This work shows that not only sugar but also nitrogen and phosphate starvation signals converge with GA signaling to promote the co-nuclear import of MYBS1 and MYBGA and expression of a large set of GA-inducible but functionally distinct hydrolases, transporters, and regulators active in nutrient mobilization required for seedling growth in rice and barley.
Abstract
Germination is a unique developmental transition from metabolically quiescent seed to actively growing seedling that requires an ensemble of hydrolases for coordinated nutrient mobilization to support heterotrophic growth until autotrophic photosynthesis is established. This study reveals two crucial transcription factors, MYBS1 and MYBGA, present in rice (Oryza sativa) and barley (Hordeum vulgare), that function to integrate diverse nutrient starvation and gibberellin (GA) signaling pathways during germination of cereal grains. Sugar represses but sugar starvation induces MYBS1 synthesis and its nuclear translocation. GA antagonizes sugar repression by enhancing conuclear transport of the GA-inducible MYBGA with MYBS1 and the formation of a stable bipartite MYB-DNA complex to activate the α-amylase gene. We further discovered that not only sugar but also nitrogen and phosphate starvation signals converge and interconnect with GA to promote the conuclear import of MYBS1 and MYBGA, resulting in the expression of a large set of GA-inducible but functionally distinct hydrolases, transporters, and regulators associated with mobilization of the full complement of nutrients to support active seedling growth in cereals.
INTRODUCTION
Germination followed by seedling growth constitutes two essential steps in the initiation of a new life cycle in plants, and completion of these steps requires the coordination of developmental and biochemical processes, including mobilization of seed reserves and elongation of the embryonic axis. Active mobilization of stored nutrients upon germination contributes to seedling vigor and has a profound impact on plant growth and eventual productivity (Karrer et al., 1993). In cereals, the stored reserves (starch, proteins, lipids, nucleic acids, and mineral complexes) in the endosperm are degraded and mobilized by a battery of enzymes and transporters acting in concert, and gibberellin (GA) is the major hormone that initiates these processes (Fincher, 1989). In barley (Hordeum vulgare) and rice (Oryza sativa) aleurone cells surrounding the starchy endosperm, up to 1300 genes encoding hydrolases and other proteins involved in general metabolism, transcription, nutrient transport, and programmed cell death were found to be upregulated after treatment with GA3 (one of the active GAs) (Chen and An, 2006; Tsuji et al., 2006). Several regulators in the GA signaling pathway have been identified (Gómez-Cadenas et al., 2001; Sun and Gubler, 2004; Schwechheimer and Willige, 2009; Sun, 2010). However, the detailed mechanism coordinating the expression of a large number of essential proteins within a limited time window after germination is not well understood.
Starch, which constitutes ∼75% of cereal grain dry weight (Kennedy, 1980), provides the major carbon source for generating energy and metabolites during germination and seedling growth. Consequently, among all hydrolases, α-amylases are the most abundant and play a central role in the mobilization of starch and thus the rate of germination and seedling growth. The regulatory mechanism of α-amylase expression, which is induced by both GA and sugar demand/starvation, has served as a model for extensive studies for decades (Yu, 1999a, 1999b; Lu et al., 2002, 2007; Sun and Gubler, 2004; Woodger et al., 2004; Chen et al., 2006; Lee et al., 2009). Upon imbibition of cereal grains, sugars in the embryo are rapidly depleted, leading to activation of α-amylase synthesis in the scutellum, where starch degradation is initiated (Ranjhan et al., 1992; Yu et al., 1996). Meanwhile, the embryo synthesizes GA that is diffused to aleurone cells to activate the synthesis and secretion of α-amylases and an array of other hydrolases. The stored starch and other complex nutrients in the endosperm are hydrolyzed by these enzymes to sugars, reduced nitrogen, and other soluble nutrients that are adsorbed by the scutellum and transported to the embryonic axis to support seedling growth (Akazawa and Hara-Nishimura, 1985; Beck and Ziegler, 1989; Woodger et al., 2004).
The α-amylase promoter is activated by sugar starvation through the sugar response complex (SRC), in which the TA box is the key element (Lu et al., 1998). The TA box is the most highly conserved motif and controls the promoter activity of at least 20 genes upregulated by sugar starvation in rice suspension cells (Wang et al., 2007). The α-amylase promoter is also activated by GA through the gibberellin response complex (GARC), in which the adjacent gibberellin response element (GARE) and the TA/Amy box are key elements and act synergistically (Lanahan et al., 1992; Rogers et al., 1994; Gubler et al., 1999; Gómez-Cadenas et al., 2001). The TA/Amy box and GAREs have been identified in promoters of a variety of GA-inducible hydrolase genes (Sun and Gubler, 2004; Tsuji et al., 2006).
MYBs are a group of transcription factors with conserved DNA binding domains that consist of three imperfect repeats (designated R1, R2, and R3) of ∼51 to 52 amino acids, each in the mammalian cellular myb protooncogene (c-Myb) (Sakura et al., 1989). In plants, MYBs are involved in diverse physiological and biochemical processes. The rice genome contains 183 MYB genes, but only 5 MYBs contain all three repeats (R1R2R3 type), while 109 MYBs contain two repeats (R2R3 type), and ∼70 MYBs have only one repeat (R1 type) (Yanhui et al., 2006).
MYBGA (also called GAMYB) is a GA-inducible R2R3 MYB that binds to the GARE and activates promoters of α-amylases and other hydrolases in rice and barley aleurone cells in response to GA, and the function of MYBGA is interchangeable in both plant species (Gubler et al., 1995, 1997, 1999). MYBGA is regarded as a major transcription factor in the GA signaling pathway (Tsuji et al., 2006). The rice protein MYBS1 is a sugar-repressible R1 MYB that binds to the TA box and activates α-amylase promoters in rice suspension cells and embryos under sugar starvation (Lu et al., 2002, 2007).
The GA and sugar starvation signaling pathways regulating α-amylase expression have long been regarded as independent pathways. In the GA signaling pathway, GA binds to the receptor GID1, and the GA-GID1 complex binds to the DELLA domain repressor (homologs SLR1 in rice and SLN1 in barley). The GA-GID1-SLR1/SLN1 complex then binds to an F-box protein, the SCFGID2 ubiquitin ligase, to target SLR1/SLN1 for degradation by the 26S proteosome, thus allowing the expression of GA-inducible genes (Sun and Gubler, 2004; Ueguchi-Tanaka et al., 2007). On the other hand, in the sugar starvation signaling cascade, a calcineurone B–like protein-interacting protein kinase (CIPK15) induces the accumulation of the global energy sensor, SNF1-related kinase 1A (SnRK1A) protein kinase, which in turn promotes the MYBS1–TA box interaction and turns on α-amylase transcription (Lee et al., 2009). The GA signal was later found to interfere with sugar repression of the α-amylase promoter in rice endosperm, which indicates possible crosstalk between the sugar and GA signaling pathways (Chen et al., 2006). MYBS1 is a positive activator of α-amylase promoters in response to GA and sugar starvation (Lu et al., 2002), which further indicates that MYBS1 may serve as a module of crosstalk between the GA and sugar starvation signaling pathways.
Unraveling the mechanisms globally regulating the coordinated expression of a large number of genes involved in nutrient mobilization may facilitate the development of strategies aimed at promoting seedling growth in both normal and unfavorable environments. Extensive studies have advanced our knowledge of upstream regulators in the GA signaling pathway (Sun and Gubler, 2004). However, how the two crucial transcription factors MYBGA and MYBS1 connect the distinct GA and sugar starvation signaling pathways and coordinate the transcription of over 1000 hydrolase and related genes remains elusive. In this study, we discovered that not only starvation of sugar/carbon but also of other nutrients, such as nitrogen and phosphate, activates a common mechanism to regulate the expression of α-amylase and a variety of other hydrolases, transporters, and transcription factors that are responsible for coordinated mobilization of the full complement of diverse nutrients in the rice endosperm. This process involves unique interactions between MYBS1 and MYBGA, their conuclear import, and the coactivation of target gene promoters in response to the balance of nutrient availability and the germination-promoting hormone GA.
RESULTS
Glc Inhibits the Nuclear Localization of MYBS1
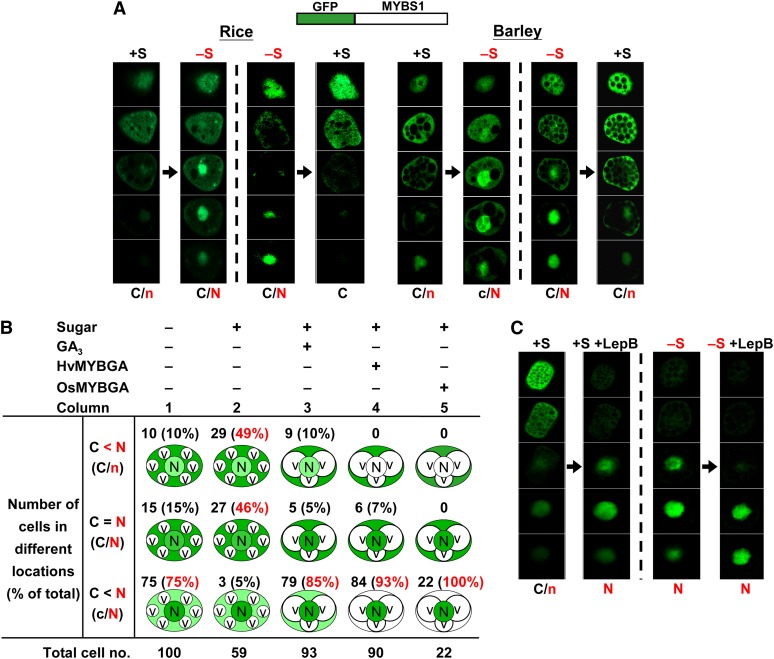
The barley genome has not been completely sequenced, but a barley MYBS1 gene closest to the rice MYBS1 gene, sharing 71% homology throughout its coding region and 100% homology in the R1 repeat, could be identified from the database. We first investigated the spatial dynamics of the subcellular distribution of the rice MYBS1 in rice and barley aleurone cells. A construct encoding Os-MYBS1 tagged with green fluorescent protein (GFP) was transfected into rice and barley aleurone cells using the particle bombardment–mediated transient expression system. The nuclear localization of MYBS1 induced by sugar starvation was detected regardless of GFP fusion orientation. As the GFP-MYBS1 signal was distributed throughout different focal planes in aleurone cells, we prepared 30 optical sections for each cell. Five regularly spaced sections representing the distributions of GFP throughout each cell are shown in each figure, and all 30 sections examined are shown in the corresponding supplemental figures. Similar results were obtained with both rice and barley aleurones. The rice aleurone has a single layer of cells and is fragile, while the barley aleurone has three to four layers and is relatively stronger and easier to manipulate using the microscope. Therefore, barley aleurones were used more frequently in the determination of MYB cellular localization, and rice aleurones were used in all transient expression assays.
