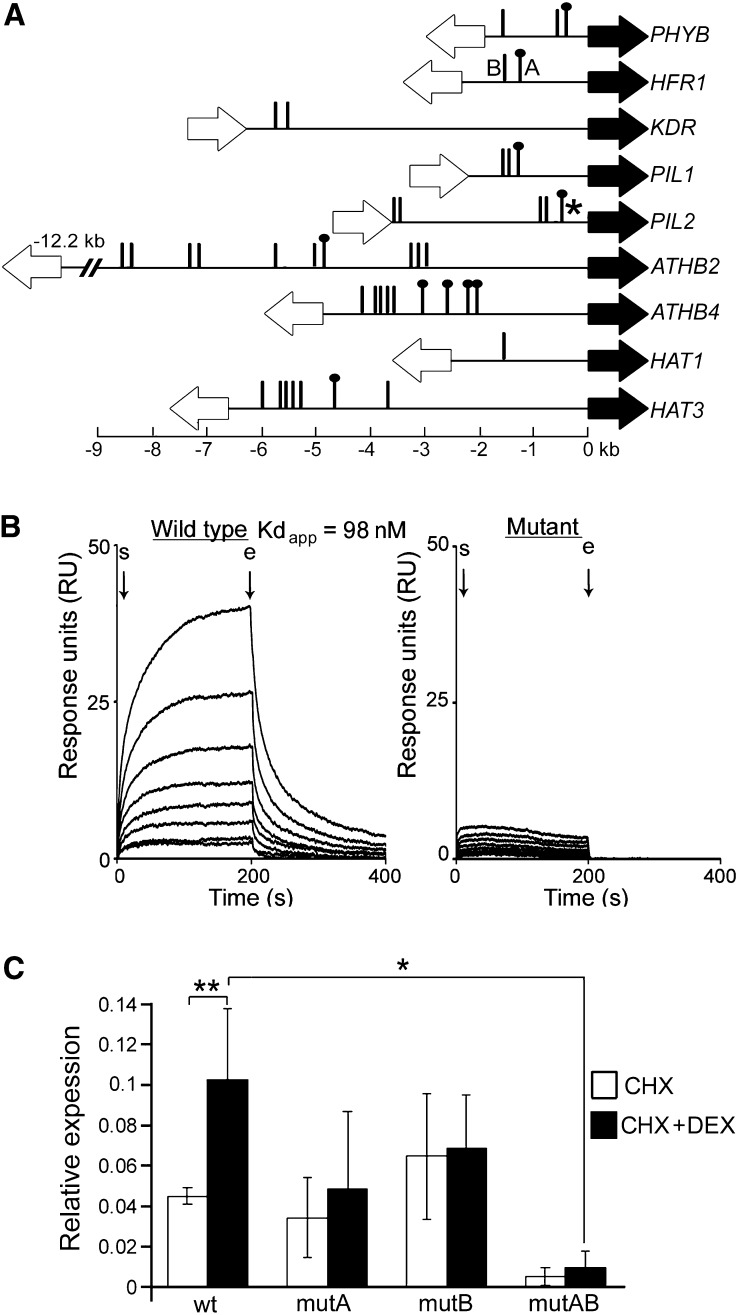
Comparison of the nine SPT targets known for their roles in shade avoidance revealed the presence of multiple G-box motifs (CACGTG) in each of their 5′-flanking sequences (Figure 4A). In seven of these targets, between one and four of the G-boxes present occur in the context of an 8-bp motif containing an additional two G-residues on one DNA strand (CACGTGGG), which might suggest binding to a heterodimeric complex of transcription factors. Multiple G-box motifs occur also in the promoters of the remaining SPT targets identified in this study (see Supplemental Data Set 2 online).
Figure 4.
Interactions of SPT with G-Box Motifs in Shade Avoidance Gene Promoters.
(A) Positions of G-boxes (CACGTG; vertical lines) and extended G-boxes (CACGTGGG; dotted vertical lines) in the promoters of SPT targets. Coding sequences of SPT targets and their adjacent upstream genes are shown as closed and open arrows, respectively. G-boxes in target gene promoters that are analyzed experimentally in (B) and (C) are indicated by an asterisk, and by the letters A and B, respectively.
(B) SPR analyses showing specific binding of recombinant SPT to a wild-type PIL2 promoter fragment containing the G-box shown by an asterisk in Figure 4A and lack of binding to a control fragment in which the central two positions of the G-box had been mutated (CAATTG). The start (s) and end (e) points of protein injection are indicated with arrows. Protein concentrations vary in equal proportional increments from 39 to 394 nM from lowest to highest interaction curves. The equilibrium constant for dissociation (Kd) is shown for the wild-type interaction.
(C) qRT-PCR analysis of pHFR1:mVENUS reporter gene expression in inflorescence tissues of transformants also containing a 35S:SPT-GR construct, following induction of SPT activity by CHX+DEX treatment and in CHX-treated controls. Four reporter constructs analyzed (wild type [wt], mutA, mutB, and mutAB) contained all possible combinations of wild-type and mutated G-boxes at sites A and B shown in Figure 4A. This figure is a summary of the complete data set shown in Supplemental Figure 3A online, in which each bar represents the mean expression of the mVENUS reporter, normalized to GAPDH expression, in four independent transgenic lines analyzed for each construct tested. The error bars shown were calculated as the root mean squares of the sd values for each group of four transgenic lines analyzed (see Supplemental Figure 3A online). Asterisks indicate significant differences in paired t tests between treatments or in unpaired t tests between genotypes following CHX +DEX treatment, at *P < 0.05 and **P < 0.01.
