An animal-like cryptochrome (aCRY) functions as a sensory blue light receptor in the green alga Chlamydomonas; in addition, this flavoprotein unexpectedly acts as a sensory red light receptor. For plant cryptochromes, the dark form is proposed to contain an oxidized flavin, whereas for aCRY, the broad spectral responses point to the neutral radical state in the dark.
Abstract
Cryptochromes are flavoproteins that act as sensory blue light receptors in insects, plants, fungi, and bacteria. We have investigated a cryptochrome from the green alga Chlamydomonas reinhardtii with sequence homology to animal cryptochromes and (6-4) photolyases. In response to blue and red light exposure, this animal-like cryptochrome (aCRY) alters the light-dependent expression of various genes encoding proteins involved in chlorophyll and carotenoid biosynthesis, light-harvesting complexes, nitrogen metabolism, cell cycle control, and the circadian clock. Additionally, exposure to yellow but not far-red light leads to comparable increases in the expression of specific genes; this expression is significantly reduced in an acry insertional mutant. These in vivo effects are congruent with in vitro data showing that blue, yellow, and red light, but not far-red light, are absorbed by the neutral radical state of flavin in aCRY. The aCRY neutral radical is formed following blue light absorption of the oxidized flavin. Red illumination leads to conversion to the fully reduced state. Our data suggest that aCRY is a functionally important blue and red light–activated flavoprotein. The broad spectral response implies that the neutral radical state functions as a dark form in aCRY and expands the paradigm of flavoproteins and cryptochromes as blue light sensors to include other light qualities.
INTRODUCTION
The green alga Chlamydomonas reinhardtii serves as a model system for both vascular plants and algae with respect to photosynthetic function and for animals with respect to an understanding of the structure, assembly, and function of eukaryotic cilia (Merchant et al., 2007). Furthermore, C. reinhardtii represents a model for studying features typical of flagellate algae, including light-driven phototactic processes. Light regulates a variety of processes in C. reinhardtii, including its sexual cycle (Huang and Beck, 2003), cell cycle (Oldenhof et al., 2004), circadian clock (Kondo et al., 1991), nitrogen metabolism (Chen and Silflow, 1996), photosynthesis (Eberhard et al., 2008), and photoorientation (Hegemann, 2008). For these purposes, C. reinhardtii uses different photoreceptors. Some of these photoreceptors have already been well characterized. For example, two microbial-type rhodopsins, the channel rhodopsins, act directly as light-gated ion channels and collect light mainly in the green region of the visible spectrum. These photoreceptors are involved in photoorientation (Sineshchekov et al., 2002) and can also be used as optogenetic tools in animals (Hegemann, 2008). Phototropins (phots), which were first identified and well characterized in Arabidopsis thaliana (Christie et al., 1998, Christie, 2007), are involved in the sexual cycle of C. reinhardtii (Huang and Beck, 2003). They also regulate blue light–dependent expression of certain genes encoding light-harvesting complex (LHC) apoproteins, such as LHCBM6, or others encoding certain enzymes of the chlorophyll (glutamate-1-semialdehyde aminotransferase [GSA]) and carotenoid (phytoene desaturase [PDS]) biosynthesis pathways (Im et al., 2006). On the other hand, there is still light photoreceptor encoded on the C. reinhardtii genome, the plant cryptochrome Chlamydomonas Photolyase Homologue1 (CPH1). The responses of this photoreceptor to blue light have been thoroughly investigated in vitro (Immeln et al., 2007, 2010; Langenbacher et al., 2009). However, in vivo studies have been limited, although it was shown that the expression pattern of C. reinhardtii CPH1 tracks the diurnal light/dark cycle, with light-dependent CPH1 degradation mediated by the proteasome pathway; this degradation involves blue light, but surprisingly, red light also participates in the degradation process (Reisdorph and Small, 2004).
Data from several experiments indicate that C. reinhardtii has at least one red light–absorbing photoreceptor, although no phytochrome or other known red light–absorbing protein has been identified (Mittag et al., 2005; Merchant et al., 2007). Physiological experiments have demonstrated that red, but not far-red, light can significantly phase reset the circadian clock of C. reinhardtii (Kondo et al., 1991). Furthermore, genes encoding proteins involved in light harvesting (LHCBM6) and chlorophyll synthesis (GSA) appear to be regulated by both blue and red light of 650, 720, and 740 nm (Im et al., 2006). DCMU, an inhibitor of photosynthetic electron transport, did not have any significant effect on the blue and red light–stimulated expression of these genes. This lack of effect of a photosynthetic inhibitor on red light–activated gene expression suggests that signaling occurs via a red light–absorbing photoreceptor. Red light is also involved in regulating the expression of the PsbA Binding Protein gene (Alizadeh and Cohen, 2010).