The distribution of GFP-MYBS1 in the same rice or barley aleurone cell was traced continuously during consecutive incubations in +S (with Glc) followed by −S (without Glc) medium as well as incubation in −S followed by +S medium. MYBS1 mainly accumulated in the cytoplasm of cells cultured in +S medium, while cells cultured in −S medium exhibited nuclear accumulation; conversely, MYBS1 accumulation decreased in the nucleus and increased in the cytoplasm when the same aleurone cell was switched from −S to +S medium (Figure 1A; see Supplemental Figure 1A online). This study indicates that the Glc inhibition of nuclear localization of MYBS1 is reversible. Staining with the red fluorescent dye SYTO17 confirmed the nuclear localization in aleurone cells (see Supplemental Figure 1B online). To ensure that the GFP-MYBS1 fusion protein was as active as MYBS1 alone, we verified the ability of GFP-MYBS1 to activate the Glc-repressible promoter containing αAmy3 SRC (Lu et al., 1998) (see Supplemental Figure 1C online).
Figure 1.
Glc Inhibits the Nuclear Localization of MYBS1.
Aleurones were transfected with Ubi:GFP-OsMYBS1 and then incubated in +S or −S medium . Thirty optical sections of 0.7 to 0.9 µm thickness each were prepared for each cell; only five regularly spaced sections (sections 3, 9, 15, 21, and 27) are shown here. C and N indicate higher GFP signals and c and n indicate lower GFP signals in the cytoplasm and nucleus, respectively. See also Supplemental Figures 1A and 1D online.
(A) Transfected rice or barley aleurones were incubated in +S or −S medium for 24 h, then shifted from +S to −S medium or from −S to +S medium, and incubated for another 24 h.
(B) Barley aleurones were transfected with Ubi:GFP-OsMYBS1 only or with Ubi:GFP-OsMYBS1 plus Ubi:OsMYBGA and incubated in +S or −S medium. GFP was detected after 24 h. Percentages indicate the number of cells with GFP distribution in the indicated category divided by the total number of cells examined. C, cytoplasm; N, nucleus; V, vacuole. Vacuolation was observed in cells treated with GA3 or overexpressing MYBGA.
(C) Transfected rice aleurones were incubated in +S or −S medium for 24 h. LepB was then added, and aleurones were incubated for another 24 h.
[See online article for color version of this figure.]
Because the subcellular distribution of GFP-MYBS1 in +S and −S media is normally not an all-or-none pattern, cells exhibiting different patterns of GFP-MYBS1 distribution were assessed visually. In −S medium, MYBS1 accumulation was higher in the nucleus than in the cytoplasm in 75% of cells (Figure 1B, column 1); by contrast, only 5% of cells in +S medium displayed higher MYBS1 accumulation in the nucleus than in the cytoplasm (Figure 1B, column 2).
GFP by itself was distributed mostly in the cytoplasm and some in the nucleus of aleurone cells, and the pattern was unchanged by the medium employed (see Supplemental Figures 2A and 2B online). To determine whether Glc promotes the nuclear export of MYBS1, aleurones transfected with GFP-MYBS1 were incubated with leptomycin B (LepB), a specific nuclear export inhibitor (Toyoshima et al., 1998). MYBS1 accumulated exclusively in the nucleus in both +S and −S medium in the presence of LepB (Figure 1C; see Supplemental Figure 1D online), indicating that MYBS1 was actively exported to the cytoplasm in the presence of sugar. Collectively, these results demonstrate that although MYBS1 is shuttling between the nucleus and cytoplasm, it is preferentially localized to the nucleus under sugar starvation conditions.
Multiple Subcellular Targeting Signals Regulate the Sugar-Dependent Nucleocytoplasmic Shuttling of MYBS1
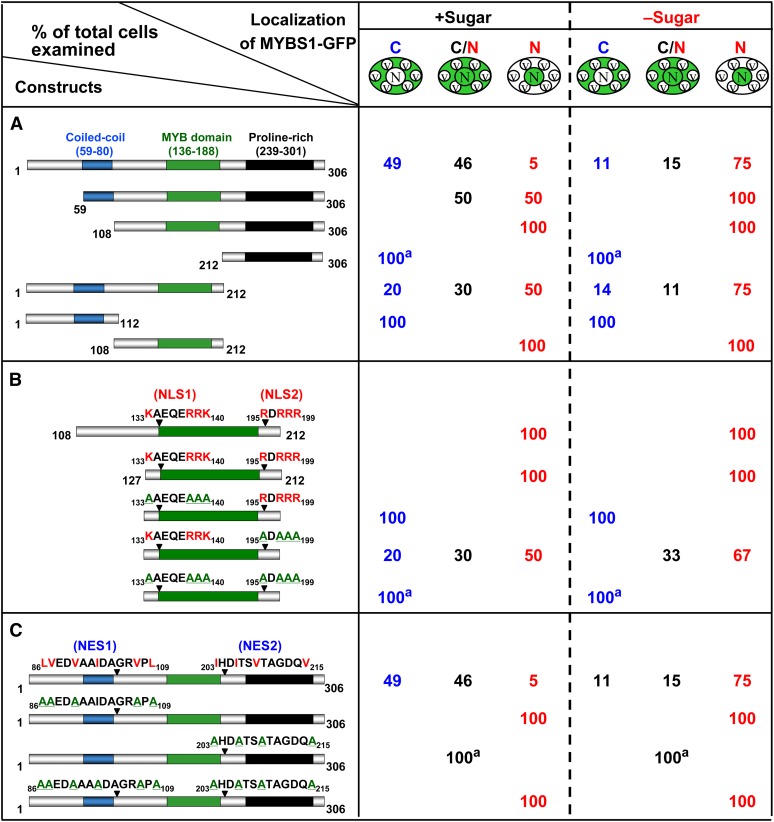
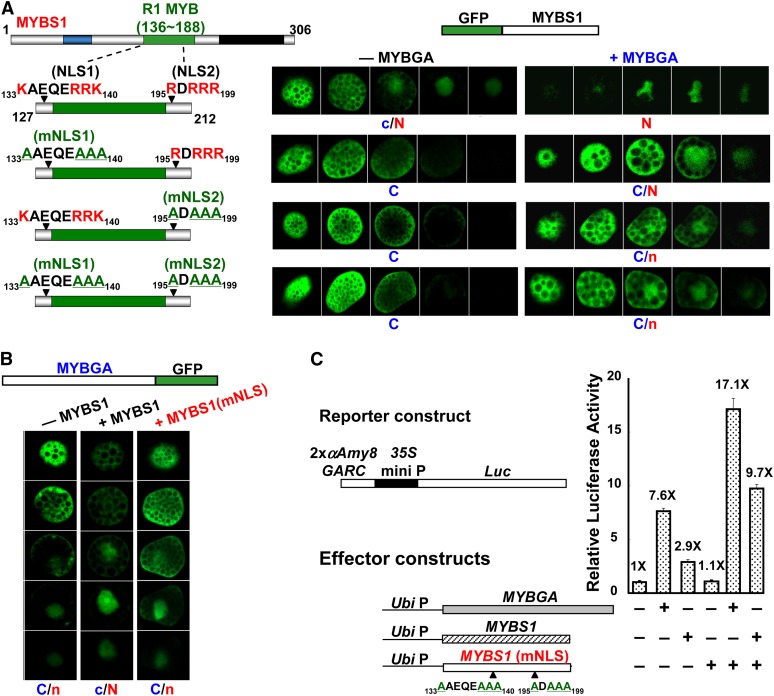
To study how the nucleocytoplasmic shuttling of MYBS1 is regulated, various constructs encoding truncated MYBS1 fused to GFP were transfected into barley aleurones. As shown in Figure 2A, the truncated MYBS1 lacking N-terminal amino acid residues 1 to 58, MYBS1(59-306), was distributed preferentially in the nucleus in +S medium but was exclusively present in the nucleus in −S medium (see Supplemental Figure 3A online). MYBS1(108-306) became localized exclusively in the nucleus (see Supplemental Figure 3B online), whereas MYBS1(212-306) was localized exclusively in the cytoplasm in both +S and −S media (see Supplemental Figure 3C online). A C-terminal truncated variant of MYBS1, MYBS1(1-212), accumulated in both nucleus and cytoplasm (see Supplemental Figure 3D online); by contrast, MYBS1(1-112) was localized exclusively in the cytoplasm in both +S and −S media (see Supplemental Figure 3E online). A truncated protein containing only the central region of MYBS1, MYBS1(108-212), was localized exclusively in the nucleus in both +S and −S media (see Supplemental Figure 3F online). These results indicate that residues 1 to 108 of MYBS1 may contain a sequence signaling for cytoplasmic retention in the presence of Glc, while residues 108 to 212 may contain a nuclear localization signal (NLS).
Figure 2.
Bipartite NLS and NES Are Involved in the Sugar-Regulated Cellular Targeting of MYBS1.
Barley aleurones were transfected with Ubi:GFP-OsMYBS1 (full length, truncated, or mutated) and incubated in +sugar or −sugar medium for 24 h. Amino acids highlighted in red are conserved residues for NLS or NES, and those underlined in green are substituting residues. a, GFP with a punctate pattern; C, cytoplasm; C/N, nucleus and cytoplasm; N, nucleus; V, vacuole.
(A) Nucleocytoplasmic partitioning of different forms of MYBS1 fused to GFP.
(B) MYBS1(108-212) contains a bipartite NLS.
(C) MYBS1 contains a bipartite NES.
No canonical consensus sequence of NLS motifs was found in MYBS1(108-212); however, two Arg (R)- and Lys (K)-rich domains, amino acids 133KAEQERRK140 (designated NLS1) and 195RDRRR199 (designated NLS2), were selected to test their putative role as a NLS. As shown in Figure 2B, MYBS1(127-212)-GFP containing these two sequences was detected exclusively in the nucleus in both +S and −S media. Substitution of conserved Arg and Lys residues with Ala (A) in NLS1 alone or in both NLS1 and NLS2 induced exclusive cytoplasmic localization of the truncated MYBS1, while mutations in NLS2 alone led to MYBS1 distribution in both the cytoplasm and nucleus. These results indicate that both NLS1 and NLS2 are sufficient and necessary for directing the nuclear import of MYBS1 and that NLS1 has a stronger nuclear import activity than NLS2.
