This study analyzes the genetic and protein interactions between the two LOB domain proteins JLO and AS2. Both proteins physically interact with each other but are also able to build homomeric complexes. Genetically, functions were identified that are seemingly common to JLO and AS2 in the regulation of KNOX and PIN expression. JLO depends for some of these functions on AS2.
Abstract
Organ initiation requires the specification of a group of founder cells at the flanks of the shoot apical meristem and the creation of a functional boundary that separates the incipient primordia from the remainder of the meristem. Organ development is closely linked to the downregulation of class I KNOTTED1 LIKE HOMEOBOX (KNOX) genes and accumulation of auxin at sites of primordia initiation. Here, we show that Arabidopsis thaliana JAGGED LATERAL ORGANS (JLO), a member of the LATERAL ORGAN BOUNDARY DOMAIN (LBD) gene family, is required for coordinated organ development in shoot and floral meristems. Loss of JLO function results in ectopic expression of the KNOX genes SHOOT MERISTEMLESS and BREVIPEDICELLUS (BP), indicating that JLO acts to restrict KNOX expression. JLO acts in a trimeric protein complex with ASYMMETRIC LEAVES2 (AS2), another LBD protein, and AS1 to suppress BP expression in lateral organs. In addition to its role in KNOX regulation, we identified a role for AS2 in regulating PINFORMED (PIN) expression and auxin transport from embryogenesis onwards together with JLO. We propose that different JLO and AS2 protein complexes, possibly also comprising other LBD proteins, coordinate auxin distribution and meristem function through the regulation of KNOX and PIN expression during Arabidopsis development.
INTRODUCTION
Organ formation and growth requires a continuous supply of new cells. The shoot apical meristem (SAM) of higher plants can provide these through a pool of pluripotent stem cells in the central zones. When these stem cells divide, daughter cells are displaced toward the periphery where they can be recruited to form organ primordia. There, cells undergo rapid divisions, expansion, and ultimately differentiation. Cell fate appears to be determined mostly by a cell’s position within the SAM. Emerging organs are separated from the remainder of the meristem by morphological boundaries with distinct cell division and gene expression patterns (reviewed in Rast and Simon, 2008).
Organ initiation in the peripheral zone is regulated by two critical events: the accumulation of auxin in a group of founder cells and a simultaneous change in gene expression programs. Such local auxin maxima are generated by active polar transport mediated through auxin efflux carriers of the PINFORMED (PIN) family (Galweiler et al., 1998; Paponov et al., 2005; Zazimalova et al., 2007). The direction of auxin flux within the L1 layer of the SAM is mainly determined by the subcellular localization of PIN1 (Benkova et al., 2003; Friml et al., 2003; Reinhardt et al., 2003). Live imaging of PIN1-GFP (for green fluorescent protein) revealed the formation of an expression focus at the flanks of the SAM that raises auxin levels in organ founder cells and depletes auxin from the vicinity. As primordia growth starts, PIN1 polarity reverses to form a new auxin peak at a distant position. Thus, phyllotactic pattering requires dynamic PIN1 polarity changes to generate new auxin peaks (Heisler et al., 2005).
Meristematic and organ founder cells are further distinguished by the expression of specific gene sets. These contrasting patterns depend on the mutual repression between meristem and organ-specific genes. For example, SHOOT MERISTEMLESS (STM), a member of the class I KNOTTED LIKE HOMEOBOX (KNOX) family, is specifically expressed in meristematic tissues and excluded from organ primordia. stm mutants lack a functional SAM due to ectopic expression of the MYB domain protein ASYMMETRIC LEAVES1 (AS1), which is normally confined to organ primordia (Byrne et al., 2000). AS1 in turn restricts the KNOX genes BREVIPEDICELLUS (BP), KNAT2, and KNAT6 from organ initials (Belles-Boix et al., 2006; Byrne et al., 2000, 2002; Ori et al., 2000). This repression of KNOX genes depends on the molecular interaction between AS1 and AS2, a protein that belongs to the LATERAL ORGAN BOUNDARY DOMAIN (LBD) family of Arabidopsis thaliana that shares the plant-specific LOB domain (Shuai et al., 2002; Xu et al., 2003; Guo et al., 2008). All LBD proteins analyzed localize to the nucleus (Iwakawa et al., 2002; Borghi et al., 2007; Naito et al., 2007), and the LOB domain was shown to bind to DNA in vitro (Husbands et al., 2007). Heteromers of AS1 and AS2 can directly interact with the promoter regions of BP and KNAT2. This binding is suggested to recruit the chromatin remodeling factor HIRA, resulting in stable repression of KNOX genes in lateral organs (Phelps-Durr et al., 2005; Guo et al., 2008).
The downregulation of KNOX genes in organ primordia and auxin-regulated gene expression programs are interconnected. Auxin activity appears to act in parallel with the AS1/AS2 module to exclude BP expression from lateral organs (Hay et al., 2006). Furthermore, leaf defects of plants that misexpress KNOX genes are in part caused by the disruption of local auxin gradients in the leaf margins (Tsiantis et al., 1999; Zgurski et al., 2005). Correct cell fate allocation therefore requires the combined activities of auxin and AS1/AS2 activity in the cells of lateral organs and an antagonistic activity of KNOX genes in meristematic cells.
An important part of this regulation may take place at the boundaries between organ primordia and the meristem (Aida and Tasaka, 2006). A number of boundary-specific genes were shown to contribute to meristem homeostasis and organ development. Among them are CUP-SHAPED COTYLEDON1 (CUC1), CUC2, and CUC3, which are already required during the early stages of embryogenesis to activate and later delineate STM expression (Aida et al., 1999). Furthermore, boundary-specific expression of LATERAL ORGAN BOUNDARIES (LOB), the founding member of the LBD gene family, is promoted by BP and AS2 (Lin et al., 2003).
The LBD family gene JAGGED LATERAL ORGANS (JLO/LBD30) was previously shown to be required for auxin-mediated development from the earliest stages of embryogenesis onwards. JLO loss-of-function mutants (jlo-1 and jlo-2) arrest during embryogenesis or early seedling stages due to aberrant cell division patterns and meristem cell differentiation. These defects are at least in part caused by effects on the BODENLOS/MONOPTEROS pathway and a resulting failure in auxin signaling. As a consequence, expression of members of the PIN and PLETHORA gene families is severely reduced in jlo mutant roots (Bureau et al., 2010). During shoot development, JLO is expressed at sites of organ initiation and later in meristem-to-organ boundaries. Ectopic high-level expression of JLO in organ primordia causes leaf lobing and misexpression of both STM and BP in developing organs. This indicates that JLO could act from the boundary to orchestrate gene expression patterns (Borghi et al., 2007). However, all jlo mutant alleles described so far grossly disturbed embryogenesis and arrested growth already at early stages, so that JLO functions during postembryonic development were not yet understood.
Here, we characterized an allelic series of jlo alleles, which allowed us to uncover the role of JLO during organ development in shoot and floral meristems. We find that JLO integrates the promotion of PIN transcription with the regulation of KNOX expression. We demonstrate that the JLO and AS2 proteins interact molecularly and form multimeric complexes with AS1 to suppress KNOX expression. Furthermore, we uncover a previously unsuspected role for AS2, together with JLO, in regulating auxin transport in seedling roots.
RESULTS
Isolation of Novel jlo Alleles
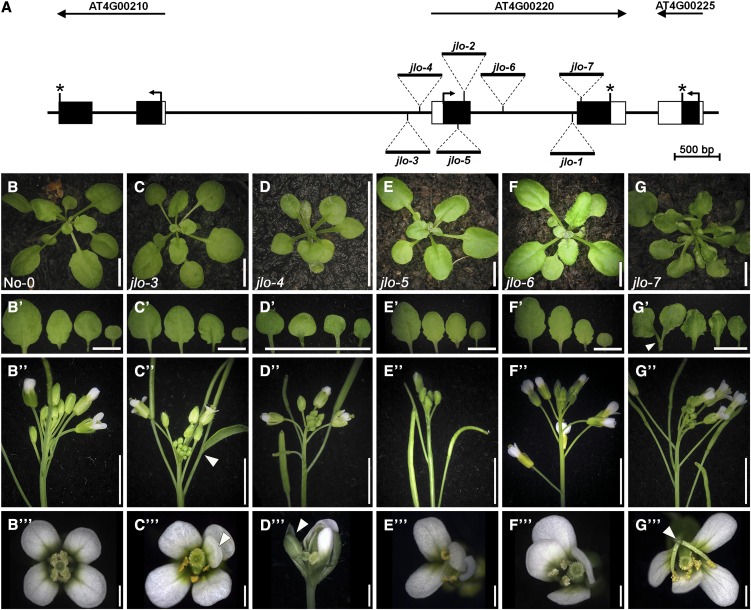
The embryonic or early seedling lethality of jlo-1 and jlo-2 mutants interfered with any functional analysis during later stages of development (Borghi et al., 2007; Bureau et al., 2010). Thus, most of our conclusions regarding the function of JLO in the shoot were drawn from misexpression experiments. We now characterized a series of novel jlo alleles that revealed phenotypically milder defects (jlo-3 to jlo-7, Figure 1A) and allowed the dissection of JLO functions during later development. RT-PCR analyses of RNA isolated from seedlings showed that insertions located 5′ to the JLO transcriptional start (jlo-3 and jlo-4) cause a reduction in RNA levels, which are more severe in the jlo-4 allele. Homozygous jlo-5 to jlo-7 mutants produced shortened transcripts that were truncated 3′ to the insertions (see Supplemental Figure 1A online). Sequencing of theses transcripts confirmed that the jlo-5 to jlo-7 alleles encode proteins that lack parts of the conserved LOB domain (see Supplemental Figures 1B and 1C online).