Initial functional analyses of C. reinhardtii clock components did not identify an associated photoreceptor (Mittag et al., 2005). After the first draft of the C. reinhardtii genome sequence became available, homology searches were performed with clock-relevant components, including photoreceptors from other model organisms such as Neurospora, Arabidopsis, Drosophila, and mouse. These analyses demonstrated that the C. reinhardtii genome encodes a cryptochrome (CRY) that has homology to animal but not plant CRYs. This difference is of interest with regard to the clock system because plant CRYs, such as the ones from Arabidopsis (CRY1 and CRY2), and the animal type I CRYs, such as the one from Drosophila, mediate the input of blue light into clock function (Somers et al., 1998; Stanewsky et al., 1998; Harmer, 2009). On the other hand, animal type II CRYs, represented by mammalian CRYs or monarch butterfly (Danaus plexippus) CRY2, are part of the circadian oscillator (Etchegaray et al., 2003; Zhu et al., 2008). However, not all CRYs are clock-related components. DASH CRYs are present in diverse organisms, including Arabidopsis (CRY3), and function in the repair of photodamaged, single-stranded, and loop-structured double-stranded DNA in vitro (Selby and Sancar, 2006; Pokorny et al., 2008). This DNA-repair ability is reminiscent of the close CRY homologs, the photolyases. Photolyases are classified according to their ability to repair either cyclobutane pyrimidine dimers or (6-4) photoproducts (Sancar, 2003) that can accumulate in DNA as a consequence of the absorbance of UV light. Recently, algal CRY/Photolyase Family1 (CPF1) proteins have been identified in Phaeodactylum tricornutum and Ostreococcus tauri (Coesel et al., 2009; Heijde et al., 2010). CPF1 proteins are difficult to classify because they seem to function both in blue light signaling and DNA repair of (6-4) photoproducts, and additionally, they have been shown to act as transcriptional repressors of clock components in COS7 cells. All CRYs and photolyases bind the chromophore flavin adenine dinucleotide (FAD) in the ∼500-amino acid photolyase homology region (PHR) (Lin et al., 1995; Sancar, 2003). A C-terminal extension of variable length and with limited sequence similarity is present in both CRYs and (6-4) photolyases but is absent in cyclobutane pyrimidine dimer photolyases.
In addition to the CRYs, several proteins have been characterized in the past few years that are either partially or closely connected to the central oscillator of C. reinhardtii; a knockout or knockdown of these genes may cause phase shifts, arrhythmicity, or period lengthening or shortening (Schulze et al., 2010; Matsuo and Ishiura, 2011). These clock-related proteins include the two subunits of the RNA binding protein CHLAMY1 (C1 and C3 subunits; Iliev et al., 2006), several Rhythm Of Chloroplast (ROC) proteins (Matsuo et al., 2008), Casein Kinase1 (CK1; Schmidt et al., 2006), and Constans (CO), which is also involved in photoperiodic signaling (Serrano et al., 2009). Among the ROCs are a number of putative transcription factors (ROC15, ROC40, ROC66, and ROC75), a putative subunit of E3 ubiquitin ligase (ROC114), and a protein of unknown function that contains leucine-rich repeats (ROC55). Up to now, it was not known which of the photoreceptors transmits light information to components of the circadian oscillator in C. reinhardtii. These photoreceptors do not necessarily include a member of the CRY family, since in the fungus Neurospora crassa, the light, oxygen, or voltage (LOV) domain–containing proteins White Collar1 as well as Vivid (VVD) play important roles in circadian oscillation (summarized in Chen et al., 2010). Moreover, in mouse, rod-cone and melanopsin-driven light inputs are transmitted to the brain by retinal ganglion cells (Altimus et al., 2008).
Here, we have characterized, both in vitro and in vivo, the light-signaling properties of the animal-like cryptochrome (aCRY) from C. reinhardtii. Spectroscopic analysis of heterologously expressed aCRY demonstrates photoreduction from the oxidized to the neutral radical state by blue light and then to the fully reduced state by red light. Based on analysis of an acry mutant in C. reinhardtii, a number of genes are significantly influenced by aCRY function and their expression is modulated by the absorbance of blue (465 nm), yellow (590 nm), or red (635 nm) light. These broad spectral aCRY-dependent responses are in accord with the spectral qualities of the aCRY neutral radical state observed in vitro, suggesting that C. reinhardtii aCRY acts as a blue and red light photoreceptor in vivo.