We used surface plasmon resonance (SPR) analysis to test the in vitro interaction of recombinant SPT protein with the G-box closest to the coding sequence in the SPT target PIL2 (Figure 4A). SPT bound this promoter fragment with high affinity and showed no binding following mutation of the central two positions of the G-box (Figure 4B), indicating a specific interaction with the wild-type G-box motif.
We furthermore tested whether G-box motifs are necessary for the in vivo induction of SPT targets in inflorescence tissues. For these studies, we focused on the SPT target HFR1, as the promoter region of this gene contains only two G-boxes (Figure 4A). We generated a series of pHFR1:mVENUS constructs containing all four possible combinations of wild-type and mutated G-boxes and introduced these separately into an Arabidopsis line containing a DEX-inducible 35S:SPT-GR construct. Wild-type HFR1 promoter activity was significantly induced by CHX +DEX treatment compared with CHX-treated controls, though the activity of HFR1 promoters in which one or both G-boxes had been mutated was not significantly increased by CHX +DEX treatment (Figure 4C, summarizing results obtained for individual lines presented in Supplemental Figure 3A online). Mutation of both of the G-boxes present led to a significant reduction in HFR1 promoter activity in CHX +DEX-treated plants (Figure 4C). These results indicate that the activation by SPT of HFR1 occurs through both of the G-boxes present in the HFR1 promoter. SPT thus appears to interact physically with the G-box motifs present in its target genes, and these interactions are necessary for the positive regulation of these targets by SPT.
Simulated Vegetation Shade Restores Wild-Type Gynoecium Development to spt Mutants by Acting through PHYB, PIF4, and PIF5
The identification of genes that mediate shade avoidance as SPT targets suggested that gynoecium development might be influenced by light quality. In agreement with this possibility, we found that low R/FR ratios, characteristic of vegetation shade, restored wild-type gynoecium development to spt-2 mutants (Figure 5A). Transcript levels of seven and eight of the nine SPT targets previously analyzed were significantly increased under low R/FR conditions in wild-type and spt-2 mutant inflorescences, respectively (Figures 5B and 5C), consistent with a role for these genes in the complementation of the spt mutant phenotype by simulated vegetation shade. In addition, we found that the two G-boxes in the HFR1 promoter that are necessary for the induction of HFR1 expression by SPT (Figure 4C; see Supplemental Figure 3A online) are similarly necessary for the upregulation of this gene in the leaves of plants grown under low R/FR ratios (see Supplemental Figure 3B online).
Figure 5.
Effects of Light Quality on the spt Mutant Phenotype and Expression of SPT Targets.
(A) Restoration of wild-type gynoecium development to spt-2 mutants grown under low R/FR ratios. Ler, Landsberg erecta; s, septum. Bars = 100 µm in stigma/style scanning electron microscopy images and 200 µm in ovary transverse sections.
(B) and (C) qRT-PCR analyses showing the upregulation of SPT targets in inflorescence tissues of wild-type Ler (B) and spt-2 mutant (C) plants grown under low R/FR ratios. Four technical replicates were performed, and sd error bars are shown. Asterisks indicate significant differences in t tests at *P < 0.05, **P < 0.01, and ***P < 0.001.
The genetic interaction between SPT and PHYB recently reported by Foreman et al. (2011) has been independently confirmed in our studies. Accordingly, the mutational inactivation of PHYB restores wild-type gynoecium development to spt-2 mutants (Figure 6A). These data indicate phyB to be the principal photoreceptor mediating the effect, demonstrated in this work (Figure 5A), of simulated vegetation shade on gynoecium development. While cell elongation in the style is reduced in spt mutants (Alvarez and Smyth, 2002), both the overexpression of SPT and the inactivation of PHYB cause an increase in style cell elongation (see Supplemental Figure 4 online), in agreement with the antagonistic effects (Figure 6A) of spt and phyB mutations on gynoecium development.
Figure 6.
Light Quality Effects on the spt Mutant Phenotype Are Mediated by PHYB, PIF4, and PIF5.
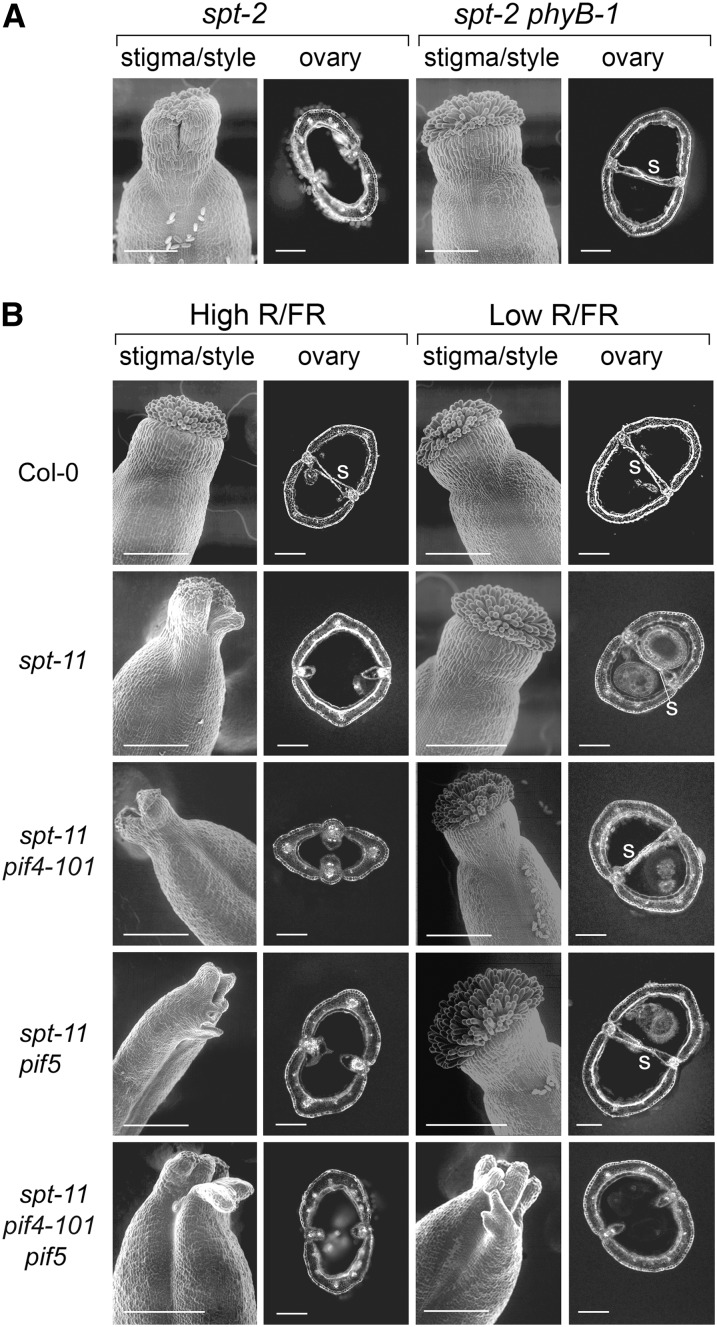
(A) Restoration of wild-type gynoecium development by inactivation of PHYB in spt2 phyB-1 double mutants. s, septum. Bars = 100 µm in stigma/style scanning electron microscopy images and 200 µm in ovary transverse sections.