Figure 1.
Analysis of jlo Mutant Alleles.
(A) Gene structure of JLO and the neighboring genes on chromosome 4. The positions of the Ds element (jlo-2 to jlo-7) or T-DNA (jlo-1) insertions are indicated. The jlo-1 and jlo-2 alleles have been described previously. Black boxes, exons; white boxes, untranslated regions; black arrows, start codon; asterisk, stop codon.
(B)/(B’) to (G)/(G’) Three-week-old wild-type plants (No-0) compared with homozygous jlo-3 to jlo-7 mutants and the corresponding first four leaves. jlo-4 mutants are small and develop misshapen leaves ([D]/[D’]). jlo-7 leaves curl upwards and are occasionally fused ([G]/[G’]; arrowhead).
(B’’) to (G’’) and (B’’’) to (G’’’) Inflorescences and flowers of the wild type and jlo mutants. Phenotypes comprised floral meristem identity defects ([C’’] and [D’’’] arrowheads), homeotic transformations ([C’’’]; arrowhead), reduced number of floral organs ([E’’’] and [F’’’]), and organ fusions ([G’’’]; arrowhead). Bars = 1 cm.
[See online article for color version of this figure.]
Allelism tests were subsequently performed. As expected for allelic mutations, transheterozygosis of jlo-2 with jlo-3 to jlo-7 failed to complement the embryo mutant phenotypes and seedling lethality of homozygous jlo-2 mutants (see Supplemental Table 1 and Supplemental Figure 2 online). We conclude that the phenotypic defects observed in the different jlo alleles solely are due to reduced or missing JLO function and not to previously unnoticed mutations in other genes.
Developmental Defects in jlo-3 to jlo-7 Mutants
Plants homozygous for the jlo-4 mutation showed phenotypic alterations from early stages of seedling development onwards. Similar to the previously described jlo-2 allele (Bureau et al., 2010), jlo-4 seedlings were smaller than wild-type seedlings with a disorganized root and narrow cotyledons. However, although growth and leaf development was strongly impaired, homozygous jlo-4 mutants were eventually able to bolt (Figures 1D to 1D’’’). jlo-7 seedlings generated curled and fused leaves (Figures 1G and 1G’, arrowhead). The other jlo mutants studied here displayed normal vegetative development.
After the transition to flowering, all novel jlo alleles displayed related defects that were categorized into three classes. Class I includes floral meristem identity defects (e.g., flower meristems that show characteristics of inflorescence meristems) (Figure 1D’’’, arrowhead). Some of these flowers were subtended by cauline leaves, further supporting this assumption (Figure 1C’’, arrowhead). This phenotype appeared with a low frequency (4.4%; n = 18/405) and mainly within the first four flowers of jlo-3 and jlo-4 mutants (see Supplemental Figure 3 and Supplemental Table 2 online).
Class II comprises homeotic transformations of petals and stamen, which appeared mostly on the first flowers of jlo mutants (25.4%; n = 103/405; Figure 1C’’’; see Supplemental Figure 3 and Supplemental Table 2 online). However, the majority of mutant flowers exhibited a reduced number and size of sepals, petals, and stamens or displayed organ fusions (55.8%; n = 226/405; Figures 1E’’’ to 1G’’’). Together, these phenotypes were classified as class III.
The jlo Mutant Phenotype Is Dosage Dependent
jlo loss-of-function mutants arrest during embryogenesis or early seedling stages (jlo-1 and jlo-2), whereas reduced JLO activity causes leaf and floral defects (jlo-3 to jlo-7). In addition, we previously found altered target gene expression in heterozygous jlo-2/+ plants (Bureau et al., 2010), suggesting that plant development may be sensitive to the level of JLO activity. We therefore compared shoot development of wild-type, jlo-2/+, and jlo-2 plants to test this notion. Heterozygous jlo-2/+ mutants appeared aphenotypic during vegetative development. However, after floral transition, jlo-2/+ flowers displayed defects that were comparable to those of jlo-3 to jlo-7 mutants, namely, homeotic transformations of second and third whorl organs, reduction in floral organ number, and organ fusions. Floral buds opened prematurely due to smaller sepals and petals, and stamen size was notably reduced (see Supplemental Figure 4 online).
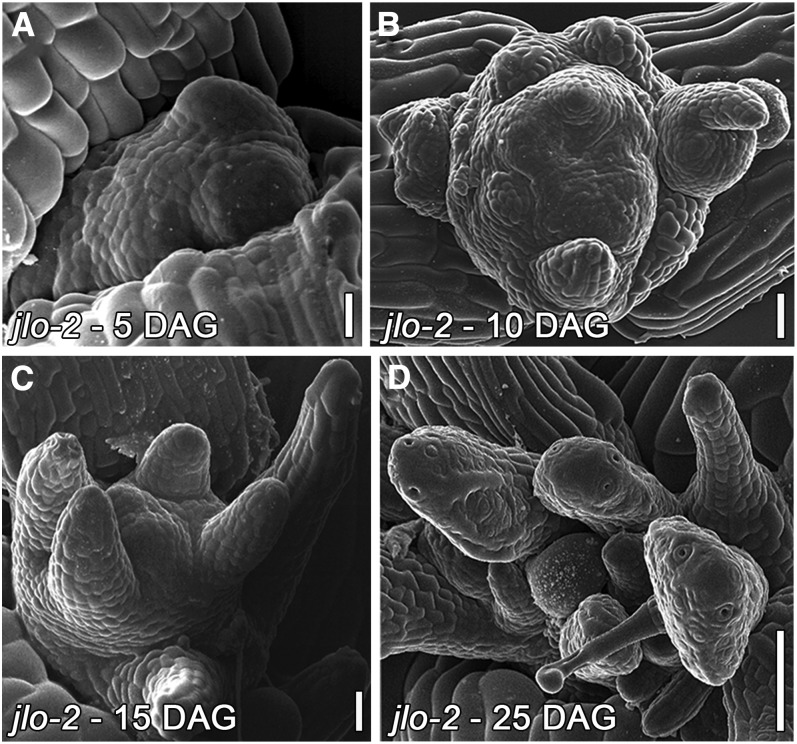
jlo-2 homozygous mutants show a severe retardation in shoot growth. Scanning electron micrographs revealed that mutant meristems initiated primordia at arbitrary positions, indicating phyllotactic defects (Figures 2A to 2D). Most of these organs failed to grow out, and the remaining primordia gave rise to radialized organs (Figures 2B to 2D). By 25 d after germination (DAG), the shoot meristem had stopped further growth. Notably, this phenotype resembles that caused by inducible misexpression of a dominant-negative version of JLO (JLO-DN; Borghi et al., 2007). Thus, both jlo-2/+ and jlo-2 plants exhibit defects in organ development, albeit with different severity.
Figure 2.
Shoot Development of jlo-2 Mutants.
Scanning electron micrographs of jlo-2 SAMs. Bars = 20 µm in (A) and (B), 30 µm in (C), and 100 µm in (D).
(A) Meristems reveal phyllotactic defects at 5 DAG.
(B) and (C) At 10 and 15 DAG, the shoot apex is expanded and organs are initiated but fail to grow out or appear radialized.
(D) Some organs display leaf-like structures at 25 DAG.
Class I KNOX Genes Are Upregulated and Ectopically Expressed in jlo Mutants
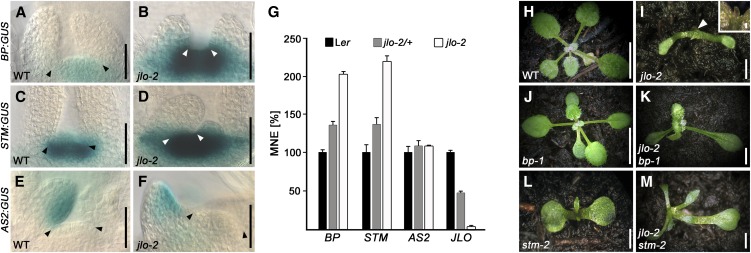
Meristem development requires expression of class I KNOX genes, such as STM or BP, and their downregulation in lateral organs. The arrest of meristem activity in the jlo-2 mutants could be caused by changes in the activity or expression levels of these genes. Thus, we examined BP and STM expression in homozygous jlo-2 loss-of-function mutants. In the wild type, both BP:GUS (for β-glucuronidase) and STM:GUS were expressed only in meristematic tissue and downregulated in organ primordia at 5 DAG (Figures 3A and 3C). By contrast, BP:GUS and STM:GUS signal intensity increased in jlo-2 meristems and expanded to the basis of lateral organ primordia (Figures 3B and 3D). At 15 DAG, BP and STM were expressed throughout the enlarging apex and at the basis of organ primordia (see Supplemental Figure 5 online). Using quantitative RT-PCR (qRT-PCR) assays, we found that transcript levels of BP and STM are at least twofold increased in jlo-2 mutant seedlings. Furthermore, we observed an upregulation of both genes in jlo-2/+ and in jlo-4 to jlo-7 mutant seedlings (Figure 3G; see Supplemental Figure 1B online). To further test whether JLO regulates organ development via repression of BP and STM, we generated the double mutants jlo-2 bp-1 and jlo-2 stm-2. As previously published (Douglas et al., 2002), bp-1 mutants appear aphenotypic during vegetative development (Figure 3J), but produce shorter internodes and pedicels, together with downward-pointing siliques after floral induction. In stm-2 single mutants, a shoot meristem is initiated but arrested after generating a few leaves (Figure 3L; Clark et al., 1996). When we combined jlo-2 with bp-1 or stm-2, primary leaves were visible at 14 DAG in jlo-2 bp-1 and jlo-2 stm-2 (Figures 3K and 3M) seedlings, before leaf development and meristem activity was eventually arrested at 25 DAG. By contrast, jlo-2 single mutants initiated only radialized organs (Figure 3I, inset), revealing that the reduction in BP and STM function in jlo-2 bp-1 and jlo-2 stm-2 double mutants can partially rescue the leaf growth defects of jlo-2 (see Supplemental Table 3 online).