Next, we investigated MYBS1 with regard to the presence of nuclear export signals (NESs). As was the case for the NLS, no canonical NES sequence was found in MYBS1. However, two sequences, 86 to 109 (designated NES1) and 203 to 215 (designated NES2), contain hydrophobic amino acid residues similar to other known NESs (Sharma et al., 2011). Conserved hydrophobic amino acids (Leu, Val, and Ile) in two putative NESs in MYBS1 were substituted with Ala. As shown in Figure 2C, mutations in NES1 alone or both NES1 and NES2 led to exclusive localization of MYBS1 in the nucleus (see Supplemental Figures 4B and 4C online), and mutations in NES2 alone resulted in localization of MYBS1 in both the nucleus and cytoplasm (see Supplemental Figure 4D online), irrespective of whether +S or −S medium was used. Additionally, MYBS1(1-212) containing two NLSs and NES1 was localized in both the nucleus and cytoplasm, while MYBS1(108-306) containing two NLSs and NES2 was exclusively localized in the nucleus (Figure 2A). These results indicate that both NESs are necessary for directing the nuclear export of MYBS1, particularly in the presence of sugar, but NES1 has a stronger nuclear export activity than NES2.
GA and MYBGA Antagonize Sugar Repression of MYBS1 Nuclear Import and α-Amylase Expression
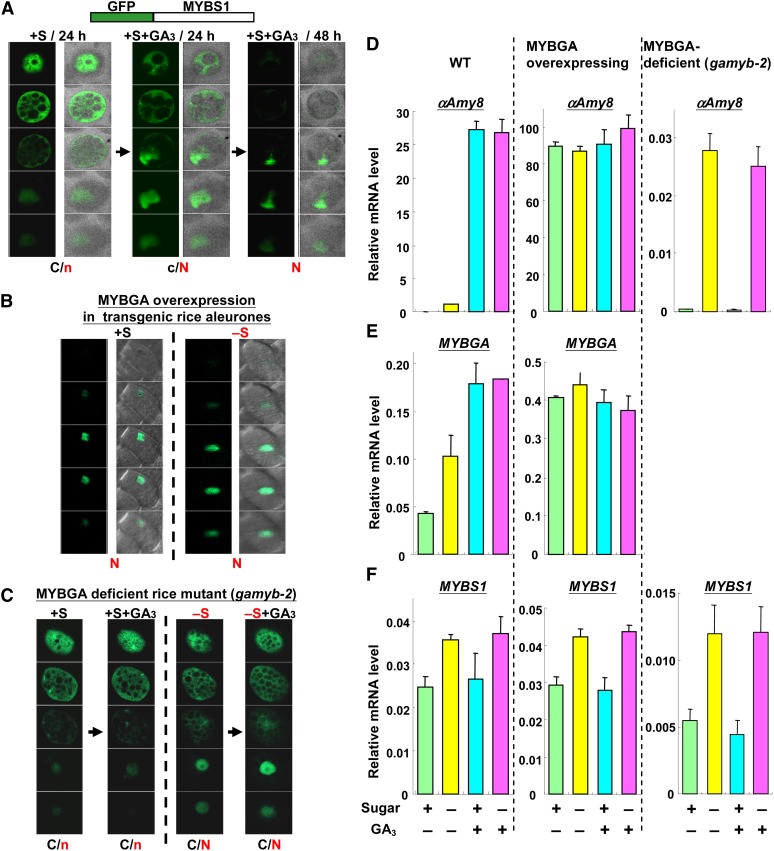
The rice and barley MYBGA genes share 88% homology throughout their coding regions and 99% homology in the R2 and R3 repeats (Gubler et al., 1997). In rice, the MYBGA–GARE interaction promotes α-amylase promoter activity in the endosperm in the presence of sugar (Chen et al., 2006). To determine whether GA3 affects the nuclear localization of MYBS1, barley aleurones were transfected with GFP-MYBS1 and incubated with GA3. In +S medium, MYBS1 was detected mainly in the cytoplasm; by contrast, MYBS1 was detected predominantly in the nucleus upon addition of GA3 and exclusively in the nucleus after prolonged incubation (Figure 3A; see Supplemental Figure 5A online). In addition to α-amylase expression, vacuolation caused by programmed cell death is also a hallmark of the GA response in aleurone cells (Bethke et al., 1997; Zentella et al., 2002). Vacuolation of aleurone cells was observed after GA3 treatment, and this led to distortion of the nucleus (Figure 3A). Upon incubation of barley aleurones with GA3 in +S medium, GFP signals were higher in the nucleus than in the cytoplasm in 85% of cells (Figure 1B, column 3). This result demonstrated that GA3 interferes with the sugar-promoted nuclear export of MYBS1. The distribution of GFP by itself was unchanged by the medium employed (see Supplemental Figures 2C and 2D online).
Figure 3.
GA and MYBGA Antagonize the Sugar Repression of Nuclear Import of MYBS1 and α-Amylase Expression.
(A) Barley aleurones were transfected with Ubi:GFP-OsMYBS1 and incubated in +S medium for 24 h. GA3 was then added, and aleurones were incubated for another 24 and 48 h.
(B) Transgenic rice aleurones carrying Ubi:OsMYBGA were transfected with Ubi:GFP-OsMYBS1 and incubated in +S or −S medium for 24 h.
(C) Aleurones of gamyb-2 mutant seeds were transfected with Ubi:GFP-OsMYBS1 and incubated in +S or −S medium for 24 h. Next, GA3 was added, and aleurones were incubated for another 24 h.
Images of fluorescent fields and composites of fluorescent and bright fields are shown in (A) and (B). Vacuolation was observed in GA3-treated or MYBGA-overexpressing aleurone cells but not in GA3-treated gamyb-2 aleurone cells. The nucleus is distorted due to overlap with enlarged vacuoles. C and N indicate higher GFP signals and c and n indicate lower GFP signals in the cytoplasm and nucleus, respectively. See also Supplemental Figure 5 online.
(D) to (F) Accumulation of αAmy8, MYBGA, and MYBS1 mRNA in aleurones of wild-type (WT), MYBGA-overexpressing transgenic, and gamyb-2 mutant rice. Rice aleurone layers were incubated in +S or −S medium with or without GA3 for 24 h. Total RNA was purified and subjected to quantitative RT-PCR analysis. MYBGA mRNA was undetectable in the gamyb-2 mutant.
[See online article for color version of this figure.]
To determine whether the GA-inducible MYBGA is sufficient for promoting the nuclear retention of MYBS1, barley aleurones were cotransfected with Ubi:HvMYBGA or Ubi:OsMYBGA and Ubi:GFP-OsMYBS1. GFP-MYBS1 was detected exclusively in the nucleus in cells cultured in either +S or −S medium (see Supplemental Figure 5B online). Upon transient overexpression of Hv-MYBGA and Os-MYBGA in +S medium, GFP-MYBS1 accumulated at higher levels in the nucleus than in the cytoplasm in 93% and 100% of cells, respectively (Figure 1B, columns 4 and 5). We next transfected transgenic rice constitutively overexpressing Os-MYBGA (Chen et al., 2006) with Ubi:GFP-OsMYBS1. Similarly, GFP-MYBS1 was detected exclusively in the nuclei of transgenic rice in both +S and −S media (Figure 3B; see Supplemental Figure 5C online). Since the interaction of Hv-MYBGA with Os-MYBS1 is similar to Os-MYBGA in rice and barley aleurone cells, only Os-MYBGA was used for the following studies.
The expression of αAmy8 is not induced by GA3 in the aleurone/endosperm of a MYBGA knockout rice mutant, gamyb-2 (Kaneko et al., 2004; Chen et al., 2006). To determine whether MYBGA is necessary for the nuclear retention of MYBS1 in +S medium, aleurones of gamyb-2 were transfected with Ubi:GFP-OsMYBS1. We detected MYBS1 mainly in the cytoplasm of cells in +S medium despite the addition of GA3 (Figure 3C; see Supplemental Figure 5D online, left panel) and in both the nucleus and cytoplasm of cells cultured in −S medium with the addition of GA3 (Figure 3C; see Supplemental Figure 5D online, right panel).
The expression of αAmy8 and αAmy32b in rice and barley aleurones, respectively, was repressed by sugar and activated by sugar starvation in the absence of GA3, and both were significantly activated by GA3 concomitantly with the expression of MYBGA regardless of the presence or absence of sugar (Figure 3D, left panel; see Supplemental Figures 6A and 6B online). The expression of αAmy8 in MYBGA-overexpressing transgenic rice was constitutively activated (Figure 3D, middle panel; see Supplemental Figure 6C online), while in the gamyb-2 mutant, it was only activated by sugar starvation, albeit at significantly lower levels, regardless of the presence or absence of GA3 (Figure 3D, right panel; see Supplemental Figure 6D online). The accumulation of MYBS1 mRNA in rice was increased under sugar starvation, independent of the presence or absence of GA3 and MYBGA transcript levels in the wild type and MYBGA-overexpresssing transgenic rice (Figure 3F, left and middle panels) but at significantly lower levels in the gamyb-2 mutant (Figure 3F, right panel). Taken together, these results demonstrate that the activation of α-amylase gene expression is tightly associated with the nuclear localization of MYBS1 under sugar starvation and that GA3 or MYBGA promotes both processes irrespective of the sugar status.