RESULTS
aCRY and the Plant CRY from C. reinhardtii Differ in Sequence, Spectral Characteristics, and Diurnal Expression
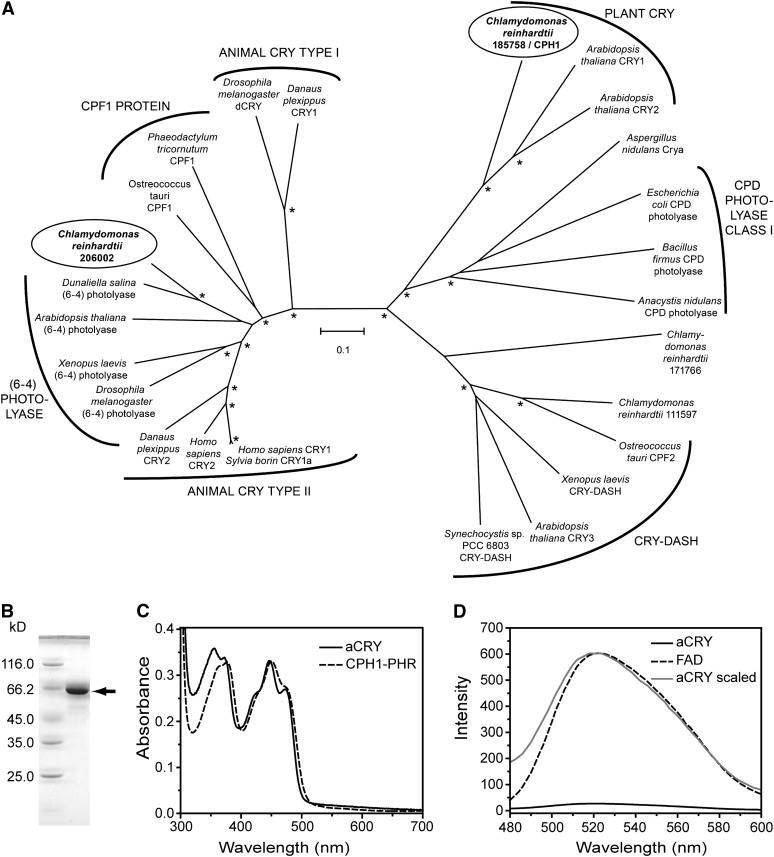
Sequences of four potential members of the CRY and photolyase families (protein identifiers 111597, 171766, 185758, and 206002) are encoded in the C. reinhardtii genome, based on the version 4.0 genome sequence (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). A phylogenetic analysis of C. reinhardtii CRY proteins and well-characterized representatives of CRYs and photolyases from other organisms is presented in Figure 1A and Supplemental Data Set 1 online. For clarity, the most distantly related members, such as class II photolyases, as well as the recently identified proteobacterial CRYs (Hendrischk et al., 2009; Geisselbrecht et al., 2012) were omitted from the analysis. This analysis shows that the protein with identifier 111597 clusters with the DASH CRYs, whereas 171766 cannot be assigned to a specific family. The aCRY (206002) protein clusters with the superfamily of animal type II CRYs, (6-4) photolyases, and CPF1 proteins, while it is highly diverged from CPH1 (185758), the plant CRY in C. reinhardtii.
Figure 1.
Phylogenetic Position of aCRY and Characterization of Heterologously Expressed aCRY.
(A) aCRY from C. reinhardtii (gene 206002; circled) groups with type II animal CRYs, (6-4) photolyases, and CPF1 proteins but not with the CPH1 plant CRY from C. reinhardtii (gene 185758; circled). For simplicity, photolyases other than members of cyclobutane pyrimidine dimer (CPD) class I and (6-4) were omitted. Bootstrap values larger than 95% are indicated by asterisks. Numbering of uncharacterized sequences from C. reinhardtii is according to Joint Genome Institute Chlre version 4.0. The scale bar indicates amino acid substitutions per site.
(B) SDS-PAGE of heterologously expressed, full-length aCRY with a His tag. A single band is detected and marked with an arrow. This band migrates to a position that aligns with the marker protein BSA at a molecular mass of 66.2 kD.
(C) UV/Vis spectrum of purified aCRY. The absorption spectrum shows the characteristic pattern of protein-bound, oxidized flavin with absorbance maxima at ∼360 and 447 nm. The spectrum differs from that of the PHR domain of CPH1, the plant CRY of C. reinhardtii (dashed line), which suggests a different hydrogen bonding environment of the flavin.