(B) Restoration of wild-type gynoecium development to spt-11 single mutants and to spt-11 pif4-101 and spt-11 pif5 double mutants, but not to spt-11 pif4-101 pif5 triple mutants, by low R/FR light ratios. The pif5 mutation used in these experiments corresponds to the pil6-1 allele (Fujimori et al., 2004; Lorrain et al., 2008). Bars = 100 µm in stigma/style scanning electron microscopy images and 200 µm in ovary transverse sections.
The regulation by light quality of several of the shade avoidance genes identified here as SPT targets is known to be mediated by PIF4 and PIF5 (de Lucas et al., 2008; Lorrain et al., 2008; Hornitschek et al., 2009). We therefore tested the potential involvement of these factors in the light quality–dependent restoration of wild-type gynoecium development to spt mutants by combining the probable null spt-11 allele (Ichihashi et al., 2010) with the pif4-101 and pif5 (pil6-1) loss-of-function alleles (Lorrain et al., 2008; Figure 6B). Wild-type gynoecium development could be restored to spt-11 single mutants and to spt-11 pif4-101 and spt-11 pif5 double mutants by low R/FR ratios, though not to spt-11 pif4-101 pif5 triple mutants. These data indicate that PIF4 and PIF5 can redundantly replace the role of SPT in the gynoecium in plants grown under low R/FR ratios. Hence, SPT targets involved in shade avoidance appear to be regulated in the gynoecium by light quality, acting through PHYB, PIF4, and PIF5. We propose that the R/FR ratio may affect PIF4 and PIF5 stability in the gynoecium, as it does during shade avoidance responses in vegetative tissues (Lorrain et al., 2008). However, we have not yet experimentally verified this hypothesis, which would require the measurement of PIF4 and PIF5 protein levels in gynoecium tissues. These data suggest that PIF4 and PIF5 are expressed in the same tissues as SPT in the gynoecium. In agreement with this prediction, publicly available microarray data (Schmid et al., 2005) indicate PIF4 and PIF5 to be generally active in flower tissues, though at lower levels than SPT in the mature, wild-type gynoecium (see Supplemental Figure 5 online).
DISCUSSION
A Model for the Parallel Regulation of Gynoecium Development by SPT and Light Quality
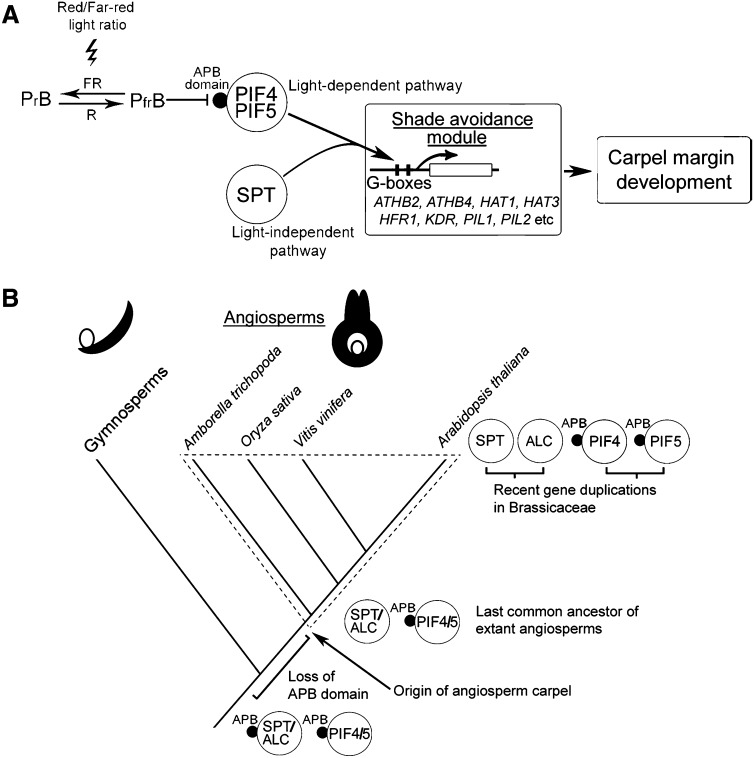
In this work, we show the development of the stigma, style, and septum of the Arabidopsis gynoecium to be regulated in parallel by SPT and an alternative pathway that is derepressed under low R/FR light ratios, resembling vegetation shade (Figure 5). This light quality–dependent pathway is negatively regulated by phyB (Figure 6A), whose active Pfr form is known to negatively regulate the bHLH factors PIF4 and PIF5. We show that PIF4 and PIF5 participate redundantly in the restoration of wild-type carpel development that occurs in spt mutants grown under low R/FR ratios (Figure 6B).