Figure 3.
JLO Regulates Class I KNOX Gene Expression.
(A) to (F) BP:GUS ([A] and [B]), STM:GUS ([C] and [D]), and AS2:GUS ([E] and [F]) expression in wild-type (WT) and jlo-2 mutants. In (B) and (D), BP and STM expression is increased and ectopic expression is detectable at the base of lateral organs compared with (A) and (C). Arrowheads mark meristem boundaries.
(G) qRT-PCR of whole seedlings confirms reduction of JLO transcript levels in jlo-2 mutants, while expression of BP and STM is upregulated compared with the wild type (5 DAG). Note that gene expression is already altered in jlo-2/+ seedlings. AS2 transcription is unaffected. MNE, mean normalized expression. Bars indicate se (n ≥ 3)
(H) to (M) Genetic interactions between JLO and the class I KNOX genes BP and STM. Pictures of the wild type (H), jlo-2 (I), bp-1 (J), jlo-2 bp-1 (K), stm-2 (L), and jlo-2 stm-2 (M) mutants were taken 14 DAG. Homozygous jlo-2 initiate radialized organs (arrowhead; inset shows a close-up; [I]). bp-1 mutants exhibit a wild type–like appearance (J), while stm-2 mutants initiate a SAM, which arrests growth after initiation of a few leaves (L). jlo-2 bp-1 (K) and jlo-2 stm-2 (M) double mutants show partial rescue of the jlo-2 phenotype and produce primary leaves before the meristem finally arrests activity.
Bars = 50 μm in (A) to (F), 50 mm in (H) and (J), and 10 mm in (I) and (K) to (M).
[See online article for color version of this figure.]
Genetic Interaction between JLO and AS2
Plants defective in AS2 function grow lobed leaves that accumulate BP transcripts (Semiarti et al., 2001). Furthermore, as2 mutants develop flowers that open prematurely due to reduced petal and sepal sizes (Ori et al., 2000). Thus, jlo and as2 mutants share several characteristics. Because expression of both genes overlaps in newly initiated organ primordia, we hypothesized that they might act in a common pathway to direct organ development and regulate KNOX expression (Borghi et al., 2007; Iwakawa et al., 2007; Soyano et al., 2008).
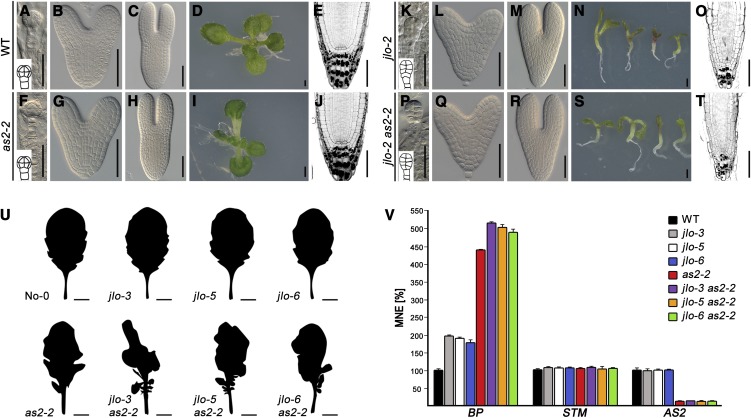
We generated jlo-2 as2-1 and jlo-2 as2-2 double mutants to analyze their genetic interactions. Both double mutant combinations displayed similar genetic interactions (see below), and only the jlo-2 as2-2 double mutants will be further discussed. Compared with the wild type, jlo-2 embryogenesis is strongly impaired with aberrant patterning from the first cell division of the proembryo onwards and an overall delay in development (Figures 4K to 4M; Bureau et al., 2010), whereas as2-2 single mutants are aphenotypic during embryo development (Figures 4F to 4H). jlo-2 as2-2 mutant embryos were indistinguishable from jlo-2 mutant embryos (Figures 4P to 4R), and the strong jlo-2 seedling phenotype was unaltered in jlo-2 as2-2 double mutants (Figures 4N, 4O, 4S, and 4T). To analyze a genetic interaction during later stages of development, we also combined the as2-1 and the as2-2 mutation with the weaker jlo-3, jlo-5, and jlo-6 alleles (Figure 4U). Again, the combination of jlo-3, jlo-5, and jlo-6 with either as2-1 or as2-2 caused similar phenotypes, and only the jlo as2-2 double mutants will be further described. Compared with either single mutant, we observed increased leaf lobing and ectopic leaflet formation in all double mutant combinations, indicating enhanced KNOX misexpression. Using qRT-PCR analysis (Figure 4V), we found a moderate increase of BP, but not of STM, transcript levels in jlo-3, jlo-5, and jlo-6 mutant leaves. As we did not observe altered leaf morphology in the jlo-3, jlo-5, or jlo-6 single mutants (Figure 4U), the level of ectopic BP activity might be too low to affect leaf development. The expression of BP was higher in as2-2 mutant leaves and substantially higher in leaves of each jlo as2-2 double mutant combination. Notably, the ectopic expression of BP in all jlo mutant leaves was not accompanied by any reduction in AS2 transcripts. These results suggest that both JLO and AS2 are involved in the repression of BP during leaf morphogenesis.
Figure 4.
Genetic Interaction of JLO and AS2.
(A) to (T) Embryonic and seedling development (5 DAG) of the wild type (WT) ([A] to [E]), as2-2 ([F] to [J]), jlo-2 ([K] to [O]), and jlo-2 as2-2 mutants ([P] to [T]). Embryonic stages: 16-cell stage ([A], [F], [K], and [P]), heart stage ([B], [G], [L], and [Q]), and torpedo stage ([C], [H], [M], and [R]). as2-2 embryos are aphenotypic; jlo-2 and jlo-2 as2-2 embryos reveal altered cell division planes at the early proembryo stage ([K] and [P]). These embryos did not develop beyond the heart stage. Insets in (A), (F), (K), and (P) show the respective embryo schematically. From the heart stage onwards, jlo-2 and jlo-2 as2-2 embryos displayed reduced hypocotyl diameter and length and showed an overall developmental delay. Postembryonically, as2-2 mutants developed lobed leaves (I) but normal roots (cf. [E] and [J]). jlo-2 and jlo-2 as2-2 seedlings developed asymmetric and atrophic cotyledons, short hypocotyls, and a defective root organization ([N] and [S], and [O] and [T]). Roots were stained via the modified pseudo-Schiff propidium iodide method; black dots indicate the starch granules in differentiated columella cells.
(U) Silhouettes of mature leaves of the wild type (No-0), jlo-3, jlo-5, jlo-6, and as2-2 single mutants as well as jlo-3 as2-2, jlo-5 as2-2, and jlo-6 as2-2 double mutants.
(V) qRT-PCR analysis of BP, STM, and AS2 expression in mature leaves. BP expression is elevated in jlo-3, jlo-5, and jlo-6 leaves, further upregulated in as2-2 leaves, and increased even more in each jlo as2-2 double mutant combination. No change in STM transcript level is detectable in mature leaves of each mutant background. AS2 expression remains unaltered in jlo-3, -5, and -6 mutant leaves but is strongly decreased in as2-2 single and jlo as2-2 double mutant leaves. MNE, mean normalized expression. Bars indicate se (n ≥ 3).
Bars = 20 µm in (A), (F), (K), and (P), 50 µm in (B), (C), (E), (G), (H), (J), (L), (M), (O), (Q), (R), and (T), 1 mm in (D), (I), (N), and (S), and 0.5 cm in (U).
[See online article for color version of this figure.]
We further analyzed flowers of jlo-2/+ as2-2 double mutants. With respect to either single mutant, jlo-2/+ as2-2 sepals, petals, and stamens were reduced in size, and cell length was decreased (see Supplemental Figure 4 online). Taken together, these genetic data indicate that JLO and AS2 can function partially independently to direct leaf and flower development.
AS2 Function Is Required for the JLO Overexpression Phenotype
The fact that STM and BP gene expression is upregulated in jlo mutant seedlings suggested that JLO normally acts to downregulate these two homeobox genes. However, this is in contrast with our previous observation that inducible misexpression of a fusion between JLO and the hormone binding domain of the glucocorticoid receptor (GR) causes a drastic upregulation of STM and BP expression (Borghi et al., 2007). To exclude that fusion to the GR domain interferes with the normal KNOX repressing function of JLO, we designed an estradiol-inducible i35S:JLO-FLAG transgene (Bureau et al., 2010). JLO-FLAG–misexpressing plants revealed strongly lobed leaves, resembling the JLO-GR misexpression phenotype (see Supplemental Figure 6F online) and showed enhanced transcript levels of BP and STM upon JLO-FLAG induction (see Supplemental Figures 6G and 6H online). Within 1 h after induction (HAI), both JLO protein and RNA were strongly upregulated (see Supplemental Figures 6I and 6J online). KNOX expression levels significantly increased within 4 HAI, suggesting an indirect mechanism of upregulation (see Supplemental Figures 6G and 6H online). We therefore speculated that the transgenic high-level expression of JLO might interfere with the regulatory pathways that normally restrict KNOX expression. In line with this, we found that JLO requires AS2 for this activity because induction of JLO-FLAG in as2-2 mutants did not alter the typical as2 leaf phenotype in 96% of all F2 plants analyzed (n = 53; see Supplemental Figure 6D online). Moreover, expression levels of BP and STM remained unaffected by JLO-FLAG expression in an as2-2 mutant background (see Supplemental Figures 6G and 6H online). From these data, we conclude that JLO regulates KNOX expression together with AS2. Ectopic JLO expression could then either inhibit AS2 transcription or interfere with AS2-dependent regulation at the protein level. Because qRT-PCR analysis showed that AS2 transcript levels are not altered in jlo mutants or upon JLO misexpression (Figures 3G and 4V; see Supplemental Figure 6K online), we tested the possibility that both proteins physically interact.