(D) Fluorescence emission spectrum of aCRY excited at 447 nm (solid line) as compared with that of free FAD (dashed line). The weak emission of FAD is further reduced by binding to aCRY. A scaled trace of aCRY (gray) demonstrates the shift in the fluorescence maximum of free FAD by 2 nm in native aCRY.
To further characterize aCRY, a codon-adapted gene (see Supplemental Figure 1 online) was expressed in Escherichia coli, yielding up to 50 mg yellow aCRY protein/L culture. The yellow color of the isolated protein indicates flavin binding. The protein has an apparent molecular mass of ∼66 kD, which is in accord with the theoretical mass of 66.4 kD for His tag aCRY (Figure 1B). Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis of the trypsin-digested 66-kD species confirmed the identity of this protein as aCRY (see Supplemental Table 1 online). Native CRYs typically bind FAD, but heterologously expressed flavoproteins might incorporate riboflavin or flavin mononucleotide. Fluorescence spectra of the isolated chromophore of C. reinhardtii aCRY showed a rise in fluorescence upon acidification, which is typical for FAD (see Supplemental Figure 2 online; Faeder and Siegel, 1973).
A UV/visible (Vis) spectrum of the heterologously expressed aCRY was used to analyze chromophore content (Figure 1C). In our preparation, up to 40% of the protein had bound FAD. Two bands with absorbance peaks at ∼360 and 447 nm, which represent the oxidized flavin, were detected. Both bands show a vibrational fine structure at 355, 373, 423, and 473 nm, which is characteristic of a protein-bound flavin (Kotaki et al., 1970). In comparison with the PHR domain of the plant CRY in C. reinhardtii, CPH1, which has an absorbance peak at ∼368 nm (Immeln et al., 2007), aCRY is significantly blue shifted (Figure 1C). This spectral region of flavins is sensitive to solvent characteristics (Kotaki et al., 1970), indicating that the hydrogen bonding environment in the FAD binding site differs significantly between aCRY and CPH1.
The fluorescence emission of the native aCRY protein was also investigated (Figure 1D). The emission intensity is 2 orders of magnitude weaker than that of free FAD in solution, which already shows weak fluorescence, with a quantum yield of 0.04 (Bowd et al., 1968). These results suggest that the protein performs efficient ultrafast quenching of the excited FAD through processes such as electron transfer reactions. The emission spectrum of the native protein has a small but characteristic blue shift of 2 nm in comparison with free FAD in solution.
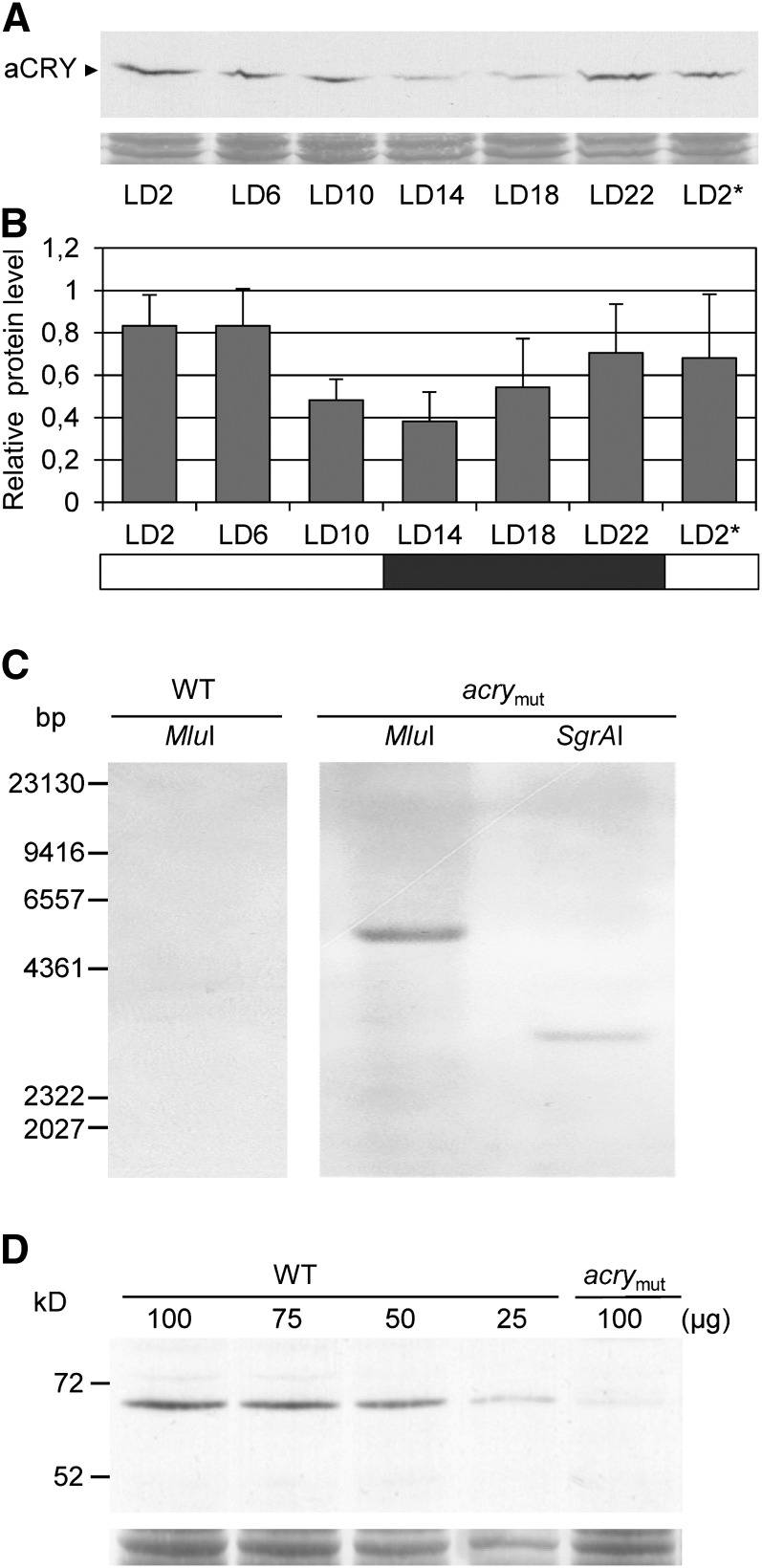