The major reason for the convergence of SPT and light quality on gynoecium development appears to be that SPT and PIF4/PIF5 regulate a common or overlapping set of targets in the gynoecium, many of which also play roles in shade avoidance in vegetative tissues, as summarized in Figure 7A. These targets include PHYB and four light-regulated transcription factors from each of the bHLH and HD-ZIP II families (Figure 2A), together with several components of auxin and brassinosteroid signaling (see Supplemental Data Set 2 online). We show that these genes are rapidly upregulated following SPT induction in plants in which protein synthesis is blocked. This finding is in agreement with those of Groszmann et al. (2008), who concluded that SPT generally acts as a transcriptional activator. We propose that some or all of the SPT targets identified here form a genetic module whose members are involved in both the formation of carpel margin tissues and shade avoidance responses. In accordance with this proposal, three of these SPT targets, ATHB2, PIL1, and HFR1, are also known to be upregulated by the redundant action of PIF4 and PIF5 in the hypocotyl in response to simulated vegetation shade (Lorrain et al., 2008). Of these targets, ATHB2 is known to promote cell elongation (Steindler et al., 1999), which forms a major component of both shade avoidance responses (Martinez-Garcia et al., 2010) and carpel margin development (Alvarez and Smyth, 2002; see Supplemental Figure 4 online). In addition, the SPT targets ATHB4 and HAT3 act redundantly to regulate both shade avoidance responses (Sorin et al., 2009) and carpel development (Figure 3), while the SPT target HAT1/JAIBA has also recently been shown to play a role in carpel development (Zúñiga-Mayo et al., 2012). It remains to be seen whether the same 24 SPT targets we identified in flower tissues are also involved in promoting the functions of SPT in cotyledon, leaf, and dehiscence zone development and seed dormancy (Penfield et al., 2005; Ichihashi et al., 2010; Sidaway-Lee et al., 2010; Girin et al., 2011; Groszmann et al., 2011; Josse et al., 2011).
Figure 7.
A Model for the Functional Evolution of SPT and PIFs in Carpel Margin Development.
(A) SPT acts in parallel with the redundant factors PIF4 and PIF5 to upregulate a genetic module containing genes, listed in Figure 2A, known for their roles in shade avoidance. This regulation occurs through physical interactions with G-boxes present in target gene promoters (Figure 4). SPT upregulates its targets independently of light quality, whereas PIF4 and PIF5 are available for this function conditionally on the inactivation of their negative regulator phyB. This latter condition can be achieved either under low R/FR light ratios (Figure 5A) or in a phyB loss-of-function mutant background (Figure 6A).
(B) The light-independent induction of SPT targets in female reproductive tissues arose later than the loss from SPT of an active phytochrome binding domain. This loss must have occurred before the last common ancestor of the extant angiosperms and may have occurred before the last common ancestor of all extant seed plants, as deduced from phylogenetic analyses in Figure 1 and Supplemental Figure 1 online.
At the biochemical level, SPT (Figure 4), PIF4, and PIF5 (de Lucas et al., 2008; Hornitschek et al., 2009) all bind to G-boxes in their target promoters, and the same two G-boxes in the HFR1 promoter are responsible for the upregulation of this gene by SPT in flower tissues (Figure 4C; see Supplemental Figure 3A online) and by simulated vegetation shade in vegetative tissues (see Supplemental Figure 3B online). All these lines of evidence point to the parallel regulation of gynoecium development by SPT and PIF4/PIF5 through interactions with G-boxes in the promoters of target genes known for their roles in shade avoidance. We propose that, whereas PIF4 and PIF5 perform this function conditionally on the inactivation of phyB, assuming that they are indeed stabilized under low R/FR light ratios in the gynoecium, SPT might do so independently of phyB and the ambient R/FR ratio that regulates its activity.
The Role of SPT in Angiosperm Evolution
Phylogenetic and structural analyses indicate Group 15 of the bHLH family to have evolved from an ancestral sequence that possessed an N-terminal APB-like domain (Figure 1; see Supplemental Figure 1 online). However, the APB domain is absent from SPT/ALC pro-orthologs throughout the extant angiosperms, and we may therefore conclude that it was lost before the initial radiation of this plant group. The absence of the APB domain from the SPT/ALC pro-ortholog of at least one gymnosperm, P. glauca, furthermore suggests that it was lost before the last common ancestor of the extant seed plants, as indicated in Figure 7B. Interaction with active phytochrome via the APB domain leads to the targeting of many PIFs, including PIF1 (Shen et al., 2005), PIF3 (Bauer et al., 2004), PIF4, and PIF5 (Lorrain et al., 2008), for destruction in the proteasome. PIF7, which groups closely in phylogenetic analyses to the SPT/ALC clade (Figure 1; see Supplemental Figure 1 online), also interacts physically with active phytochrome and antagonizes its effects (Leivar et al., 2008). We propose that the loss of the APB domain from an ancestor of SPT and ALC caused the escape of this protein from regulation by phytochrome and ultimately led to the recruitment of a downstream genetic module associated with shade avoidance to a novel, light-independent role in carpel development and other processes. Shade adaptation responses are known from gymnosperms, ferns, lycophytes, and bryophytes (Mathews, 2006; Rensing et al., 2008), all of which arose before the flowering plants, which is consistent with the proposed recruitment of a genetic module from shade avoidance to carpel development.