MYBS1 and MYBGA Interact Physically and Coactivate the α-Amylase Promoter
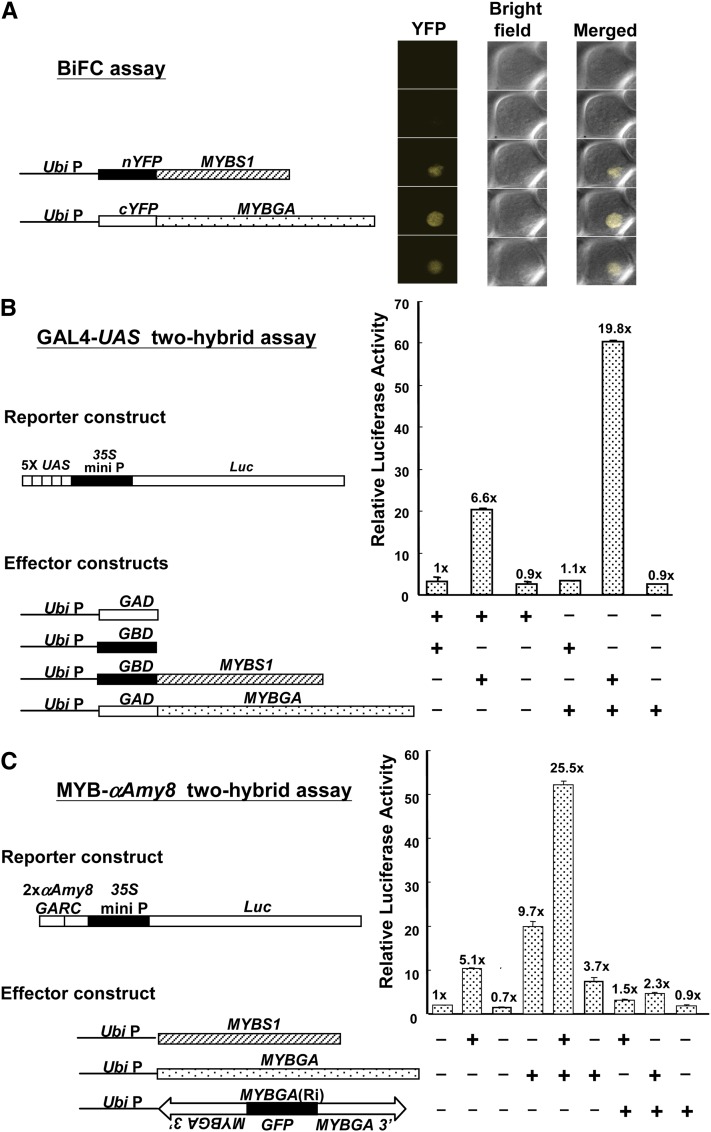
To examine whether MYBS1 and MYBGA interact in vivo, we first employed a bimolecular fluorescence complementation (BiFC) assay (Hu et al., 2002). Os-MYBS1 and Os-MYBGA were fused to the N or C terminus of yellow fluorescent protein (YFP), and barley aleurones were cotransfected with different combinations of fusion proteins. A fluorescent signal was detected in the nucleus only with the combination of Ubi:nYFP-OsMYBS1 and Ubi:cYFP-OsMYBGA constructs (Figure 4A; see Supplemental Figure 7 online), indicating MYBS1 and MYBGA interact in vivo.
Figure 4.
MYBS1 and MYBGA Interact Physically and Coactivate the α-Amylase Promoter.
(A) BiFC assay. Barley aleurones were cotransfected with Ubi:nYFP-OsMYBS1 and Ubi:cYFP-OsMYBGA and incubated in the presence of Glc for 24 h. YFP was detected by confocal microscopy. See also Supplemental Figure 7 online.
(B) GAL4-UAS two-hybrid assay. Rice aleurones were cotransfected with effectors Ubi:GBD-OsMYBS1 and Ubi:GAD-OsMYBGA and reporter (5xUAS)-CaMV35S:Luc.
(C) MYB-αAmy8 two-hybrid assay. Rice aleurones were cotransfected with effectors Ubi:OsMYBS1 and Ubi:OsMYBGA and reporter 2xαAmy8 GARC-CaMV35S:Luc.
In (B) and (C), transfected rice aleurones were incubated in the presence of Glc for 24 h, and luciferase activity was assayed. The luciferase activity in rice aleurones bombarded with effectors Ubi:GAD and Ubi:GBD in (A) or reporter 2xαAmy8 GARC:Luc in (B) was set to 1×, and other values were calculated relative to this value.
[See online article for color version of this figure.]
Two rice aleurone two-hybrid assays were employed to confirm the physical and functional interactions of MYBS1 and MYBGA in vivo. In the GAL4-UAS two-hybrid assay, construct 5xUAS:Luc, which contains five tandem repeats of the GAL4 upstream activation sequence (5xUAS) fused upstream of luciferase cDNA (Luc) (Lu et al., 2002), was used as a reporter (Figure 4B). Constructs Ubi:GBD-OsMYBS1 and Ubi:GAD-OsMYBGA, in which MYBS1 is fused at the C terminus of the GAL4 DNA binding domain (Lu et al., 2002) and MYBGA is fused at the C terminus of the GAL4 activation domain, served as effectors (Figure 4B). In transfected rice aleurone cells, overexpression of GAD-MYBGA had no effect while overexpression of GBD-MYBS1 enhanced luciferase activity by 6.6-fold, likely due to the transactivation capability of MYBS1 by itself; co-overexpression of GAD-MYBGA and GBD-MYBS1 enhanced luciferase activity by 19.8-fold (Figure 4B). These data suggest that MYBS1 and MYBGA interact and coactivate the 5xUAS promoter.
In the MYB-αAmy8 two-hybrid assay, overexpression of MYBS1 and MYBGA each alone enhanced αAmy8 GARC–controlled luciferase activity by 5.1- and 9.7-fold, respectively, and co-overexpression of both MYBSs enhanced luciferase activity by 25.5-fold (Figure 4C). An RNA interference construct, OsMYBGA(Ri), significantly reduced the MYBS1- and MYBGA-activated luciferase activity. These results also demonstrate that MYBS1 and MYBGA interact physically to coactivate αAmy8 GARC.
MYBS1 and MYBGA Are Conuclear Imported, Mutually Promote Nuclear Localization, and Synergistically Activate Promoters of Distinct Hydrolases and Transporters
To determine whether MYBS1 and MYBGA are first transported individually into the nucleus and then interact, or whether they interact first in the cytoplasm and then are cotransported into the nucleus, barley aleurones were transfected with GFP-fused nuclear import–competent or nuclear import–deficient MYBS1 with or without MYBGA. As shown in Figure 5A and Supplemental Figure 8 online, in the presence of sugar, GFP-MYBS1 was localized in both the cytoplasm and nucleus in the absence of MYBGA but was found exclusively in the nucleus in the presence of MYBGA. GFP-MYBS1 with mutations in NLS1 and/or NLS2 became localized exclusively in the cytoplasm in the absence of MYBGA but appeared in the nucleus when coexpressed with MYBGA. These results indicate that MYBS1 and MYBGA interact in the cytoplasm and are subsequently cotransported into the nucleus; however, active nuclear import of MYBS1 is necessary for its complete localization in the nucleus.
Figure 5.
MYBS1 and MYBGA Are Conuclear Imported and Mutually Promote Nuclear Localization.
(A) Barley aleurones were transfected with Ubi:GFP-OsMYBS1 (wild type or with mutation in NLS1 and/or NLS2) alone or with Ubi:OsMYBGA by particle bombardment and incubated in −S medium for 24 h. See also Supplemental Figure 8 online.
(B) Barley aleurones were transfected with Ubi:OsMYBGA-GFP alone or with Ubi:OsMYBS1 or Ubi:OsMYBS1(mNLS) and incubated in +S or −S medium for 24 h. C and N indicate higher GFP signals and c and n indicate lower GFP signals in the cytoplasm and nucleus, respectively. See also Supplemental Figure 9 online.
(C) Rice aleurones were cotransfected with effectors Ubi:OsMYBGA and Ubi:OsMYBS1 or Ubi:OsMYBS1(mNLS) and reporter 2xαAmy8 GARC-CaMV35S:Luc, incubated in the presence of Glc for 24 h, and luciferase activity was assayed.
The luciferase activity in aleurones transfected with reporter only was set to 1×, and other values were normalized relative to this value.
MYBGA has been shown to promote the nuclear localization of MYBS1, so we investigated whether MYBS1 also promotes the nuclear localization of MYBGA. In the presence of sugar, MYBGA-GFP by itself was imported into the nucleus but accumulated preferentially in the cytoplasm rather than in the nucleus in transfected barley aleurones (Figure 5B; see Supplemental Figure 9 online). MYBS1 significantly enhanced the nuclear localization of MYBGA-GFP, with an increase from none to 71% (54% + 17%) in +S medium and to 80% (70% + 10%) in −S medium (see Supplemental Figure 10 online). The import-deficient MYBS1 [MYBS1(mNLS)] only slightly increased the nuclear localization of MYBGA (Figure 5B; see Supplemental Figure 9 online). The importance of the nuclear colocalization of MYBS1 and MYBGA in activation of the αAmy8 promoter was also determined. Overexpression of MYBS1 and MYBGA individually enhanced luciferase activity by 2.9- and 7.6-fold, respectively, but MYBS1(mNLS) did not have any effect (Figure 5C). Co-overexpression of MYBGA and MYBS1 enhanced luciferase activity by 17.1-fold, but MYBGA and MYBS1(mNLS) enhanced the activity by only 9.7-fold, indicating that import-competent MYBS1 is important for the nuclear localization of MYBGA and coactivation of αAmy8 GARC by the two MYBs (Figure 5C).
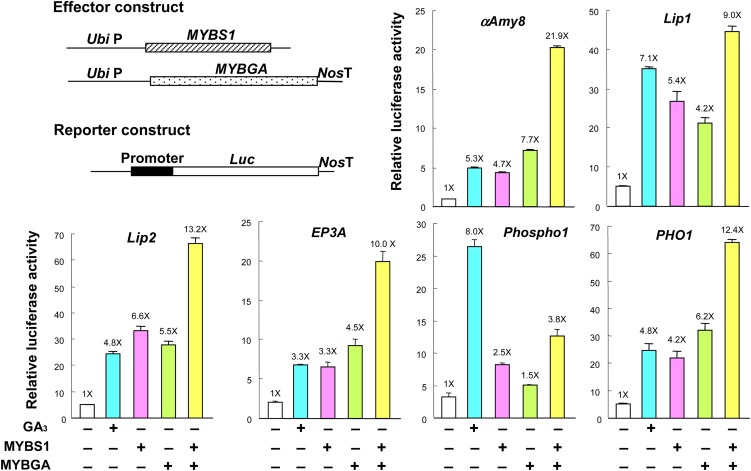
An examination of promoters of genes that are highly sensitive to GA3 in rice aleurones (Ho et al., 2000; Diaz et al., 2002; Tsuji et al., 2006) revealed the presence of canonical or conserved TA box and GARE sequences (Gubler and Jacobsen, 1992; Gubler et al., 1999; Sutoh and Yamauchi, 2003; Shin et al., 2011) in single or multiple copies, and those of 23 genes encoding hydrolases, transporters, and regulatory factors are shown here as examples (see Supplemental Figure 11 online). To determine whether MYBS1 and MYBGA coactivate promoters that contain putative TA box and GARE sequences, promoters of some GA3-inducible genes, including α-amylase (αAmy8), lipases (Lip1 and Lip2), Cys protease (EP3A), phosphatase (Phospho1), and phosphate transporter (PHO1) (Ho et al., 2000; Tsuji et al., 2006), were isolated from rice and fused to Luc. In transfected rice aleurones, these promoters were upregulated by GA3, MYBS1, or MYBGA alone and, except for Phospho1, to a greater extent by the synergistic action of MYBS1 and MYBGA (Figure 6).
Figure 6.
MYBS1 and MYBGA Synergistically Activate Promoters of Distinct Hydrolases and Transporters.