JLO and AS2 Can Physically Interact in Yeast
We performed GAL4-based yeast two-hybrid experiments to assay a potential interaction between AS2 and JLO. AS2 was fused to the GAL4 activation domain (GAL4-AD) and used as bait. Since full-length JLO was unstable and the JLO C terminus was activating transcription by itself, we used only the N-terminal LOB domain fused to the GAL4 DNA binding domain (GAL4-BD) as prey. In this assay, AS2 was able to interact with the JLO LOB domain, as demonstrated by growth on selective medium lacking Leu, Trp, His, and Ade (Figure 5B).
Figure 5.
Mapping the JLO–AS2 Interaction Domains.
(A) Protein structure of JLO and AS2. The LOB domain at the N terminus of each protein consists of a C-BLOCK (turquoise/orange), GAS-BLOCK (purple/green), and coiled coil (CC; gray/black) domain.
(B) and (C) GAL4-based yeast two-hybrid study. Mating with empty pGADT7 and pGBKT7 vectors excludes autoactivation. Growth on -Leu/Trp media was used to select for both plasmids.
(B) Growth on selective media (-Leu/Trp/His/Ade) was only detected for JLO versions, including the GAS-BLOCK and coiled coil domain. The AS2 C terminus fused to Gal4-AD appeared to be toxic.
(C) AS2 full-length or a C-terminal truncation was able to interact with JLO. All results were verified via calculation of Miller units in a liquid culture assay (black bars). Mating with pGADT7 (white bars) and pGBKT7 (gray bars) was used to calculate the background (gray shadowed). Asterisks indicate a significant difference to background (P ≥ 0.05; analyzed by Student’s t test). Bars indicate se (n ≥ 3).
[See online article for color version of this figure.]
To map the domains relevant for the interaction, we tested several truncations of both proteins. Interaction was observed when AS2 was combined with JLO versions carrying the GAS BLOCK and coiled coil domain, which are highly conserved amino acid regions within the LOB domain (Figure 5B; Shuai et al., 2002). None of the AS2 truncations, including only parts or the complete LOB domain, were able to interact with JLO. This suggests that domains within the AS2 C-terminal region mediate the interaction. However, the AS2 C terminus fused to the GAL4-AD appeared to be toxic for yeast (Figure 5B). Thus, we performed yeast two-hybrid experiments with AS2 GAL4-BD fusions as prey (Figure 5C). Interaction with JLO was obtained with full-length AS2 or only the C-terminal domain of AS2. In yeast, both JLO and AS2 can also interact with LBD31, an LBD protein closely related to JLO, opening up the possibility that LBD proteins can form higher order complexes and act in a combinatorial fashion. However, the interactions are not random between LBD proteins, as, for instance, LBD2 was not able to bind JLO or AS2 in yeast assays (see Supplemental Figure 7 online).
JLO Interaction with AS1 Is Mediated by AS2
Complex formation between AS2 and AS1 was shown to be required for BP repression in lateral organs (Xu et al., 2003; Guo et al., 2008). Coexpression of AS2 and AS1 allowed yeast growth on selective media, thus verifying the previously published data. However, we did not observe any direct interaction between JLO and AS1 (Figure 6). We therefore performed a yeast three-hybrid assay to test whether JLO, AS2, and AS1 have the potential to form a multimeric complex. Yeast was able to grow on selective medium when all three proteins were expressed, showing that JLO can interact indirectly with AS1 through AS2. The observation of such higher order complexes between JLO, AS2, and AS1 could help explain the effects of JLO misexpression on KNOX expression (see Supplemental Figure 6F online). Here, high level expression of JLO in a wild-type background may interfere with the KNOX-restricting activity of the AS1/AS2 complex, possibly by sequestering AS2 into JLO/AS2 or multimeric complexes.
Figure 6.
Yeast Three-Hybrid Assay.
(A) Protein structure of JLO, AS2, and AS1. JLO and AS2: C-BLOCK (turquoise/orange), GAS-BLOCK (purple/green), and coiled coil domain (CC; gray/black). AS1: MYB domain (blue) and coiled coil domain (red).
(B) GAL4-based yeast studies revealed an interaction between AS2 and AS1 but not between JLO and AS1. A yeast three-hybrid assay showed that AS2 can bridge the interaction between JLO and AS1. AS2 was cloned in the pTFT1 vector and cotransformed with AS1-GAL4-AD and JLO(LOB)-GAL4-BD vectors into yeast. Growth on -Leu/Trp media was used to select for GAL4-AD and GAL4-BD constructs. Growth on selective media (-Leu/Trp/His/Ade) was used to monitor interactions. Cotransformation with empty pGADT7 and pGBKT7 vectors exclude autoactivation.
(C) All results were verified via calculation of Miller units in a liquid culture assay (black bars). Cotransformation with pGADT7 (white bars) and pGBKT7 (gray bars) was used to calculate the background (gray shadowed). Asterisks indicate a significant difference to background (P ≥ 0.05, analyzed by Student’s t test). Bars indicate se (n ≥ 3).
[See online article for color version of this figure.]
Intracellular Localization of Fluorescent Protein–Tagged JLO, AS2, and AS1
To analyze protein interaction in planta, JLO, AS2, and AS1 were fused to the fluorescent proteins (FPs) GFP or mCherry and transiently expressed in Nicotiana benthamiana leaf epidermal cells. We used a β-estradiol expression system to limit overexpression artifacts and unspecific interactions (Bleckmann et al., 2010). Integrity of the different fusion proteins was confirmed by immunoblotting using an anti-GFP antibody (see Supplemental Figure 8G online). JLO and AS2 fusion proteins were found in the cytoplasm and enriched in the nucleoplasm (Figures 7A and 7A’). AS1 was localized to the nucleus with higher protein abundance in the nucleolus (Figure 7B’). Consistent with previously published results, the presence of AS2 caused relocalization of AS1 to the nucleoplasm (Zhu et al., 2008; Figures 7C and 7C’’). By contrast, JLO did not affect AS1 localization, supporting the notion that JLO and AS1 do not directly interact (Figures 7B and 7B’’). We used inducible misexpression in stably transformed Arabidopsis plants to test the fusion proteins for functionality. In all cases, we obtained the previously described gain-of-function phenotypes (Iwakawa et al., 2002; Xu et al., 2003; Borghi et al., 2007; Zhu et al., 2008), indicating that the fusion proteins are fully active. The observed subcellular localizations in Arabidopsis root epidermal cells (Figures 7D to 7F) resembled those in N. benthamiana leaf epidermal cells (Figures 7A to 7C). Similarly, coexpression of AS2/AS1 (Figures 7F and 7F’’) but not of JLO/AS1 (Figures 7E and 7E’’) resulted in a relocalization of AS1 to the nucleoplasm. These findings are again consistent with a direct interaction of JLO/AS2 and AS2/AS1 and no direct binding of JLO to AS1.
Figure 7.
Intracellular Protein Localization and FRET-Based Protein Interaction Analysis.
(A) to (C) Colocalization of FP-tagged proteins in N. benthamiana epidermis cells: colocalization of JLO-GFP and AS2-mCherry ([A] to [A″]), colocalization of JLO-GFP and AS1-mCherry ([B] to [B″]), and colocalization of AS2-GFP and AS1-mCherry ([C] to [C″]). Bars in (A) and (B) indicate se.
(D) to (F) Colocalization of FP-tagged proteins in Arabidopsis root epidermis cells: colocalization of JLO-GFP and AS2-mCherry ([D] to [D″]), colocalization of JLO-GFP and AS1-mCherry ([E] to [E″]), and colocalization of AS2-GFP and AS1-mCherry ([F] to [F″]). Bars = 10 μm in (A) to (F).
(G) and (H) Protein interaction revealed by percent EFRET. Intramolecular EFRET obtained by direct fusion of GFP to mCherry in a single molecule (gray bars) and GFP background fluctuation (white bars) were calculated as positive and negative controls. Control measurements were performed for all proteins tested and were similar in all experiments. Gray shaded area indicates background fluctuation level of GFP. Asterisks mark a significant difference from background (*P ≤ 0.05 and **P ≤ 0.01; analyzed by Student’s t test).
(G) EFRET measured with a transient expression of FP-tagged protein in epidermis cells of N. benthamiana (n ≥ 35 for each combination) AS2 (red): EFRET measured with coexpression of FP-tagged JLO and AS1 together with untagged AS2 protein.
(H) EFRET measured in root epidermis cells of stable transformed Arabidopsis plants (n ≥ 25 for each combination).
[See online article for color version of this figure.]
Fluorescence Resonance Energy Transfer–Based Protein Interaction Analysis Supports the Formation of JLO/AS2/AS1 Complexes
Next, we measured fluorescence resonance energy transfer efficiencies (EFRET) between the GFP and mCherry pairs (Albertazzi et al., 2009) in planta. EFRET was calculated as the percentage increase of GFP (donor) fluorescence after photobleaching of mCherry (acceptor) (Bleckmann et al., 2010). All photobleaching experiments and EFRET measurements were performed in the nucleus. In N. benthamiana leaf epidermal cells, fluorescent signals were first detectable at 1 HAI and remained stable over 12 h (see Supplemental Figure 8E online). Upon extended induction (≥24 HAI), some cells carried fluorescent aggregates, indicating protein overexpression (see Supplemental Figure 8F online, arrowhead). Therefore, all measurements were performed within 12 HAI. As EFRET depends on the orientation and distance of both chromophores to each other, we measured intramolecular EFRET as a control for the minimal distance. To this end, both GFP and mCherry were fused together to the C termini of all tested proteins. The intramolecular EFRET we measured for all fusion proteins ranged from 26 to 28%. Calculation of GFP fluorescence fluctuation during photobleaching in the absence of the donor revealed a maximal background of 6% in all control experiments. Thus, only EFRET significantly higher than 6% was regarded as an indication of close proximity or physical interaction of the two proteins for this set of experiments (Figure 7G).