The sudden origin and rapid diversification of the flowering plants, which fitted poorly with Charles Darwin’s strict notions of evolutionary gradualism (Friedman, 2009), are nonetheless consistent with more recent extensions to Darwinism, such as the theory of Punctuated Equilibria (Eldredge and Gould, 1972). This theory recognizes the existence of long periods of relative stasis punctuated by periods of rapid change, though the developmental mechanisms underlying such evolutionary bursts have not yet been elucidated. In this work, we show that an important molecular mechanism of carpel development in Arabidopsis depends on a structural change that happened to SPT before the last common ancestor of the flowering plants and probably before the radiation of the living seed plants (angiosperms and gymnosperms). This change represents an interesting example of the potential of modular events to generate rapid evolutionary change through a switch in function of an entire, preformed genetic module. Careful analysis of SPT/ALC pro-orthologs in taxa occupying phylogenetically informative positions will now be necessary to reconstruct the functional evolution of this gene lineage in processes including carpel and fruit development, temperature-dependent growth, leaf and cotyledon size, and seed dormancy.
A Potential Role for Light Quality Perception in Internal Plant Tissues
SPT functions in the gynoecium both to promote cell elongation and to generate transmitting tissue. While the former of these processes occurs also in shade avoidance responses, the latter is specific to the gynoecium and might therefore require the activity of SPT targets other than those involved in shade avoidance. In addition, the Group 19 bHLH factors (Toledo-Ortiz et al., 2003), HEC1-3 and IND, which are proposed to dimerize with SPT in the gynoecium (Gremski et al., 2007; Girin, 2011), are not known to be involved in shade avoidance. Both of these considerations pose the question of why the shade avoidance factors PIF4 and PIF5 should be capable, conditionally on the inactivation of phyB, of fully replacing the role of SPT in the gynoecium. To explain this capacity, we might postulate that SPT and PIF4/PIF5 have conserved an ancestral capacity to interact with any necessary cofactors, such as Group 19 bHLH factors, and that any novel targets required for transmitting tissue development were acquired by changes to the promoters of those targets, rather than by changes to SPT or PIF4/PIF5. By contrast, an alternative hypothesis can be proposed in which SPT and PIF4/PIF5 evolved in parallel to acquire novel targets and/or cofactors involved in gynoecium development. However, for this second hypothesis to be correct, the role of PIF4/PIF5 in the gynoecium must surely have been exposed to selective pressure alongside that of SPT, which might have required the local inactivation of phyB. Interestingly, this condition may be fulfilled in the gynoecium by the vegetation shade of surrounding bracts or perianth organs. Much is known about the effect on plant development of shade from nearby vegetation. However, the case of the carpel, whose development may be modulated by the shade of surrounding floral organs, poses the question of whether shade avoidance responses also occur within a given plant structure to contribute to wild-type development.
METHODS
Gene Identification and Phylogenetic Analyses
A complete list of putative orthologs of Arabidopsis thaliana Group 15 bHLH family members (Toledo-Ortiz et al., 2003) from the sequenced genomes of grape (Vitis vinifera) and rice (Oryza sativa), and the available putative orthologs of these genes from gymnosperms, were obtained by BLAST searching of online databases at http://blast.ncbi.nlm.nih.gov/blast.cgi. In addition, EST assemblies of Group 15 bHLH sequences were identified from Amborella trichopoda and Zamia vasquezii by BLAST searching of databases at http://ancangio.uga.edu/blast/blast.html. The EST assembly of a putative SPT/ALC pro-ortholog A. trichopoda was extended using rapid amplification of cDNA ends by PCR from a female flower cDNA library (Fourquin et al., 2005) to obtain the corresponding full-length cDNA, designated as Atr-SPT. The predicted amino acid sequences of all data collected were aligned using MUSCLE in the SEAVIEW program (Gouy et al., 2010). Well-aligned amino acid sites were selected for phylogenetic analyses and converted to the corresponding nucleotide alignments using TRANALIGN (http://emboss.sourceforge.net/). Maximum likelihood phylogenetic analyses were performed in PhyML (Guindon et al., 2009), incorporating 1000 bootstrap replicates and using HKY85 and LG substitution models for nucleotide and amino acid sequence data, respectively.
Plant Material and Growth Conditions
Arabidopsis wild-type and mutant seeds were obtained from the ABRC and Nottingham Arabidopsis Stock Centre, except for pif4-101 pif5, which was obtained from Christian Fankhauser (University of Lausanne). This double mutant, which showed no marked gynoecium phenotype or impaired female fertility, was pollinated using pollen from spt-11 plants, and F2 progeny were genotyped by PCR to identify double and triple mutant combinations that included the spt11 mutation. All plants were grown from prechilled seed under 8/16-h light/dark cycles for 4 weeks, prior to transfer to 18/6-h cycles to induce flowering. The temperature of plant growth chambers was 21°C. Standard fluorescent illumination gave a fluence rate of ∼150 µmol⋅m−2⋅s−1 at a R/FR ratio of ∼5. To modify this ratio, arrays of 24 light-emitting diodes (LEDs; Roithner) were used to generate red (λmax = 660 nm; M3L1-660-30) or far-red (λmax = 740 nm; M3L1-740-30) wavelengths at fluence rates of ∼60 µmol⋅m−2⋅s−1. Combined fluorescence and LED sources gave high and low R/FR ratios of 11 and 0.7, respectively.