Rice aleurones were transfected with reporter containing various promoters fused to Luc and untreated or treated with GA3 or cotransfected with effectors Ubi:OsMYBS1 and Ubi:OsMYBGA. Transfected aleurones were incubated in the presence of Glc for 24 h, and luciferase activity was assayed. The luciferase activity in aleurones transfected with reporter only and without GA3 was set to 1×, and other values were normalized relative to this value.
[See online article for color version of this figure.]
Carbon, Nitrogen, and Phosphate Starvation Cross-Induce the Conuclear Import of MYBS1 and MYBGA and the Expression of Hydrolases and Related Proteins
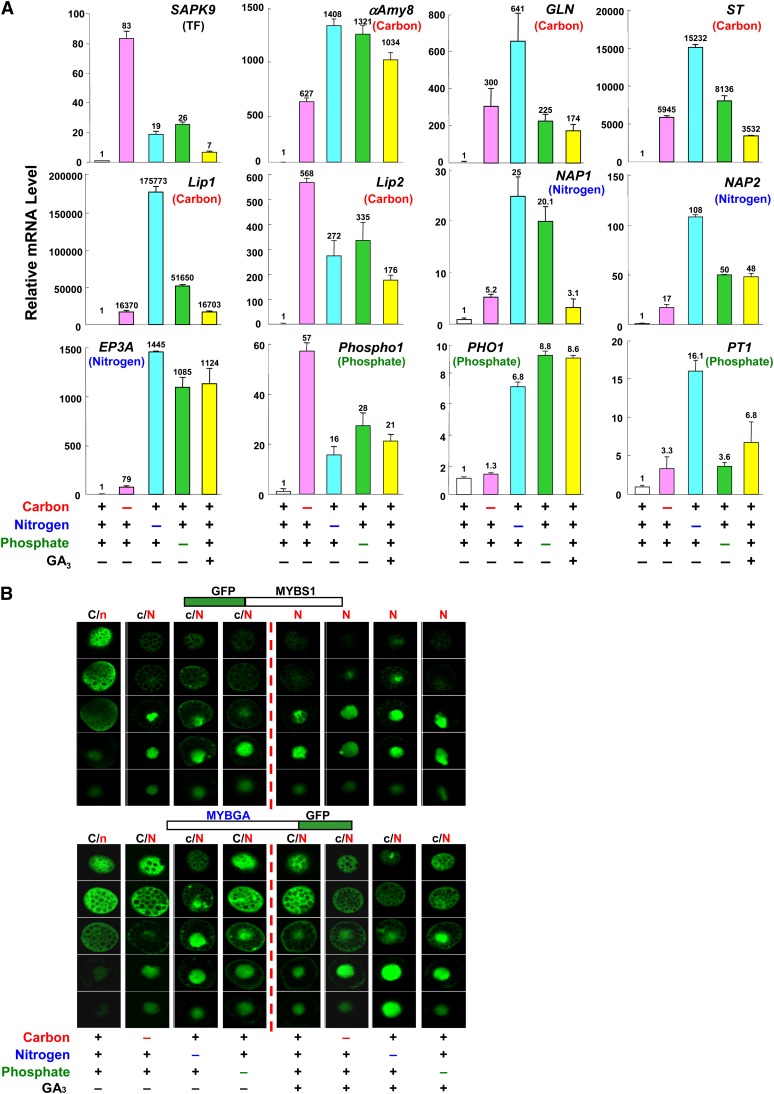
The above study showed that promoters of genes involved in the mobilization of different nutrients were coactivated by MYBS1 and MYBGA, and we asked next whether the expression of GA3-inducible genes with distinct functions could be activated by the starvation of any nutrient. Rice aleurones were incubated in media containing all or lacking the individual nutrients, and mRNAs were purified. We then performed quantitative (real-time) RT-PCR analyses of the genes encoding a transcription factor (SAPK9), endo-β-1,3-glucanase (GLN), sugar transporter (ST), nepenthesin-1 aspartic protease precursor (NAP1 and NAP2), and phosphate transporter (PT1) (Ho et al., 2000; Tsuji et al., 2006) in addition to the six genes used in the study in Figure 6. The accumulation of mRNA of all tested genes was induced by starvation of any one of the nutrients, although with different degrees of induction, as well as by GA3 despite the presence of all nutrients (Figure 7A).
Figure 7.
Carbon, Nitrogen, and Phosphate Starvation Cross-Signaling Induces the Nuclear Import of MYBS1 and MYBGA and the Expression of Hydrolases and Related Proteins.
(A) Rice aleurones were incubated in medium containing or lacking carbon, nitrogen, or phosphate sources with or without GA3 for 24 h. Total RNA was purified and subjected to quantitative (real-time) RT-PCR analysis for various hydrolase, transporter, and transcription factor genes. The parentheses indicate that the tested gene functions as a transcription factor (TF) or is involved in carbon, nitrogen, or phosphate nutrient mobilization.
(B) Barley aleurones were transfected with Ubi:GFP-OsMYBS1 or Ubi:OsMYBGA-GFP and incubated in medium containing or lacking carbon, nitrogen, or phosphate sources with or without GA3 for 24 h. C and N indicate higher GFP signals and c and n indicate lower GFP signals in the cytoplasm and nucleus, respectively. See also Supplemental Figure 12 online.
[See online article for color version of this figure.]
To further investigate the mechanism, we asked whether cross-activation of an array of unrelated genes by diverse nutrient starvation are linked to the nuclear import of MYBS1 and/or MYBGA. As shown in Figure 7B and Supplemental Figure 12A online, in the absence of GA3, GFP-MYBS1 was mostly localized in the cytoplasm when the medium contained carbon, nitrogen, and phosphate but was largely imported into the nucleus when the medium lacked any of these nutrients. In the presence of GA3, GFP-MYBS1 was almost exclusively localized in the nucleus regardless of whether the medium was deficient in any of the nutrients. The cellular localization of MYBGA was similarly regulated by nutrients and GA3, but relatively more MYBGA remained in the cytoplasm than MYBS1. The distribution of GFP by itself was unchanged regardless of the medium employed (see Supplemental Figure 12B online).
DISCUSSION
Seed germination is a unique developmental transition during which the metabolically quiescent seed is transformed to an actively growing seedling. Mobilization of stored nutrients is crucial in supporting the heterotrophic growth in the germinated seed until the autotrophic photosynthetic process is fully established. Two key regulatory factors are in play in this process: the hormone GA, promoting hydrolase production, and the mobilization of nutrients needed for seedling growth. In this study, we have observed that deprivation of an individual nutrient, such as carbon, nitrogen, or phosphate, cross-activates the expression of α-amylase and a variety of other hydrolases, transporters, and regulatory proteins that are responsible for coordinated mobilization of the full complement of different nutrients in the rice and barley endosperms. This process involves unique interactions between MYBS1 and MYBGA, which are induced by nutrient starvation and GA, respectively, resulting in their conuclear import and interaction on target gene promoters in response to the balance of nutrient availability and hormone GA.
Multiple Subcellular Targeting Signals Regulate the Nucleocytoplasmic Shuttling of MYBS1 in Response to Sugar Availability
A sugar-regulated nucleocytoplasmic shuttling of regulatory protein has not been observed in plants previously. Here, we showed that MYBS1 is preferentially imported into and retained in the nucleus under sugar depletion conditions and is exported and retained in the cytoplasm in the presence of sugar. In yeast, the catalytically inactive Snf1 protein kinase interacting with the C terminus of the scaffold protein Gal83 is cytoplasmic in the presence of Glc, whereas under Glc depletion, Snf1 becomes catalytically active and the Snf1-Sal83 complex is translocated to the nucleus (Hedbacker and Carlson, 2006). MYBS1 is a potential substrate of SnRK1A, a yeast Snf1 homolog in rice (Lu et al., 2002, 2007). Whether SnRK1A plays a role in the nuclear import of MYBS1 remains for further study.
We found that a bipartite NLS is sufficient and necessary for the nuclear import of MYBS1. Bipartite NLS motifs have been proposed to function cooperatively for efficient nuclear transport (Kosugi et al., 2009). MYBS1 contains a bipartite NLS separated by a single DNA binding domain, and NLS1 overlaps with the N terminus of the DNA binding domain by five amino acids (Figure 2B). NLS1 is more active than NLS2, as mutations in NLS1 completely impaired nuclear import of the cargo GFP, while mutations in NLS2 led to partial impairment (Figure 2B).
A bipartite NES is also necessary for the sugar-dependent nuclear export of MYBS1. LepB inhibited the nuclear export of MYBS1 in the presence of sugar (Figure 1C), suggesting that the reduced MYBS1 accumulation in the nucleus is due to the active export of MYBS1 to the cytoplasm in the presence of sugar, despite the fact that MYBS1 contains an NLS. The sensitivity to LepB furthermore suggests that the nuclear exclusion of MYBS1 is a Crm1 export receptor–dependent pathway (Yoshida and Horinouchi, 1999). The N-terminal 110 amino acids of MYBS1 were sufficient and necessary to act as a cytoplasmic retention signal (CRS), as MYBS1(108-306), lacking the CRS, was exclusively localized in the nucleus regardless of the presence or absence of sugar, whereas MYBS1(1-306), MYBS1(1-212), and MYBS1(1-112), which all contain the CRS, were actively targeted to the cytoplasm (Figure 2A). NES1 has a stronger export activity than NES2 and happens to reside at the C terminus of the CRS (Figure 2, compare A with C).
GA Antagonizes Sugar Repression by Enhancing the Conuclear Import of MYBS1 and MYBGA and the Formation of a Stable Bipartite MYB-DNA Complex
In this study, we showed that under sugar starvation, MYBS1 was predominantly localized in the nucleus (Figure 1A) but was equally distributed in the nucleus and cytoplasm in gamyb-2 aleurones (Figure 3C). This result indicates that although MYBS1 is actively imported into the nucleus under sugar starvation, MYBGA is necessary for the stable nuclear retention of MYBS1 in aleurone cells. Interaction of two different types of MYBs has not been reported in the literature. Here, we showed that MYBGA and MYBS1 interact physically and synergistically to coactivate the αAmy8 GARC (Figure 4C). Consequently, the bipartite protein (MYBS1-MYBGA)-DNA (GARE-TA box) complex effectively sequesters MYBS1 in the nucleus and prevents it from being exported to the cytoplasm, which in turn activates α-amylase expression regardless of the presence or absence of sugar (Figures 3D to 3F, left panels). MYBGA could be imported into the nucleus, but it is also prone to export to the cytoplasm in the presence of all nutrients (Figures 5B and 7B). MYBS1 promotes the nuclear localization of MYBGA (Figure 5B; see Supplemental Figure 10 online), indicating that the bipartite MYB-DNA complex also sequesters MYBGA in the nucleus.