The results we obtained confirmed our yeast GAL4 interaction studies and showed a clear JLO/AS2 (EFRET = 15% ± 0.5%) and AS2/AS1 (EFRET = 23% ± 2%) interaction in both reciprocal GFP/mCherry combinations. By contrast, we did not observe a significant EFRET for JLO/AS1 (4.3% ± 0.8%). However, when we coexpressed untagged AS2, significant EFRET between JLO and AS1 was recorded (14.5% ± 1%). Measurements performed at 1, 2, 4, and 12 HAI revealed that EFRET remained stable over time (see Supplemental Figure 8H online), indicating that EFRET as a measure of protein interaction did not depend on protein overexpression. Interestingly, JLO (EFRET = 11% ± 1.0%), AS2 (EFRET = 17% ± 1.7%), and AS1 (EFRET = 8.9% ± 1.5%) showed significant homomerization (Figure 7G), suggesting that various homomers and heteromers may coexist within the nucleus.
FP-tagged LBD31 and LBD2 proteins were used to further test the specificity of the observed interactions. Both proteins localized to the nucleus and cytoplasm, thus resembling subcellular localizations of JLO and AS2 (see Supplemental Figures 8A to 8D online). In line with our yeast studies, we detected an interaction between JLO/LBD31 (11.1% ± 1.0%) and AS2/LBD31 (15.0% ± 1.9%), whereas EFRET values for JLO/LBD2 (6.8% ± 0.7%) and AS2/LBD2 (7.1% ± 1.0%) were close to background (Figure 7G).
Overall, we noted a reduction of EFRET in root epidermal cells of stably transformed Arabidopsis plants compared with N. benthamiana (Figure 7H). The intramolecular EFRET we measured ranged only from 15 to 17%, with a GFP background fluctuation of 1.5%. We verified heteromerization of JLO/AS2 (4.1% ± 0.2%) and AS2/AS1 (8.6% ± 0.4%) as well as homomerization of JLO (3.1 ± 0.3%), AS2 (4.4 ± 0.3%), and AS1 (2.4 ± 0.1%). EFRET values for JLO/AS1 (1.6% ± 0.4%) were in the range of background level.
We conclude that JLO and AS2 specifically interact in yeast, N. benthamiana, and Arabidopsis.
AS2 and JLO Regulate Auxin Transport
Our studies revealed the capacity of JLO and AS2 to form protein complexes in Arabidopsis and that both proteins can act together in the regulation of KNOX expression in the shoot. JLO was previously shown to regulate auxin transport and signaling from embryogenesis onwards, and we therefore asked if JLO and AS2 share functions during early stages of development. Expression of AS2 was analyzed using an AS2:GUS reporter gene construct (Jun et al., 2010). GUS signals appeared on the adaxial side of cotyledons and organ primordia during embryonic and postembryonic development. In addition, AS2:GUS was expressed in cells of the suspensor and the embryonic root tip (Figures 8F to 8H). After germination, AS2 is expressed in the tips of seedling roots (Figure 8I). JLO expression in embryos was analyzed through whole-mount RNA in situ hybridization. At the early heart stage, JLO is expressed in the entire embryo with a stronger transcript accumulation in the basal domain (Figure 8A). Later on, expression is confined to the embryo axis, the developing vasculature, and the root pole (Figures 8B and 8C). Thus, the JLO and AS2 expression domains overlap at the heart stage and in the basal root tip of later embryo stages. The expression pattern of JLO during postembryonic development was analyzed using a JLO:GUS reporter construct. In the shoot, JLO is expressed in the vasculature of cotyledons and leaves, and weaker signals were also detected in the leaf blade (Figure 8E). In seedling roots, JLO:GUS is strongly expressed in the stele and in the root tip, including the columella cells (Figure 8D), thus overlapping extensively with the expression of AS2 in these tissues. Notably, the JLO expression levels are unaltered in as2-2 mutants (Figure 8Q). Similarly, neither expression domain nor expression levels of AS2 are altered in the jlo-2 mutant background (Figures 8J and 8Q).
Figure 8.
Gene Expression Analyses during Embryonic and Root Development.
Embryonic stages: heart stage ([A], [F], and [K] to [P]), torpedo stage ([B] and [G]), and bent cotyledon stage ([C] and [H]).
(A) to (C) Embryonic expression pattern of JLO analyzed by whole-mount RNA in situ hybridization.
(A) At the heart stage, JLO transcripts are detectable in provascular cells. WT, wild type.
(B) and (C) Later on, expression becomes restricted to the basal root tip ([C]; arrowhead) and the stele.
(D) and (E) Expression of a JLO:GUS reporter in wild-type seedlings (5 DAG). JLO is expressed in the stele as well as in columella cells of primary roots (D). In mature leaves, the JLO:GUS reporter is strongly expressed in the vascular system and hydathodes. Weaker expression is also detectable in the leaf blade (E).
(F) to (H) During embryogenesis, AS2:GUS is expressed at the adaxial side of cotyledons and the basal root tip (arrowhead in [H]), including the suspensor.
(I) AS2 expression at the tip of wild-type roots.
(J) jlo-2 roots reveal an unaltered AS2 expression.
(K) to (M) DR5rev:GFP expression in heart stage embryos.
(K) In the wild type, auxin concentrates in the root primordium and suspensor as well as in the tips of the cotyledons.
(L) jlo-2 embryos reveal only a weak signal at the root pole.
(M) as2-2 embryos show unaltered DR5rev activity.
(N) to (P) PIN1:PIN1-GFP expression in heart stage embryos.
(N) PIN1 concentrates to the cotyledon tips and provascular cells in the wild type.
(O) and (P) jlo-2 (O) and as2-2 (P) mutants show similar PIN1 distributions but reduced expression levels.
(Q) qRT-PCR analysis of JLO and AS2 transcript levels (5 DAG). JLO expression is reduced in jlo-2 mutants but unaltered in the as2-2 mutant background. Similarly, AS2 expression is downregulated in as2-2 mutants but unchanged in jlo-2 seedlings. MNE, mean normalized expression. Bars indicate se (n ≥ 3).
Bars = 50 μm.
[See online article for color version of this figure.]
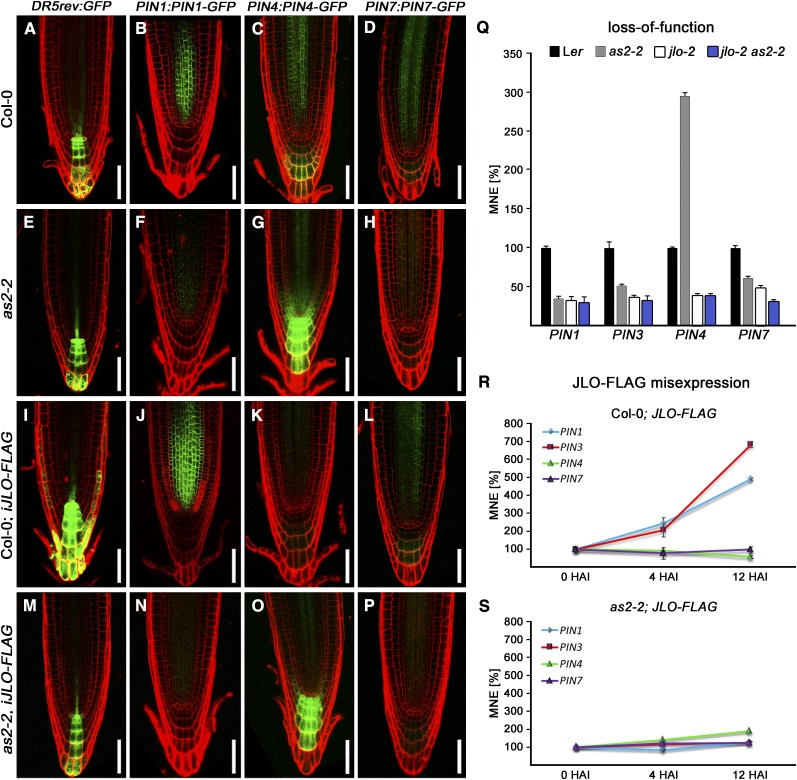
The severe patterning defects of jlo-2 embryos and roots result from a failure in auxin signaling and transport (Bureau et al., 2010). Thus, we examined auxin distribution in as2-2 mutants using the synthetic auxin response reporter DR5rev:GFP (Ulmasov et al., 1997). In wild-type heart stage embryos, auxin accumulates at the root pole with an intensity maximum in the uppermost suspensor cell (Figure 8K). Unlike jlo-2 mutants, as2-2 embryos displayed an unaltered DR5rev:GFP activity (Figures 8L and 8M). Postembryonic roots exhibit a maximum of DR5rev:GFP expression in the quiescent center, the adjacent stem cells, and columella cells. Again, DR5rev activity was unaltered in seedling roots of as2-2 mutants (Figures 9A and 9E). Because auxin accumulation is regulated by several PIN proteins, we analyzed their expression in more detail (Vieten et al., 2005). We previously identified JLO as an important regulator of these efflux carriers (Bureau et al., 2010). Consistent with this, expression levels of PIN1/3/4 and PIN7 were reduced in the novel jlo-3 to jlo-7 alleles (see Supplemental Figure 1B online). We also analyzed the expression of PIN1:PIN1-GFP, PIN4:PIN4-GFP, and PIN7:PIN7-GFP reporters in the as2-2 mutant background. We found a decreased PIN1-GFP signal in as2-2 embryos and roots (Figures 8P and 9F). PIN7-GFP expression in as2-2 roots was also reduced, whereas PIN4-GFP signals were strongly increased (Figures 9G and 9H). Thus, AS2 and JLO are similarly required for PIN1 and PIN7 expression, but they differ in their capacity to regulate PIN4 expression.