Transgenic Plants and Methods for Transcriptome Analysis
The SPT coding sequence was fused to the 5′-extremity of a sequence encoding the 75 C-terminal residues of the viral VP16 transcriptional activator (Sadowski, 1998) and inserted into the pG0229-35S:GR plant transformation vector (Yu et al., 2004) between the cauliflower mosaic virus 35S promoter and sequences encoding the rat GR, so as to conserve the entire reading frame. The resulting plasmid was transferred to Agrobacterium tumefaciens C58/pMP90 cells and used to transform Arabidopsis Col-0 plants by standard methods. A homozygous, single-copy transformant was identified, from which two populations of 10 T3 descendents were grown and treated by dipping of inflorescences for 2 min in CHX (10 µg/mL) containing Silwet L-77 surfactant (0.01% [v/v]). This treatment reduced translation to ∼5% of its native level in inflorescence tissues, as measured by the in vivo incorporation of [35S]Met into proteins (see Supplemental Figure 6 online). One hour later, inflorescence tissues of the two plant populations were dipped for 2 min in CHX solution, as described above, with and without DEX (10 µM). After a further 2 h, treated inflorescences, excluding open flowers, were harvested and pooled prior to RNA extraction for global gene expression analyses.
Microarray analyses were performed using Complete Arabidopsis Transcriptome Microarrays, each containing 31,776 gene-specific tags corresponding to 22,089 Arabidopsis genes (Crowe et al., 2003; Hilson et al., 2004). Three biological replicates were performed, based on three entirely separate induction experiments involving T3 plants from the same T2 parent. A transformed line containing a construction in which SPT had been replaced by an initiation codon and nuclear localization signal (Borrell et al., 2002) was used to verify that the targets identified did not respond to the nuclear translocation of VP16-GR protein alone (35S:NLS-VP16-GR plants). In this nuclear localization signal control experiment, two biological replicates were performed. One technical replicate with fluorochrome reversal was performed for each biological replicate. The labeling of cRNAs with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization of these cRNAs to microarrays, and the subsequent scanning of microarrays were performed as described by Lurin et al. (2004).
Statistical Analyses of Transcriptomic Data
For each microarray analysis, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green). No background subtraction was performed. An array-by-array normalization was performed to remove systematic biases. Spots considered as badly formed features were excluded from the analysis. A global intensity-dependent normalization using the loess procedure (see Yang et al., 2002) was then performed to correct for dye bias. Finally, for each block, the log-ratio median calculated over the values for the entire block was subtracted from each individual log-ratio value to correct print tip effects.
Differential analysis was based on the mean log ratios of the dye-swap analysis of each biological replicate. These technical replicates were thus averaged to derive one log-ratio per biological replicate, and the values obtained were then used in combination to perform a paired t test for each set of biological replicates (i.e., a set of three biological replicates for 35S:SPT-VP16-GR transformants and a set of two biological replicates for 35S:NLS-VP16-GR transformants). In this test, variance modeling is based on trimmed variance, calculated from genes that do not display extreme variance. Genes are excluded if they have a specific variance/common variance ratio smaller than the α-quantile or greater than the (1-α)-quantile, of a χ2 distribution of two degrees of freedom with α equal to 0.0001. The raw P values obtained were adjusted by the Bonferroni method, which controls the family-wise error rate, to strongly limit false positives in a multiple comparison context. The null hypothesis that DEX treatment had no effect on gene expression was tested (see Supplemental Data Set 2 online). Genes with a Bonferroni P value of < 0.05 were considered to be differentially expressed.
qRT-PCR Analyses
First-strand cDNA was prepared from RNA samples using RevertAid M-MuLV reverse transcriptase (Fermentas) and amplified by PCR to incorporate fluorescent markers using a Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen) and primers shown in Supplemental Table 1 online, according to the manufacturers’ protocols. Amplification reactions were subjected to initial incubations at 50°C for 2 min and 95°C for 2 min followed by 45 thermal cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s in an Opticon 2 DNA engine (MJ Research). All experiments incorporated three or four technical replicates, as indicated, in addition to one or three biological replicates, as indicated. The better control gene of two candidates (see Supplemental Table 1 online) was assessed for each experiment using BESTKEEPER (Pfaffl et al., 2004) and used to normalize the data obtained. Data were processed to reveal relative transcript abundances using Opticon Monitor software. t tests were performed on all data to reveal statistically significant expression differences.
In Vivo Measurement of SPT Target Gene Induction
A 35S:SPT-GR construct, similar to that described above for microarray experiments, but lacking the VP16 transcriptional activation domain, was generated and used to transform Col-0 plants. A T3 transformant containing this construct was crossed with multiple Col-0 lines containing constructs consisting of the HFR1 promoter (a fragment from −1977 bp to the transcriptional start site) fused to the mVENUS coding sequence (Kremers et al., 2006) and nopaline synthase terminator. The HFR1 promoter in these constructs had been mutagenized by standard methods such that neither, one, or both of the G-boxes present (at Sites A and B in Figure 4A) were mutated from CACGTG to CAATTG. Progeny of crosses to 35S:SPT-GR plants, containing four independent transformation events for each of the four mutant and wild-type versions of pHFR1:mVENUS generated, were used to measure the effects of mutations to G-boxes on the regulation of mVENUS expression following SPT induction. These assays were performed on DEX +CHX- and CHX-treated plants, as described above for microarray experiments, and expression data were obtained by qRT-PCR of inflorescence tissues, as described above. The same 16 parent lines containing pHFR1:mVENUS constructs (in the absence of a 35S:SPT-GR transgene) were also used to test the effect of mutations to G-boxes on mVENUS expression in leaf tissue at high and low R/FR ratios. These light conditions were generated using the combination of fluorescent lamps and LEDs described above.