It is worthwhile to note that MYBS1 and MYBGA interact in the cytoplasm prior to cotransport into the nucleus (Figure 5A). However, active nuclear import of MYBS1 is necessary for the MYBGA-assisted complete nuclear retention of MYBS1 (Figure 5A), for assisting the nuclear localization of MYBGA (Figure 5B), and for coactivation of αAmy8 GARC (Figure 5C). Our studies represent three important discoveries. First, the R1 MYBS1 (with one DNA binding domain) and the R2R3 MYBGA (with two DNA binding domains) interact directly. Second, MYBS1 and MYBGA interact in the absence of their target promoter DNA in the cytoplasm. Third, MYBS1 and MYBGA are coimported into the nucleus.
Diverse Starvation Signals Crosstalk with GA Signal to Coordinate the Expression of Hydrolases and Related Proteins
Although several regulatory factors in the GA signaling pathway have been studied (Sun and Gubler, 2004; Woodger et al., 2004; Hirano et al., 2008; Schwechheimer, 2008), how such a wide a range of genes are coordinately upregulated by GA is not well understood. The presence of conserved but slightly varied nucleotide sequences of GARE and TA boxes in the promoters of hundreds of GA and MYBGA upregulated genes (examples shown in Supplemental Figure 11 online) provides an important clue that these genes might be coactivated by MYBGA and MYBS1. This was confirmed by the MYBGA and MYBS1 coactivation of promoters of several randomly selected genes (Figure 6). This study indicates that most, if not all, of the GA-inducible genes are targets of coactivation by MYBS1 and MYBGA, and the MYB-DNA bipartite complex constitutes one key component in the GA signaling pathway. Various nutrient starvation signals converge to a common mechanism, and crosstalk with the GA signal offers an economic and effective strategy for the coordinated regulation of up to thousands of genes within a limited time window of postgermination growth.
A few genes have previously been shown to be activated by respective nutrient starvation, such as αAmy8 by Suc or Glc (carbon) starvation (Chan et al., 1994; Sheu et al., 1996; Yu et al., 1996), EP3A by nitrogen starvation (Ho et al., 2000), and phosphate transporters PHO1;2 and PT1 by phosphate starvation (Ai et al., 2009; Secco et al., 2010). Interestingly, we found that starvation of any nutrient activates the expression of not only corresponding but also functionally distinct hydrolases and transporters that target other nutrients, indicating that the starvation signal of an individual nutrient turns on a common mechanism to control the expression of a wide variety of genes necessary for the coordinated mobilization of all nutrients stored in the endosperm at the onset of germination. We noticed that each gene responded to the deprivation of different nutrients differentially and was not necessarily more responsive to the starvation of a corresponding nutrient; for example, nitrogen and phosphate starvation resulted in greater fold induction of αAmy8 than was induced by carbon starvation, and carbon starvation led to greater fold induction of Phospho1 than was induced by nitrogen and phosphate starvation (Figure 7A). Additionally, nitrogen starvation followed by phosphate starvation appears to be the most potent in activating these genes (Figure 7A). Aleurone cells are packed with specialized protein bodies known as aleurone grains and other less critical nutrients, including sugars, lipids, phosphate, and metal ions; however, starch granules are absent from the aleurone (Fincher, 1989). This could be part of the reason that aleurone cells are more sensitive to nitrogen and phosphate than to carbon starvation signals.
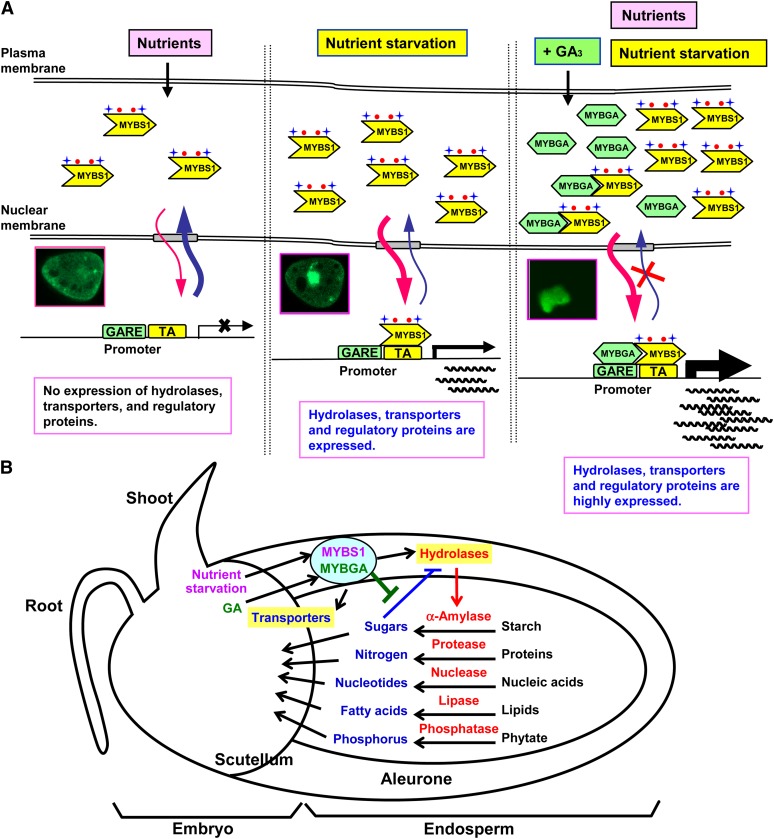
A Model Converging Starvation Signals and Hormone Crosstalk in Regulating Nutrient Mobilization upon Germination
A model concerning the detailed mechanism of nutrient regulation of the nucleocytoplasmic partitioning of MYBS1 and its interaction with MYBGA that leads to maximal expression of hydrolases, transporters, and regulatory factors is illustrated in Figure 8A. The bipartite NLS and NES allow the dynamic shuttling of MYBS1 between the nucleus and cytoplasm. In the presence of nutrients, MYBS1 is actively exported from the nucleus and possibly anchored by a yet unidentified mechanism through the N-terminal CRS in MYBS1, leading to the silencing of target gene transcription. At the very beginning of postgermination seedling growth, nutrient demand/starvation signals induce the expression and nuclear import of MYBS1. MYBS1 binds to the TA box in target gene promoters. However, this interaction may not be tight enough, as some MYBS1 is exported to the cytoplasm and the transcription of target genes is at moderate levels. At later stages, GA is produced and induces the expression of MYBGA. MYBGA interacts with MYBS1 in the cytoplasm, and they are coimported into the nucleus. There they bind to the TA box and GARE, respectively, and form a bipartite MYB-DNA complex that, in turn, effectively sequesters MYBS1 and MYBGA in the nucleus. In this context, the target gene promoters are highly activated and are no longer sensitive to nutrient feedback repression, despite the fact that a relatively high concentration of nutrient products may accumulate in the endosperm as a result of hydrolysis by the coordinated action of a battery of hydrolases (Yu et al., 1996; Woodger et al., 2004). Consequently, all stored nutrients in the endosperm are codigested to small molecules, and these are then transported to the embryo to support seedling growth (Figure 8B).
Figure 8.
A Model Converging Starvation Signals and Hormone Crosstalk in Regulating Nutrient Mobilization upon Germination.
(A) Nutrients and GA regulate the nucleocytoplasmic shuttling and interaction of MYBS1 and MYBGA. Nutrients inhibit while nutrient starvation promotes the nuclear import of MYBS1, which activates target gene transcription. GA induces the expression of MYBGA, which then interacts with MYBS1 in the cytoplasm and nucleus to coactivate target gene transcription.
(B) Diverse nutrient starvation signals converge to crosstalk with GA signal and coordinate the expression of an orchestra of hydrolases and transporters to mobilize nutrients stored in the endosperm to small molecules to support seedling growth.
[See online article for color version of this figure.]
MYBS1 is capable of nuclear import in response to any nutrient starvation signals and activates target gene expression by itself (Figures 6 and 7B), and it is thus a major player in the common nutrient starvation signaling pathway during early postgermination growth. Whether this common pathway acts through the CIPK15-SnRK1A–dependent sugar starvation signaling pathway (Lu et al., 2007; Lee et al., 2009) remains for further study, as does the mechanism of the CRS-dependent cytoplasmic anchoring of MYBS1 in the presence of nutrients. Unraveling the common mechanisms of nutrient starvation–GA cross-signaling may provide information important in the breeding of cereals with the ability to germinate and grow vigorously under normal and unfavorable environments.
In summary, this study has revealed a novel process in which nutrient starvation and hormone signals crosstalk in regulating the developmental transition from metabolic quiescent seed to actively growing seedling. Demands for diverse nutrients converge to a common signaling cascade leading to the induction of MYBS1, whose translocation to the nucleus is regulated by its interactions with the GA-induced MYBGA. Since GA is a key hormone promoting seed germination, MYBS1–MYBGA interactions integrate physiological nutrient demands with a crucial developmental regulation. This unique coordinated process ensures maximal expression of a whole battery of hydrolases and related proteins to efficiently utilize nutrients in support of active seedling growth at the beginning of the life cycle of cereals.
METHODS
Plant Materials
Rice (Oryza sativa cv Tainung 67) and barley (Hordeum vulgare cv Himalaya) were used for all experiments, except that the Tos17-tagged mutant gamyb-2, in which the retrotransposon Tos17 is inserted into the fourth exon of MYBGA, was derived from rice cv Nipponbare (Kaneko et al., 2004). Transgenic rice carrying the ubiquitin gene promoter (Ubi) linked to Os-MYBGA cDNA (Ubi:OsMYBGA) was generated previously (Chen et al., 2006).
Plasmids
Plasmid pUbi-OsMYBGA-Amy contains the rice MYBGA cDNA downstream of the Ubi promoter (Chen et al., 2006). Plasmid pαAmy8SRC/GARC-Luc contains αAmy8 SRC (−318 to −89), the cauliflower mosaic virus (CaMV) 35S minimal promoter, the alcohol dehydrogenase 1 intron (In), the luciferase (Luc) cDNA, and the nopaline synthase gene (Nos) terminator (Chen et al., 2006). Plasmid pUG contains the Ubi promoter, β-glucuronidase (GUS) cDNA, and a Nos terminator (Christensen and Quail, 1996). pAHC18 contains the Luc cDNA between the Ubi promoter and the Nos terminator (Bruce et al., 1989).
Primers
Sequences of all primers used for PCR, quantitative RT-PCR, and RT-PCR analyses are listed in Supplemental Table 1 online. Primer pairs used for plasmid construction are listed in Supplemental Table 2 online.