Figure 9.
PIN Expression Is Regulated by a JLO/AS2 Complex.
(A) to (P) Expression of DR5rev:GFP ([A], [E], [I], and [M]), PIN1:PIN1-GFP ([B], [F], [J], and [N]), PIN4:PIN4-GFP ([C], [G], [K], and [O]), and PIN7:PIN7-GFP ([D], [H], [L], and [P]) in the wild type ([A] to [D]) and as2-2 ([E] to [H]) mutants and after induced expression of JLO-FLAG in the wild type ([I] to [L]) and as2-2 mutant background ([M] to [P]) (5 DAG). Auxin accumulation is unaltered in as2-2 mutants (E). PIN1-GFP and PIN7-GFP signals are severely reduced ([F] and [H]), while PIN4-GFP signal is strongly increased (G) in as2-2 roots. Col-0, Columbia-0. Bars = 50 μm.
(I) to (L) Compared with the wild type ([A] and [B]), DR5rev promoter activity and PIN1-GFP expression is upregulated upon JLO-FLAG induction (12 HAI), while PIN4-GFP signal is reduced (K) and the PIN7-GFP signal remains unaltered (L).
(M) to (P) In as2-2 mutants, induction of JLO-FLAG expression does not alter DR5rev:GFP (cf. [E] and [M]) expression. Similarly, PIN1:PIN1-GFP (N), PIN4:PIN4-GFP (O), and PIN7:PIN7-GFP (P) expression remains unaffected by high level JLO misexpression in as2-2 mutant background (cf. with [F] to [H]).
(Q) qRT-PCR analysis reveals a downregulation of PIN1, PIN3, and PIN7 and an upregulation of PIN4 in as2-2 roots. By contrast, transcript levels of all PIN genes studied are strongly reduced in jlo-2 and jlo-2 as2-2 roots. Note that PIN1, PIN3, and PIN4 expression is similar in jlo-2 and jlo-2 as2-2 mutants, while PIN7 transcript levels are further reduced in the double mutant. MNE, mean normalized expression.
(R) and (S) Analysis of PIN1, PIN3, PIN4, and PIN7 transcript levels in roots after induced misexpression of JLO-FLAG in the wild type (R) and as2-2 mutants (S). Expression levels were normalized to uninduced controls prepared at the same time points. In the wild type (R), PIN1 and PIN3 are upregulated within 12 HAI, while PIN4 is downregulated and PIN7 expression remained unaltered. No significant response of PIN1, PIN3, and PIN7 expression is detectable in as2-2 mutants upon JLO-FLAG induction, while PIN4 expression is slightly upregulated. Bars in (Q) to (S) indicate se (n ≥ 3).
[See online article for color version of this figure.]
The JLO-AS2 Complex Regulates PIN Expression
We sought to analyze whether JLO and AS2 function together in the transcriptional regulation of PIN1,3,4 and 7. To this end, we first performed qRT-PCR analysis on jlo-2 and as2-2 single and double mutant roots (Q). PIN1 and PIN3 transcript levels are reduced in jlo-2 mutants, and we found a similar downregulation of these genes in the as2-2 mutant background. jlo-2 as2-2 double mutants revealed no further decrease in PIN1 and PIN3 transcription, suggesting that JLO and AS2 act together to promote PIN1 and PIN3 expression. However, PIN4 expression was reduced in jlo-2 mutants but nearly threefold upregulated in as2-2 mutants. In jlo-2 as2-2 double mutants, PIN4 expression levels resembled those of jlo-2 single mutants. This indicates that JLO is essential for PIN4 expression and that JLO/AS2 heteromers may act to restrict PIN4 levels. PIN7 transcription is notably reduced in both jlo-2 and as2-2 mutant roots but further decreased in the double mutant, indicating that JLO acts partially independently of AS2 to promote PIN7 expression.
To further identify AS2-dependent functions of JLO, we analyzed the expression of the DR5rev:GFP and PIN:PIN-GFP reporter in roots (5 DAG) upon JLO-FLAG induction (12 HAI). Increased JLO activity was able to upregulate DR5rev:GFP and PIN1:PIN1-GFP but not PIN7:PIN7-GFP expression in wild-type roots, while PIN4-GFP signals decreased within 12 HAI of JLO (Figures 9I to 9L). In the as2-2 mutant background, DR5rev:GFP and PIN1:PIN1-GFP expression did not respond to JLO-FLAG induction, suggesting that JLO requires AS2 for this function (Figures 9M and 9N). Expression of PIN4-GFP, which is already increased in as2-2 mutant roots compared with the wild type, remained unaltered upon JLO-FLAG induction (Figure 9O). Similarly, PIN7-GFP signals did not respond to increased JLO expression (Figure 9P).
Using qRT-PCR assays, we confirmed that increased JLO activity is sufficient for PIN1 as well as PIN3 but not for PIN7 upregulation in wild-type roots. Expression of PIN4 was first insensitive to JLO induction and decreased after prolonged (12 HAI) JLO induction (Figure 9R; Bureau et al., 2010). In as2-2 mutants, PIN1, PIN3, and PIN7 transcript levels did not respond to JLO-FLAG induction, while JLO-FLAG expression caused a minor increase in PIN4 RNA levels (Figure 9S). Thus, both JLO and AS2 are required for positive regulation of PIN1, 3, and 7 expression. JLO also contributes to complexes that promote PIN4 expression, which may compete with JLO/AS2 in the regulation of PIN4. This would imply that correct PIN4 expression requires a precise balance of various complexes that involve JLO. Interference via JLO misexpression may increase JLO/AS2 dimerization, thus repressing PIN4, while the absence of AS2 in as2-2 mutants would preferentially allow the formation of PIN4-promoting complexes (Figure 10).
Figure 10.
A Model for KNOX and PIN Regulation through Homomeric and Heteromeric JLO-AS2 Complexes.
Complexes containing JLO and AS2, either JLO/AS2/AS1 trimeric or JLO/AS2 heteromeric complexes, repress BP and PIN4 expression and promote PIN1, PIN3, and potentially PIN7 expression. The interaction between JLO and AS1 is mediated through AS2. JLO homomers repress STM and BP expression and promote PIN4 and PIN7 expression. AS2 homomeric complexes may promote PIN7 expression independently. Correct target gene expression requires a tight balance between the various protein complexes. LBD31 can bind to JLO and AS2, but the regulatory function of these complexes is so far unknown.
[See online article for color version of this figure.]
DISCUSSION
Organ initiation requires the accumulation of auxin and the simultaneous downregulation of class I KNOX expression in organ anlagen, as well as the establishment of boundaries that separate cells with different cell fates (reviewed in Rast and Simon, 2008). Several lines of evidence suggest that JLO plays an essential role during cell fate regulation. (1) JLO is expressed early at sites of organ initiation, in meristem-to-organ boundaries, and in mature leaves (Borghi et al., 2007; Soyano et al., 2008). (2) Compromised JLO activity causes pleiotropic defects that can be classified to affect meristem activity, organ identity, growth, and separation. (3) These defects are accompanied by ectopic expression of STM and BP and misregulated expression of several PIN genes.
We now conclude that JLO function is required for the repression of class I KNOX genes during lateral organ development because STM and BP expression were expanded into the basal domain of organ primordia in jlo-2 mutants. This KNOX gene misexpression is, at least in part, responsible for organ developmental defects in jlo-2 mutant meristems as we observed a partial rescue of the phenotype in jlo-2 bp-1 and jlo-2 stm-2 double mutants. The elevated levels of BP in mature leaves of the weaker jlo-3, jlo-5, and jlo-6 alleles, which still permit organ formation, most likely due to residual JLO activity, further indicate a role for JLO in the maintenance of BP repression. JLO shares this role with AS2, which interacts with the MYB protein AS1 to repress BP expression during leaf morphogenesis (Ori et al., 2000; Xu et al., 2003; Guo et al., 2008).
Our studies showed that JLO and AS2 can physically interact in vivo. Furthermore, we found that AS2 can mediate the interaction between JLO and AS1. Therefore, a simple scenario could be that a JLO/AS2/AS1 complex represses BP expression in developing organs. However, the synergistic effects in jlo-3,-5,-6 as2-2 and jlo-2/+ as2-2 mutants indicate that JLO can act also independently of AS2. Furthermore, STM was found to be misexpressed in jlo mutant backgrounds but not in as2-2 mutants (Ori et al., 2000). This suggests that both JLO/AS2/AS1 heteromeric complexes as well as JLO homomers may regulate KNOX expression in the shoot (Figure 10).
The evidence for JLO acting as a negative regulator of BP and STM expression is entirely based on the loss-of-function data and thus likely reflects genuine JLO activity. We found that JLO acquires the capacity to interfere with KNOX repression when overexpressed. The simplest explanation for this observation is that increased JLO expression disturbs the regulatory networks that normally serve to restrict KNOX expression from leaves. Since organ development is also highly sensitive to reduced JLO dosage, we conclude that JLO expression levels must be precisely regulated.