SPR Analyses
SPT was produced as a fusion protein containing thioredoxin and a 6× His tag from the pTrx1a (EMBL) expression vector in Escherichia coli Rosetta cells. Recombinant protein was purified from sonicated cell lysates on TALON (Clontech) columns and concentrated using spin concentration devices (Amicon Ultra). Double-stranded DNA corresponding to wild-type and mutant versions of a PIL2 promoter fragment containing a conserved G-box element (marked with an asterisk in Figure 4A) were generated by annealing the 5′-biotinylated oligonucleotides 5′-Biotin-GTTCTTCCCACAACCACGTGGGCTTTTTGGCCCGTT (wild-type form) and 5′-Biotin-GTTCTTCCCACAACCAATTGGGCTTTTTGGCCCGTT (mutant form, mutated bases underlined) to their respective nonbiotinylated, complementary sequences. Equal quantities, corresponding to 800 arbitrary SPR units (RU), of the resulting double-stranded DNA molecules were immobilized in separate channels on CM5 (Biacore) SPR chips, which had previously been coated with streptavidin (Sigma-Aldrich), as described in Biacore protocols. SPR analyses were performed using a T100 analyzer (Biacore) for a range of concentrations of SPT fusion protein in 1× HBSP+ buffer (Biacore), which also contained sonicated, denatured DNA (10 µg/mL) from herring testes (Roche). Recombinant SPT solutions were injected for 200 s followed by DNA-containing wash buffer for 200 s. Chip surfaces were regenerated between analyses by sequential washes with guanidium hydrochloride (3 M) for 60 s, and SDS (0.03% [w/v]) for 30 s. All injections were performed at a flow rate of 50 µL/min. Equilibrium constants for dissociation (Kd) were calculated using Biacore T100 Evaluation software assuming a 1:1 (Langmuir) interaction model.
Microscopy
Scanning electron microscopy was performed on fresh plant material using a Hitachi S800 environmental microscope. Fluorescence microscopy was performed on cross sections of unfixed ovary tissues stained with 0.1% (w/v) calcofluor under UV illumination on a Leica Macrofluor microscope. Low-power light microscopy was performed using a Leica MZ12 dissecting microscope equipped with a Leica DFC3200 digital camera.
Accession Numbers
Sequence data from this article, corresponding to the cDNA of Atr-SPT, can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number HE610412. The complete microarray data set described here is available at GEOmnibus under accession number GSE12913 and at CATdb (http://urgv.evry.inra.fr/CATdb/) as Project: GEN45-Carpel_development.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analysis of Group 15 bHLH Factors from Amino Acid Sequence Data.
Supplemental Figure 2. Complementation by DEX Treatment of the Gynoecium.
Phenotype of spt-2 Mutants Transformed with 35S:SPT-GR.
Supplemental Figure 3. Effects of Mutations of G-Boxes on the Regulation of SPT Target Expression.
Supplemental Figure 4. Opposite Effects of SPT and PHYB on Cell Length in the Style.
Supplemental Figure 5. Microarray Expression Data for PIF4, PIF5, and SPT.
Supplemental Figure 6. Determination of Cycloheximide Treatment Conditions for Recombinant SPT Induction Experiments.
Supplemental Table 1. Primers Used in Quantitative RT-PCR Analyses.
Supplemental Data Set 1. Alignments Used in Phylogenetic Analyses.
Supplemental Data Set 2. Summary of Microarray Expression Data.
Supplementary Material
Acknowledgments
We thank Marie Sémon for advice on phylogenetic analysis. We acknowledge funding from Génoplante and the French National Research Agency (ANR-BLAN-0211-01) to C.P.S., Spanish Ministerio de Economía, Hacienda o Finanzas and European Regional Development Fund (BIO2011-23489) funding to J.F.M.-G., and a Rhône-Alpes doctoral studentship to M.C.R. SPR analysis was performed at Unité Mixte de Service 3444-Biosciences, Gerland, Lyon, and electron microscopy at the University of St. Etienne.
AUTHOR CONTRIBUTIONS
M.C.R. and C.P.S. designed the experiments. M.C.R., G.B., A.C., J.F.M.-G., F..M., and C.P.S. performed the experiments. M.-L.M.M. analyzed the transcriptome data. M.C.R. and C.P.S. prepared the figures and wrote the article.
Glossary
- bHLH
basic helix-loop-helix
- R/FR
red/far-red
- APB
active phytochrome binding
- DEX
dexamethasone
- CHX
cycloheximide
- qRT-PCR
quantitative RT-PCR
- Col-0
Columbia-0
- SPR
surface plasmon resonance
- LED
light-emitting diodes
References
- Alvarez J., Smyth D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386 [DOI] [PubMed] [Google Scholar]
- Alvarez J., Smyth D.R. (2002). Crabs claw and Spatula genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 163: 17–41 [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C.S., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell A., Cutanda M.C., Lumbreras V., Pujal J., Goday A., Culiáñez-Macià F.A., Pagès M. (2002). Arabidopsis thaliana atrab28: A nuclear targeted protein related to germination and toxic cation tolerance. Plant Mol. Biol. 50: 249–259 [DOI] [PubMed] [Google Scholar]
- Busch M.A., Bomblies K., Weigel D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285: 585–587 [DOI] [PubMed] [Google Scholar]
- Carabelli M., Possenti M., Sessa G., Ciolfi A., Sassi M., Morelli G., Ruberti I. (2007). Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 21: 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarbelli A.R., Ciolfi A., Salvucci S., Ruzza V., Possenti M., Carabelli M., Fruscalzo A., Sessa G., Morelli G., Ruberti I. (2008). The Arabidopsis homeodomain-leucine zipper II gene family: Diversity and redundancy. Plant Mol. Biol. 68: 465–478 [DOI] [PubMed] [Google Scholar]
- Crowe M.L., et al. (2003). CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Eldredge N., Gould S.J. (1972). Punctuated equilibria: An alternative to phyletic gradualism. In Models in Paleobiology, T.J.M. Schopf, ed (San Francisco, CA: Freeman, Cooper & Co.), pp. 82–115