Construction of Plasmids
To streamline the cloning of various sequences into expression vectors, a number of expression cassettes were generated based on the Gateway cloning system (Invitrogen). Plasmid pUbi-attRI-ccdB-attR2-Nos, which contains the attR recombination sites, a bacterial selection marker (chloramphenicol-resistant gene), and a recombination selection marker ccdB gene, was inserted between the Ubi promoter and the Nos terminator in pAHC18 to replace Luc. This parental plasmid was used for the construction of effectors Ubi:OsMYBS1, Ubi:OsMYBS1(mNLS), Ubi:OsMYBGA, and Ubi:OsMYBGA(Ri) that contain cDNAs encoding Os-MYBS1, Os-MYBS1(mNLS), Os-MYBGA, and Os-MYBGA(Ri) (an RNA interference construct with GFP cDNA flanked by MYBGA cDNA in the sense and antisense orientations), respectively, for use in transient expression assays.
For cellular localization, GFP cDNA was inserted downstream of the Ubi promoter in the parental plasmid, generating pUbi-GFP-attRI-ccdB-attR2-Nos. cDNAs encoding truncated and NLS-mutagenized Os-MYBS1 were PCR amplified, cloned into pENTRY/D-TOPO, and then into pUbi-GFP-attRI-ccdB-attR2-Nos by LR Clonase (Invitrogen), generating pUbi-GFP-MYBS1∆-Nos and pUbi-GFP-MYBS1(mNLS)-Nos.
For BiFC analysis, Os-MYBS1 cDNA was fused to the 5′ end of YFP (encoding nYFP) and Os-MYBGA cDNA was fused to the 3′ end of YFP (encoding cYFP). Fusion genes nYFP-MYBS1 and cYFP-MYBGA were PCR amplified, cloned to pENTRY/D-TOPO (Invitrogen), and then cloned into pUbi-attRI-ccdB-attR2-Nos by LR Clonase (Invitrogen), generating pUbi:nYFP-MYBS1-Nos and pUbi:cYFP-MYBGA-Nos.
For GAL4-UAS two-hybrid assays, Os-MYBS1 cDNA was fused to the DNA encoding the GAL4 DNA binding domain and Os-MYBGA cDNA to the DNA encoding the GAL4 activation domain. Fusion genes were PCR amplified, cloned into pENTRY/D-TOPO, and then into pUbi-attRI-ccdB-attR2-Nos by LR Clonase (Invitrogen), generating pUbi-GBD-MYBS1-Nos and pUbi-GAD-MYBGA-Nos.
α-Amylase Activity Assay on Starch Agar Plate
Pollen development was defective, and therefore homozygous seeds could be propagated only from heterozygous lines of the gamyb-2 mutant and Os-MYBGA–overexpressing transgenic rice (Kaneko et al., 2004; Chen et al., 2006). A starch agar plate α-amylase assay method (Yamaguchi, 1998; Chen et al., 2006) was used for the identification of homozygous and heterozygous seeds from segregating populations of MYBGA-overexpressing transgenic rice and gamyb-2 mutant. Embryos and endosperms were collected separately from a population of gamyb-2 mutant seeds and Ubi:OsMYBGA transgenic seeds and placed on two 96-well plates. Each isolated endosperm and its corresponding embryo were labeled with the same identification number. Endosperms were sterilized with 1% NaOCl for 30 min, washed with distilled water, and then a small piece of endosperm was excised from the distal end of the seed (relative to the embryo end). Sterilized endosperm samples were placed with the cutting side facing down on starch agar plates, which contained 0.2% starch and 2% agar (Duchefa) with or without GA3, and incubated at 30°C in the dark for 3 d. Endosperm samples were removed, and α-amylase activity was assayed by staining agar with a solution containing 0.1% I2 and 1% KI. Clear zones appeared if α-amylases were expressed in endosperms and secreted into the starch agar. The absence of clear zones after staining of the starch agar plate containing GA3 indicated that the endosperm was derived from a gamyb-2 mutant seed. The appearance of clear zones after staining of starch agar without the supplement of GA3 indicated that the endosperm was derived from a transgenic seed overexpressing MYBGA. Transgenic endosperms giving rise to large clear zones were likely due to high levels of MYBGA expression. Aleurone layers were then isolated from the remaining portion of endosperms that corresponded to gamyb-2 mutant and MYBGA-overexpressing transgenic rice seeds for transient expression or RNA extraction.
Confocal Microscopy for Detection of GFP
For detecting the cellular localization of MYB-GFP fusion proteins, the embryoless barley half-seeds (endosperms) were sterilized and incubated for 3 d (rice) or 4 d (barley) in 20 mM sodium succinate buffer (pH 5.0) containing 20 mM CaCl2 and 10 µM chloramphenicol. Eight and 16 isolated aleurone layers of barley and rice, respectively, were arranged on a plate for one biolistic gene gun bombardment to deliver DNA constructs into aleurone cells. Bombarded aleurone layers were divided into two halves and incubated for 24 h at 28°C (rice) or 20°C (barley) in Murashige and Skoog medium containing 100 mM Glc or 100 mM mannitol. Aleurone cells expressing GFP were imaged with a Zeiss confocal microscope (LSM510META) using a 488-nm laser line for excitation and a 515- to 560-nm long-pass filter for emission. Very fast speed (performed in less than 1 min) and weak (only 20 to 25%) laser strength (wavelength of 488 nm) were used to scan the fluorescence signal to prevent GFP signal decay. Thirty optical sections of GFP scans were made from the top (the pericarp side toward the outside of the seed) toward the bottom (the starchy endosperm side) of each aleurone cell. The depth of the scan was ∼20 to 28 μm depending on the cell size, which usually spanned over half the size of each cell and included most cytoplasm and the entire nucleus. Cells expressing high, medium, and low signals (due to varied expression levels of GFP-MYB in transfected cells) were analyzed to avoid any bias. GFP signals in Figures 1B and 2 and Supplemental Figure 10 online were assessed qualitatively by visual comparisons. A GFP signal higher in the nucleus than in the cytoplasm was classified as c<N, higher in the cytoplasm than in the nucleus was classified as C>n, and similar in the cytoplasm and the nucleus was classified as C=N.
GFP signals in the cytoplasm in +S cells were more visible in sections of the upper half of cells. In contrast, the GFP signal in all sections of nucleus in −S cells always maintained similar intensity regardless of the depth of the scan. The gradient in the cytoplasmically localized fluorescence during laser sectioning through aleurone cells might be caused by photobleaching and/or reduced light penetration to the lower region of cells.
Embryo and Endosperm/Aleurone Transient Expression Assays
Aleurone/endosperm transient expression assays were performed as described (Chen et al., 2006). As the transfection efficiency may vary between samples in the transient expression assay, the enzyme activity of an internal control pUG (containing Ubi:GUS) was used to normalize the reporter enzyme activity. The ratio of test DNA to internal control plasmid DNA was 4:1. Each independent experiment consisted of three replicates, with eight endosperms for each treatment, and was repeated three to four times with similar results. Luciferase and GUS activity assays were performed as described (Chen et al., 2006). Error bars indicate the se for three replicate experiments.
Real-Time Quantitative RT-PCR
The embryoless rice half-seeds (endosperms) were sterilized and incubated in Murashige and Skoog medium containing 100 mM Glc for 3 d. Aleurone layers were isolated by scratching away starch in the endosperm with a razor blade. Thirty isolated aleurones were incubated in medium containing or lacking carbon (Glc; 19.82 g/L), nitrogen (NH4NO3; 1650 mg/L), or phosphate (KH2PO4; 170 mg/L) sources with or without GA3 for 24 h. Total cellular RNAs were extracted with the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Five micrograms of RNA was treated with 1 unit of RNase-free DNase I (Promega) at 37°C for 15 min, and the first-strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Twenty-fold dilution of reaction products was subjected to real-time quantitative RT-PCR analysis (using gene-specific primers that were designed based on the software provided in the Universal ProbeLibrary Assay Design Center [https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp?id=uplct_030000]). A 20-μL quantitative PCR solution contained 5 μL of cDNA, 400 nmol/L forward and reverse primers, 200 nmol/L UPL probe (Roche), 12.5 μL of LightCycler 480 Probes Master (Roche), and water. Thermal cycling was performed on a SYBR Green LightCycler 480 instrument (Roche Diagnostics) under the following conditions: 95°C for 10 min, and 45 cycles at 95°C for 10 s and 60°C for 20 s. All other reaction conditions were as described by the manufacturer’s instructions.
The cycle threshold (Ct) value is defined as the point at which the fluorescence rises above the background fluorescence. The relative mRNA levels of target genes shown in Figures 3D to 3F were calculated by the formula 2(−ΔCt), where the ΔCt value of the sample was determined by subtracting the average Ct value of target genes (αAmy8, MYBGA, and MYBS1) from the average Ct value of the rice UBQ5 gene (Jain et al., 2006) using the LightCycler software (see Supplemental Table 3 online). The relative mRNA levels of the target genes shown in Figure 7A represent fold change of mRNA levels of target genes that was determined from the ΔΔCt values with the equation 2(−ΔΔCt) (Livak and Schmittgen, 2001).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: OsMYBS1 (AY151042.1), OsMYBGA (AK119607.1), HvMYBS1 (AK366932.1), HvMYBGA (AY008692.1), αAmy8/RAmy3E (AK064300), EP3A encoding Cys protease (AF099203), GLN encoding putative β-1,3-glucanase (AK110902), Lip1 encoding GDSL-motif lipase (AK070261), Lip2 encoding GDSL-motif lipase (AK059511), NAP1 encoding nepenthesin-1 aspartic protease precursor 1 (AK072228), NAP2 encoding nepenthesin-1 aspartic protease precursor 2 (AK069691), Phospho1 encoding phosphatase-like (AK061237), PHO1 encoding phosphate transporter (AK100323), SAPK9 encoding Ser/Thr protein kinase (AK69697), ST encoding sugar transporter family protein (AK069132), PT1 encoding maize (Zea mays) phosphate transport protein (AK106694), and UBQ5 encoding ubiquitin 5 (AK061988).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Glc Regulates the Nucleocytoplasmic Shuttling of MYBS1, Related to Figures 1A and 1C.
Supplemental Figure 2. GFP without MYBS1 Was Distributed in Both the Cytoplasm and the Nucleus of Aleurone Cells Regardless of the Medium (+S or −S) or the Presence or Absence of GA3, Related to Figures 1A and 3A.
Supplemental Figure 3. MYBS1(108-212) Contains a NLS, and MYBS1(1-112) Contains a CRS, Related to Figure 2A.
Supplemental Figure 4. NES1 Is a Major Nuclear Export Signal of MYBS1, Related to Figure 2C.
Supplemental Figure 5. GA3 Promotes Nuclear Localization of MYBS1, and MYBGA Is Sufficient and Necessary for Promoting Nuclear Localization of MYBS1, Related to Figures 3A to 3C.