Our misexpression experiments showed that JLO induction in the as2-2 mutant background did not affect BP and STM transcript level or enhance the as2-2 leaf phenotype, suggesting that JLO acts with or through AS2. However, we previously noted that JLO overexpression phenotypes do not depend on AS1 activity (Borghi et al., 2007). We therefore propose that a high level JLO expression interferes with an AS1-independent function of AS2. Such independent activities have been previously suggested because AS1 and AS2 are expressed in largely overlapping but not identical expression patterns during lateral organ development, and AS2 overexpression phenotypes strongly differ from those caused by AS1 misexpression (Byrne et al., 2002; Iwakawa et al., 2002, 2007). Thus, JLO and AS2 may also form complexes that do not include AS1 but serve to regulate KNOX expression (Figure 10).
Our studies of JLO and AS2 function during root development further support the hypothesis that both proteins act together in heteromeric complexes. Mutant studies showed that JLO and AS2 act together to promote PIN1 and PIN3 expression and that further PIN1 and PIN3 upregulation by JLO overexpression requires the presence of AS2. We found increased PIN4 expression in as2-2 single mutants but a decrease in jlo-2 or jlo-2 as2-2 double mutants. A simple interpretation of these results is that JLO homomers strongly promote PIN4 transcription, JLO/AS2 heteromers regulate normal PIN4 levels, and AS2 homomers (or complexes involving also AS1) repress PIN4 expression (Figure 10). This notion is supported by the observation that JLO induction in the absence of AS2 further increased PIN4 transcription. To this end, it is noteworthy that PIN proteins are to some extent functionally interchangeable and that loss of PIN expression can be compensated for by other PIN genes (Blilou et al., 2005; Vieten et al., 2005). Thus, PIN4 expression may be enhanced by JLO homomers in as2-2 mutants, which may compensate for the reduced expression of PIN1, PIN3, and PIN7. Finally, based on our jlo-2 as2-2 double mutant analysis, JLO and AS2 act at least partially independently to direct PIN7 expression.
We reported previously that PIN expression is already reduced at the early stages of jlo embryogenesis (Bureau et al., 2010). Similarly, we showed here reduced PIN1 expression in as2-2 heart stage embryos. Thus, JLO and AS2 complexes likely regulate PIN expression from embryogenesis onwards. We also noted that the SAM of jlo-2 mutants initiated filamentous organs with a disturbed phyllotaxis and eventually arrested organ formation. Mutations in PIN1 also cause the formation of radialized organs and an arrest of lateral organ initiation from the shoot meristem (Vernoux et al., 2000; Reinhardt et al., 2003). Therefore, the overall reduction in PIN1 transcription, which we showed for jlo-2 mutant seedlings (Bureau et al., 2010), likely also contributes to the aberrant jlo-2 shoot development. This assumption is consistent with the observation that mutations in BP or STM are insufficient to fully restore the meristem activity of homozygous jlo-2 seedlings in the double mutant combinations. Taken together, the epistatic genetic interaction between jlo-2 and as2-2 at the embryonic and seedling stages, combined with our analysis of root development, suggests a continuous requirement for JLO homomeric and JLO/AS2 heteromeric complexes throughout plant development.
Our results show that JLO and AS2 regulate a similar set of target genes and act in a combinatorial manner. This raises the question of how the JLO/AS2 complexes are modulated to obtain their specific functions. A possible explanation is that the interaction with additional factors is responsible for the modulation of DNA binding specificity or transcriptional regulation. Similar combinatorial activities were reported for MADS box proteins, which determine the identity of floral organs (Theissen and Saedler, 2001). The formation of the multimeric JLO/AS2/AS1 complex in yeast supports this assumption. AS1 is expressed in developing organ primordia, leaves, and roots (Iwakawa et al., 2007). Thus, JLO/AS2 could function with AS1 to regulate KNOX and PIN expression in both shoots and roots. Indeed, the as2 and as1 mutant leaf phenotypes were not fully suppressed by bp mutants, suggesting that both genes regulate other targets besides BP (Ori et al., 2000; Byrne et al., 2002). In addition, auxin activity is asymmetrically distributed at the distal leaf tip of as1 or as2 mutants (Zgurski et al., 2005). These observations could be explained by postulating a role for AS2 and AS1 in PIN regulation, which is strongly supported by our data presented here. In the root system, where no as1 mutant phenotype was described so far, other MYB genes may replace AS1 function and contribute to PIN regulation.
We found that both JLO and AS2 can interact also with LBD31 in vivo. LBD31 expression is first detectable at sites of organ initiation and later in meristem-to-organ boundaries. This expression pattern overlaps with that of JLO, AS2, and AS1 during early organ development and later with that of JLO in the boundary (Borghi et al., 2007; Iwakawa et al., 2007; Xu et al., 2008; Jun et al., 2010). Whether higher order complexes with LBD31 are formed has not been analyzed so far. However, we envisage that LBD proteins can undergo a wide range of combinatorial interactions with each other. In contrast with jlo or as2 and as1 mutants, knockout alleles of LBD31 were aphenotypic, indicating that either JLO can compensate for the loss of its close homolog LBD31 or that other related LBD proteins can substitute for LBD31 in heteromeric complexes. Further experiments are required to distinguish between these possibilities.
Reduced JLO activity interfered with the establishment of boundaries, resulting in leaf and floral organ fusions in the different jlo alleles. Boundary formation requires a depletion of auxin from the cells encompassing the organ primordia (Heisler et al., 2005). Boundary cells act not only as morphological barriers, they also provide signals that regulate the development of adjacent tissues (reviewed in Aida and Tasaka, 2006). The failure to separate cells with different identities, combined with a loss of boundary-specific gene expression, will then result in aberrant organ development of jlo mutants. Somewhat similar effects were reported for the trihelix transcription factor PETAL LOSS, which is expressed in sepal boundaries. ptl loss-of-function mutants develop fused sepals but display also a reduced organ number as well as polarity defects (Brewer et al., 2006).
Taken together, our data show that JLO function is required for patterning processes throughout plant development. We demonstrate that JLO can act in homomeric as well as in heteromeric complexes with AS2 and AS1 and serves to coordinate KNOX expression and the regulation of PIN auxin efflux carriers during shoot and root development in Arabidopsis.
METHODS
Plant Material and Growth Conditions
The jlo-2 (Landsberg erecta [Ler]; Bureau et al., 2010), as2-1 (CS3117, An background), as2-2 (CS3118, ER background), stm-2 (N8137; Ler background), and bp-1 (CS30; Ler background) mutants were obtained from the Nottingham Arabidopsis Stock Centre. The jlo-3 (pst17018), jlo-4 (pst19799), jlo-5 (pst20504), jlo-6 (pst00432), and jlo-7 (pst13957) mutations are in the Nossen (No-0) background and belong to the RIKEN collection. The origins of marker lines are as follows: DR5rev:GFP (B. Scheres), PIN1:PIN1-GFP, PIN4:PIN4-GFP, and PIN7:PIN7-GFP (J. Friml), AS2:GUS (J.C. Fletcher), STM:GUS (W. Werr), and BP:GUS (M. Tsiantis). Arabidopsis thaliana plants were grown on soil under constant light conditions at 21°C. For root analyses, seeds were surface sterilized with chlorine gas, imbibed in 0.1% agarose, and plated onto GM medium (0.5× Murashige and Skoog medium with Gamborg’s No. 5 vitamins [Duchefa], 0.5 g/L MES, 1% [w/v] Suc, and 1.2% [w/v] plant agar). Plates were incubated vertically in a growth chamber. Nicotiana benthamiana plants were grown for 4 weeks in a greenhouse under controlled conditions.
Binary Constructs and Plant Transformation
For protein localization and interaction studies, attB sites were added via PCR-mediated ligation to coding regions of JLO, AS2, AS1, LBD31, or LBD2. PCR products were introduced into pDONR201 and eventually recombined into pABindGFP, pABindCherry, or pABindFRET (Bleckmann et al., 2010). Binary vectors were transformed in Agrobacterium tumefaciens GV3101 pMP90 (Koncz et al., 1984) according to the manufacturer’s instructions (Invitrogen). Abaxial leaf sides of N. benthamiana plants were infiltrated as described by Bleckmann et al. (2010). Transgene expression was induced 48 h after infiltration by spraying with 20 µM β-estradiol and 0.1% Tween 20 and analyzed within 12 HAI. Production of fusion proteins was confirmed by immunoblotting (primary antibody, anti-GFP [Roche]; secondary antibody, anti-mouse ALP conjugated [Dianova]). Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on GM medium containing hygromycin (15 mg/mL). For JLO misexpression experiments, a LexA35S:JLO-FLAG transgene (Bureau et al., 2010) was transformed into Columbia-0 or as2-2 plants. Induction of transgene expression was performed by spraying with 20 µM β-estradiol and 0.1% Tween 20 and verified by immunoblot analysis (primary antibody, anti-FLAG [Sigma-Aldrich]; secondary antibody, anti-mouse ALP conjugated antibody [Dianova]). For the analysis of the JLO expression pattern, the JLO promoter region (3273 bp upstream of the ATG) was synthesized (Life Technologies), introduced into pDONR211, and recombined into pMDC163 (Curtis and Grossniklaus, 2003).
EFRET Measurements via Acceptor Photobleaching
N. benthamiana leaf epidermal cells and Arabidopsis root epidermal cells were examined with a ×40 1.3–numerical aperture Zeiss oil-immersion objective using a Zeiss LSM 510 Meta confocal microscope. EFRET was measured via GFP fluorescence intensity increase after photobleaching of the acceptor mCherry (Bleckmann et al., 2010). The percentage change of the GFP intensity directly before and after bleaching was analyzed as EFRET = (GFPafter − GFPbefore)/GFPafter × 100. All photobleaching experiments were performed in the nucleus. A minimum of 25 measurements was performed for each experiment. Significance was analyzed using a Student’s t test.