- Foreman J., White J.N., Graham I.A., Halliday K.J., Josse E.M. (2011). Shedding light on flower development: Phytochrome B regulates gynoecium formation in association with the transcription factor SPATULA. Plant Signal. Behav. 6: 471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourquin C., Vinauger-Douard M., Fogliani B., Dumas C., Scutt C.P. (2005). Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proc. Natl. Acad. Sci. USA 102: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A., Lysak M.A., Al-Shehbaz I.A., Koch M.A., Mummenhoff K. (2011). Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Friedman W.E. (2009). The meaning of Darwin’s ‘abominable mystery’. Am. J. Bot. 96: 5–21 [DOI] [PubMed] [Google Scholar]
- Fujimori T., Yamashino T., Kato T., Mizuno T. (2004). Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 45: 1078–1086 [DOI] [PubMed] [Google Scholar]
- Garcia M.E., Lynch T., Peeters J., Snowden C., Finkelstein R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67: 643–658 [DOI] [PubMed] [Google Scholar]
- Girin T., Paicu T., Stephenson P., Fuentes S., Körner E., O’Brien M., Sorefan K., Wood T.A., Balanzá V., Ferrándiz C., Smyth D.R., Østergaard L. (2011). INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23: 3641–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Gremski K., Ditta G., Yanofsky M.F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601 [DOI] [PubMed] [Google Scholar]
- Groszmann M., Bylstra Y., Lampugnani E.R., Smyth D.R. (2010). Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J. Exp. Bot. 61: 1495–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M., Paicu T., Alvarez J.P., Swain S.M., Smyth D.R. (2011). SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 68: 816–829 [DOI] [PubMed] [Google Scholar]
- Groszmann M., Paicu T., Smyth D.R. (2008). Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. Plant J. 55: 40–52 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Hordijk W., Lefort V., Gascuel O. (2009). PhyML: Fast and accurate phylogeny reconstruction by maximum likelihood. Infect. Genet. Evol. 9: 384–385 [Google Scholar]
- Heisler M.G.B., Atkinson A., Bylstra Y.H., Walsh R., Smyth D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Hilson P., Allemeersch J., Altmann T., Aubourg S., Avon A., Beynon J., Bhalerao R.P., Bitton F., Caboche M., Cannoot B., Chardakov V., Cognet-Holliger C., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14(10B): 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Lee I. (2006). KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61: 283–296 [DOI] [PubMed] [Google Scholar]
- Ichihashi Y., Horiguchi G., Gleissberg S., Tsukaya H. (2010). The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 51: 252–261 [DOI] [PubMed] [Google Scholar]
- Josse E.M., Gan Y.B., Bou-Torrent J., Stewart K.L., Gilday A.D., Jeffree C.E., Vaistij F.E., Martínez-García J.F., Nagy F., Graham I.A., Halliday K.J. (2011). A DELLA in disguise: SPATULA restrains the growth of the developing Arabidopsis seedling. Plant Cell 23: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers G.J., Goedhart J., van Munster E.B., Gadella T.W.J., Jr (2006). Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45: 6570–6580 [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia J.F., Galstyan A., Salla-Martret M., Cifuentes-Esquivel N., Gallemi M., Bou-Torrent J. (2010). Regulatory components of shade avoidance syndrome. Adv. Bot. Res. 53: 65–116 [Google Scholar]
- Mathews S. (2006). Phytochrome-mediated development in land plants: Red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15: 3483–3503 [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Feldman L.J., Zambryski P.C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888 [DOI] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W., Tichopad A., Prgomet C., Neuvians T.P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26: 509–515 [DOI] [PubMed] [Google Scholar]
- Rajani S., Sundaresan V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11: 1914–1922 [DOI] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou J., Sorin C., Devlin P.F., Martínez-García J.F. (2006). Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 141: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou-Torrent J., Galstyan A., Carretero-Paulet L., Portolés S., Rodríguez-Concepción M., Martínez-García J.F. (2007). Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 26: 4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I. (1998). Plasmids for one- and two-hybrid analysis in mammalian cells. Anal. Biochem. 256: 245–247 [DOI] [PubMed] [Google Scholar]
- Salter M.G., Franklin K.A., Whitelam G.C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Scutt C.P., Vinauger-Douard M., Fourquin C., Finet C., Dumas C. (2006). An evolutionary perspective on the regulation of carpel development. J. Exp. Bot. 57: 2143–2152 [DOI] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman A.C. (1999). Some new terms for duplicated genes. Semin. Cell Dev. Biol. 10: 561–563 [DOI] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Sidaway-Lee K., Josse E.M., Brown A., Gan Y.B., Halliday K.J., Graham I.A., Penfield S. (2010). SPATULA links daytime temperature and plant growth rate. Curr. Biol. 20: 1493–1497 [DOI] [PubMed] [Google Scholar]
- Sorin C., Salla-Martret M., Bou-Torrent J., Roig-Villanova I., Martínez-García J.F. (2009). ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 59: 266–277 [DOI] [PubMed] [Google Scholar]
- Ståldal V., Sundberg E. (2009). The role of auxin in style development and apical-basal patterning of the Arabidopsis thaliana gynoecium. Plant Signal. Behav. 4: 83–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C., Matteucci A., Sessa G., Weimar T., Ohgishi M., Aoyama T., Morelli G., Ruberti I. (1999). Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Ito T., Wellmer F., Meyerowitz E.M. (2004). Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 36: 157–161 [DOI] [PubMed] [Google Scholar]
- Zúñiga-Mayo V.M., Marsch-Martínez N., de Folter S. (2012). JAIBA, a class-II HD-ZIP transcription factor involved in the regulation of meristematic activity, and important for correct gynoecium and fruit development in Arabidopsis. Plant J. 71: 314–326 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.