Supplemental Figure 6. GA and MYBGA Antagonize Sugar Repression of α-Amylase Expression, Related to Figures 3D to 3F.
Supplemental Figure 7. BiFC Complementation Test Indicates That MYBS1 and MYBGA Interact Physically, Related to Figure 4A.
Supplemental Figure 8. MYBS1 and MYBGA Interact in the Cytoplasm Prior to Cotransport into the Nucleus, Related to Figure 5A.
Supplemental Figure 9. Overexpression of MYBS1 Promotes the Nuclear Localization of MYBGA, Related to Figure 5B.
Supplemental Figure 10. The Nuclear Localization of MYBGA Is Enhanced by MYBS1
Supplemental Figure 11. Consensus GARE and TA Box Are Present in Promoters of GA-Inducible Genes.
Supplemental Figure 12. Carbon, Nitrogen, and Phosphate Starvation Promote the Nuclear Import of MYBS1 and MYBGA, Related to Figure 7B.
Supplemental Table 1. Primer List.
Supplemental Table 2. Primer Sets Used for Plasmid Construction.
Supplemental Table 3. Quantitative Analysis of αAmy8, MYBGA, and MYBS1 mRNA Levels in Aleurones of Wild-Type Rice, MYBGA-Overexpressing Transgenic Rice, and MYBGA-Deficient Mutant Rice.
Supplementary Material
Acknowledgments
We thank AndreAna Pena for critical review of the manuscript. This work was supported by Academia Sinica and the National Science Council (Grants NSC-97-2321-B-001-007 and NSC-98-2321-B-001-004) of the Republic of China.
AUTHOR CONTRIBUTIONS
Y.-F.H., S.-M.Y., and T.-H.D.H. designed the research. C.-F.W., S.-L.H., R.-H.Y., C.-A.L., P.-W.C., A.C., and L.-C.Y. performed the research. S.-M.Y. and T.-H.D.H. wrote the article.
Glossary
- GA
gibberellin
- SRC
sugar response complex
- GARC
gibberellin response complex
- GARE
gibberellin response element
- GFP
green fluorescent protein
- +S
with glucose
- −S
without glucose
- LepB
leptomycin B
- NLS
nuclear localization signal
- NES
nuclear export signals
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- CRS
cytoplasmic retention signal
- Ct
cycle threshold
References
- Ai P., Sun S., Zhao J., Fan X., Xin W., Guo Q., Yu L., Shen Q., Wu P., Miller A.J., Xu G. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Akazawa T., Hara-Nishimura I. (1985). Topographic aspects of biosynthesis, extracellular secretion and intracellular storage of proteins in plant cells. Annu. Rev. Plant Physiol. 70: 441–472 [Google Scholar]
- Beck E., Ziegler P. (1989). Biosynthesis and degradation of starch in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 95–117 [Google Scholar]
- Bethke P.C., Schuurink R., Jones R.L. (1997). Hormonal signalling in cereal aleurone. J. Exp. Bot. 48: 1337–1356 [Google Scholar]
- Bruce W.B., Christensen A.H., Klein T., Fromm M., Quail P.H. (1989). Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc. Natl. Acad. Sci. USA 86: 9692–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M.T., Chao Y.C., Yu S.M. (1994). Novel gene expression system for plant cells based on induction of alpha-amylase promoter by carbohydrate starvation. J. Biol. Chem. 269: 17635–17641 [PubMed] [Google Scholar]
- Chen K., An Y.-Q.C. (2006). Transcriptional responses to gibberellin and abscisic acid in barley aleurone. J. Integr. Plant Biol. 48: 591–612 [Google Scholar]
- Chen P.W., Chiang C.M., Tseng T.H., Yu S.M. (2006). Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of α-amylase genes. Plant Cell 18: 2326–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A.H., Quail P.H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5: 213–218 [DOI] [PubMed] [Google Scholar]
- Diaz I., Vicente-Carbajosa J., Abraham Z., Martínez M., Isabel-La Moneda I., Carbonero P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29: 453–464 [DOI] [PubMed] [Google Scholar]
- Fincher G.G. (1989). Molecular and cellular biology associated with endosperm mobilisation in germinating cereal grains. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 305–346 [Google Scholar]
- Gómez-Cadenas A., Zentella R., Walker-Simmons M.K., Ho T.H. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13: 667–679 [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Jacobsen J.V. (1992). Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell 4: 1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Kalla R., Roberts J.K., Jacobsen J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Raventos D., Keys M., Watts R., Mundy J., Jacobsen J.V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Gubler F., Watts R.J., Kalla R., Matthews P., Keys M., Jacobsen J.V. (1997). Cloning of a rice cDNA encoding a transcription factor homologous to barley GAMyb. Plant Cell Physiol. 38: 362–365 [DOI] [PubMed] [Google Scholar]
- Hedbacker K., Carlson M. (2006). Regulation of the nucleocytoplasmic distribution of Snf1-Gal83 protein kinase. Eukaryot. Cell 5: 1950–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Ueguchi-Tanaka M., Matsuoka M. (2008). GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 13: 192–199 [DOI] [PubMed] [Google Scholar]
- Ho S.L., Tong W.F., Yu S.M. (2000). Multiple mode regulation of a cysteine proteinase gene expression in rice. Plant Physiol. 122: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Tyagi A.K., Khurana J.P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345: 646–651 [DOI] [PubMed] [Google Scholar]
- Kaneko M., Inukai Y., Ueguchi-Tanaka M., Itoh H., Izawa T., Kobayashi Y., Hattori T., Miyao A., Hirochika H., Ashikari M., Matsuoka M. (2004). Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer E.E., Chandler J.M., Foolad M.R., Rodriguez R.L. (1993). Correlation between alpha-amylase gene expression and seedling vigor in rice. Euphytica 66: 163–169 [Google Scholar]
- Kennedy B.M. (1980). Nutritional quality of rice endosperm. In Rice: Production and Utilization. B.S. Luh , ed (Westport, CT: AVI Publishing), pp. 439–469
- Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H. (2009). Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 284: 478–485 [DOI] [PubMed] [Google Scholar]
- Lanahan M.B., Ho T.H., Rogers S.W., Rogers J.C. (1992). A gibberellin response complex in cereal α-amylase gene promoters. Plant Cell 4: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.W., Chen P.W., Lu C.A., Chen S., Ho T.H., Yu S.M. (2009). Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2: 1–9 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu C.A., Ho T.H., Ho S.L., Yu S.M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.A., Lim E.K., Yu S.M. (1998). Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 273: 10120–10131 [DOI] [PubMed] [Google Scholar]
- Lu C.A., Lin C.C., Lee K.W., Chen J.L., Huang L.F., Ho S.L., Liu H.J., Hsing Y.I., Yu S.M. (2007). The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19: 2484–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjhan S., Karrer E.E., Rodriguez R.L. (1992). Localizing alpha-amylase gene expression in germinated rice grains. Plant Cell Physiol. 33: 73–79 [Google Scholar]
- Rogers J.C., Lanahan M.B., Rogers S.W. (1994). The cis-acting gibberellin response complex in high pI α-amylase gene promoters. Requirement of a coupling element for high-level transcription. Plant Physiol. 105: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H., Kanei-Ishii C., Nagase T., Nakagoshi H., Gonda T.J., Ishii S. (1989). Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc. Natl. Acad. Sci. USA 86: 5758–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C. (2008). Understanding gibberellic acid signaling—Are we there yet? Curr. Opin. Plant Biol. 11: 9–15 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Willige B.C. (2009). Shedding light on gibberellic acid signalling. Curr. Opin. Plant Biol. 12: 57–62 [DOI] [PubMed] [Google Scholar]
- Secco D., Baumann A., Poirier Y. (2010). Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 152: 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Gaur R.K., Ikegami M. (2011). Subcellular localization of V2 protein of Tomato leaf curl Java virus by using green fluorescent protein and yeast hybrid system. Protoplasma 248: 281–288 [DOI] [PubMed] [Google Scholar]
- Sheu J.-J., Yu T.-S., Tong W.-F., Yu S.-M. (1996). Carbohydrate starvation stimulates differential expression of rice alpha-amylase genes that is modulated through complicated transcriptional and posttranscriptional processes. J. Biol. Chem. 271: 26998–27004 [DOI] [PubMed] [Google Scholar]
- Shin D., Moon S.J., Han S., Kim B.G., Park S.R., Lee S.K., Yoon H.J., Lee H.E., Kwon H.B., Baek D., Yi B.Y., Byun M.O. (2011). Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol. 155: 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P. (2010). Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 154: 567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P., Gubler F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Sutoh K., Yamauchi D. (2003). Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 34: 635–645 [DOI] [PubMed] [Google Scholar]
- Toyoshima F., Moriguchi T., Wada A., Fukuda M., Nishida E. (1998). Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 17: 2728–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Aya K., Ueguchi-Tanaka M., Shimada Y., Nakazono M., Watanabe R., Nishizawa N.K., Gomi K., Shimada A., Kitano H., Ashikari M., Matsuoka M. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47: 427–444 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Katoh E., Ohmiya H., Asano K., Saji S., Hongyu X., Ashikari M., Kitano H., Yamaguchi I., Matsuoka M. (2007). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.J., Wan A.R., Hsu C.M., Lee K.W., Yu S.M., Jauh G.Y. (2007). Transcriptomic adaptations in rice suspension cells under sucrose starvation. Plant Mol. Biol. 63: 441–463 [DOI] [PubMed] [Google Scholar]
- Woodger F., Jacobsen J.V., Gubler F. (2004). Gibberellin action in germinated cereal grains. In Plant Hormones: Biosynthesis, Signal Transduction, Action! P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 221–240
- Yamaguchi J. (1998). Analysis of embryo-specific α-amylase using isolated mature rice embryos. Breed. Sci. 48: 365–370 [Google Scholar]
- Yanhui C., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60: 107–124 [DOI] [PubMed] [Google Scholar]
- Yoshida M., Horinouchi S. (1999). Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann. N. Y. Acad. Sci. 886: 23–36 [DOI] [PubMed] [Google Scholar]
- Yu S.M.(1999a). Regulation of alpha-amylase gene expression. In Molecular Biology of Rice, K. Shimamoto, ed (Tokyo: Springer-Verlag), pp. 161–178.
- Yu S.M. (1999b). Cellular and genetic responses of plants to sugar starvation. Plant Physiol. 121: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.M., Lee Y.C., Fang S.C., Chan M.T., Hwa S.F., Liu L.F. (1996). Sugars act as signal molecules and osmotica to regulate the expression of alpha-amylase genes and metabolic activities in germinating cereal grains. Plant Mol. Biol. 30: 1277–1289 [DOI] [PubMed] [Google Scholar]
- Zentella R., Yamauchi D., Ho T.H. (2002). Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14: 2289–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.