Yeast Interaction Studies
Full-length coding sequences or fragments of genes tested in yeast interaction studies were amplified by PCR from Columbia-0 cDNA. The forward and reverse primers used for this amplification carried a restriction site that permitted cloning of the PCR product into pGADT7, pGBKT7, or pTFT1 (Egea-Cortines et al., 1999). For yeast two-hybrid studies, GAL4-BD and GAL4-AD clones were transformed into the yeast strains YST1 and AH109 (Clontech). Expression of the fusion proteins was confirmed by immunoblotting (GAL4-BD, anti-Gal4 DNA-BD [BD Biosciences]/ anti-mouse-ALP conjugated [Dianova]; GAL4-AD, anti-HA [Roche]/ anti-rat-horseradish peroxidase conjugated [Dianova]). After mating, interaction was studied by plating serial dilutions of yeasts on medium lacking Leu, Trp, His, and Ade. Three-hybrid assays were performed in yeast strain AH109. Constructs were cotransformed and selected on yeast synthetic dropout medium. Interactions were assayed on quadruple dropout medium. All other techniques were performed according to the Matchmaker protocols handbook (Clontech).
Gene Expression Analysis
Reporter gene analysis was performed in the F3 generation after genetic crossing. Detection of GUS activity was performed with GUS staining solution (0.05 M NaPO4, pH 7.0, 5 mM K3[Fe(CN)6], and 10 mM K4[Fe(CN)6]). For microscopy analysis of embryos and roots, tissue was cleared with 70% (w/v) chloral hydrate and 10% (v/v) glycerol solution. For microscopy analysis of green tissues, chlorophyll was removed using an ethanol series from 50% (v/v) to 100% (v/v). Tissues were then cleared with 50% to 100% (v/v) Roti Histol, followed by overnight incubation in immersion oil. Analysis of fluorescence reporter expression was performed using a LSM510 Meta confocal microscope. Counterstaining of root cell walls was achieved by mounting roots in 10 µM propidium iodide. The RNeasy plant mini kit (Qiagen) was used for RNA extraction. RNA was treated with DNase (Fermentas) and transcribed into cDNA using SuperScriptII (Invitrogen). qRT-PCR was performed in triplicates using the Mesa Blue Sybr Mix (Eurogentec) and a Chromo4 real-time PCR machine (Bio-Rad). Expression levels were normalized to the reference gene At4g34270 (Czechowski et al., 2005).
Phenotypic Analysis and Microscopy
Analysis of embryos was performed as described by Bureau et al. (2010). Scanning electron microscopy analysis was performed according to Kwiatkowska (2004). Root architecture was studied with the modified pseudo-Schiff propidium iodide method (Truernit et al., 2008) and imaged with a Zeiss LSM 510 Meta laser scanning microscope. For size measurements, floral organs were separated from each other and imaged with an AxioCam ICC1 camera (Zeiss) mounted onto a Zeiss Stemi 2000C. To compare cell sizes, petals were printed with 1.5% agarose, and negatives were examined and photographed using an Axiocam HR camera attached to a Zeiss Axioscope II microscope. Images were processed in ImageJ software and assembled in Adobe Photoshop.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT4g00220 (JLO), AT1G65620 (AS2), AT2G37630 (AS1), AT4G00210 (LBD31), and AT1G06280 (LBD2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular Analyses of the Novel jlo Alleles.
Supplemental Figure 2. Embryo and Seedling Development in the F1 Resulting from Crosses between jlo-2 with jlo-1 and jlo-3 to jlo-7 Mutants.
Supplemental Figure 3. Floral Phenotypes of the Novel jlo Alleles.
Supplemental Figure 4. jlo-2/+ as2-2 Double Mutant Analysis.
Supplemental Figure 5. Class I KNOX Gene Expression in jlo-2 Mutants at 15 DAG.
Supplemental Figure 6. The JLO Gain-of-Function Phenotype Requires AS2 Activity.
Supplemental Figure 7. Interaction of JLO and AS2 with LBD31 and LBD2.
Supplemental Figure 8. Intracellular Localizations of LBD31 and LBD2 and Time Course Experiment.
Supplemental Table 1. Allelism Analysis.
Supplemental Table 2. Occurrence of Phenotypic Classes I to III within the First Flowers of the jlo Alleles.
Supplemental Table 3. Suppression of the jlo-2 SAM Phenotype by the stm and bp Mutations.
Supplementary Material
Acknowledgments
We thank Cornelia Gieseler and Carin Theres for technical support and members of the R. Simon and D. Schubert laboratory for critical comments. We also thank Jennifer C. Fletcher and JiHyung Jun for providing the AS2:GUS line, Andrea Bleckmann for the modified pMDC7 vectors and for help with confocal microscopy, Yvonne Stahl for help with whole-mount RNA in situ hybridization, Marc Somssich for helping to clone the JLO promoter, and the Center for Advanced Imaging at Heinrich-Heine-Universität for help with the confocal equipment. We thank Jiri Friml, Miltos Tsiantis, and Wolfgang Werr for generously supplying plant materials. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Si 947/4-1) to R.S.
AUTHOR CONTRIBUTIONS
M.I.R. performed the experiments, and M.I.R. and R.S. designed the research and wrote the article.
Glossary
- SAM
shoot apical meristem
- GFP
green fluorescent protein
- DAG
days after germination
- GUS
β-glucuronidase
- qRT-PCR
quantitative RT-PCR
- HAI
hours after induction
- EFRET
fluorescence resonance energy transfer efficiencies
- FP
fluorescent protein
- Ler
Landsberg erecta
- No-0
Nossen
References
- Aida M., Ishida T., Tasaka M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570 [DOI] [PubMed] [Google Scholar]
- Aida M., Tasaka M. (2006). Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol. 9: 72–77 [DOI] [PubMed] [Google Scholar]
- Albertazzi L., Arosio D., Marchetti L., Ricci F., Beltram F. (2009). Quantitative FRET analysis with the EGFP-mCherry fluorescent protein pair. Photochem. Photobiol. 85: 287–297 [DOI] [PubMed] [Google Scholar]
- Belles-Boix E., Hamant O., Witiak S.M., Morin H., Traas J., Pautot V. (2006). KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bleckmann A., Weidtkamp-Peters S., Seidel C.A., Simon R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Borghi L., Bureau M., Simon R. (2007). Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19: 1795–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer P.B., Heisler M.G., Hejatko J., Friml J., Benkova E. (2006). In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nat. Protoc. 1: 1462–1467 [DOI] [PubMed] [Google Scholar]
- Bureau M., Rast M.I., Illmer J., Simon R. (2010). JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol. Biol. 74: 479–491 [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Simorowski J., Martienssen R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Clark S.E., Jacobsen S.E., Levin J.Z., Meyerowitz E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S.J., Chuck G., Dengler R.E., Pelecanda L., Riggs C.D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14: 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jurgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Galweiler L., Guan C., Muller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M.C. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Barkoulas M., Tsiantis M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133: 3955–3961 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Husbands A., Bell E.M., Shuai B., Smith H.M., Springer P.S. (2007). LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35: 6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H., Iwasaki M., Kojima S., Ueno Y., Soma T., Tanaka H., Semiarti E., Machida Y., Machida C. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51: 173–184 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., Machida Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Jun J.H., Ha C.M., Fletcher J.C. (2010). BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22: 62–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Kreuzaler F., Kalman Z., Schell J. (1984). A simple method to transfer, integrate and study expression of foreign genes, such as chicken ovalbumin and alpha-actin in plant tumors. EMBO J. 3: 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska D. (2004). Surface growth at the reproductive shoot apex of Arabidopsis thaliana pin-formed 1 and wild type. J. Exp. Bot. 55: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Lin W.C., Shuai B., Springer P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T., Yamashino T., Kiba T., Koizumi N., Kojima M., Sakakibara H., Mizuno T. (2007). A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71: 1269–1278 [DOI] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J.L., Hake S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Paponov I.A., Teale W.D., Trebar M., Blilou I., Palme K. (2005). The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci. 10: 170–177 [DOI] [PubMed] [Google Scholar]
- Phelps-Durr T.L., Thomas J., Vahab P., Timmermans M.C. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17: 2886–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast M.I., Simon R. (2008). The meristem-to-organ boundary: More than an extremity of anything. Curr. Opin. Genet. Dev. 18: 287–294 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Shuai B., Reynaga-Pena C.G., Springer P.S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Thitamadee S., Machida Y., Chua N.H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20: 3359–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthelemy J., Palauqui J.C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantis M., Brown M.I., Skibinski G., Langdale J.A. (1999). Disruption of auxin transport is associated with aberrant leaf development in maize. Plant Physiol. 121: 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., Kronenberger J., Grandjean O., Laufs P., Traas J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127: 5157–5165 [DOI] [PubMed] [Google Scholar]
- Vieten A., Vanneste S., Wisniewska J., Benkova E., Benjamins R., Beeckman T., Luschnig C., Friml J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Xu B., Li Z., Zhu Y., Wang H., Ma H., Dong A., Huang H. (2008). Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 146: 566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Xu Y., Dong A., Sun Y., Pi L., Xu Y., Huang H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Zazimalova E., Krecek P., Skupa P., Hoyerova K., Petrasek J. (2007). Polar transport of the plant hormone auxin - The role of PIN-FORMED (PIN) proteins. Cell. Mol. Life Sci. 64: 1621–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgurski J.M., Sharma R., Bolokoski D.A., Schultz E.A. (2005). Asymmetric auxin response precedes asymmetric growth and differentiation of asymmetric leaf1 and asymmetric leaf2 Arabidopsis leaves. Plant Cell 17: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li Z., Xu B., Li H., Wang L., Dong A., Huang H. (2008). Subcellular localizations of AS1 and AS2 suggest their common and distinct roles in plant development. J. Integr. Plant Biol. 50: 897–905 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.