The heterologously expressed, native aCRY was also used for the production of antibodies (see Methods). Using these antibodies, the accumulation of aCRY in C. reinhardtii was monitored over the diurnal cycle of 12 h of light and 12 h of dark (LD12:12). The accumulation of aCRY exhibits an amplitude change of about twofold within a light/dark cycle (Figures 2A and 2B). In contrast to CPH1 (Reisdorph and Small, 2004), aCRY is not rapidly degraded in the light. The highest levels of this protein are observed at the beginning of and extending to the middle of the day (LD2 and LD6), with its lowest values at the beginning of the night (LD14) (Figures 2A and 2B).
Figure 2.
Expression of aCRY over a Diurnal Cycle and Characterization of an acry Mutant.
(A) C. reinhardtii wild-type cells were grown under a LD12:12 cycle and harvested at the indicated time points. The asterisk indicates the beginning of the next light period at LD2. Equal amounts of proteins from crude extracts (either 75 or 100 µg of protein per time point, depending on the biological replicate) were separated by 10% SDS-PAGE along with molecular mass standards and immunoblotted using anti-aCRY antibodies. To control for equal amounts of proteins loaded, the PVDF membrane was stained with Coomassie Brilliant Blue R 250 after immunological detection. From this stain, selected, unspecified protein bands are shown (bottom).
(B) Quantification of aCRY protein levels using ImageMaster 2D Elite version 4.01 (GE Healthcare). The highest level of protein for each set of experiments (n = 3) was set to 1. White bar, light; black bar, darkness.
(C) DNA gel blot analysis of genomic DNA from the SAG73.72 wild-type strain (WT) and the mutant strain SAG.73.72:acry1A (acrymut). Thirty micrograms of genomic DNA was digested with MluI and SgrAI, as indicated. DNA fragments were separated on a 0.8% agarose gel and blotted onto a nylon membrane. A labeled APHVIII cassette fragment was used as the hybridization probe (see Methods). The positions of the restriction sites are shown in Supplemental Figure 3A online.
(D) Immunoblot analysis of aCRY protein in the wild type and acrymut. Cells were harvested at LD6 and used to make crude extracts. Different amounts of protein from a crude extract of wild-type cells (100, 75, 50, and 25 µg per lane) as well as 100 µg of protein from a crude extract of acrymut were separated by 10% SDS-PAGE along with molecular mass standards and immunoblotted with anti-aCRY antibodies. To control for the different protein load, the PVDF membrane was stained with Coomassie Brilliant Blue R 250 after immunological detection. From this stain, one selected, unspecified band is shown (bottom). Quantification of the aCRY protein level was done as described in (B) (n = 3).
Generation of an Insertional acry Mutant with Reduced Protein Levels
Functional analysis of aCRY in C. reinhardtii required the generation of a stable mutant that is either null or strongly reduced for the synthesis of aCRY. Since the use of RNA interference to knock down transcript levels may result in transgenic lines in which the level of the protein encoded by the target gene is not stable over time (Schroda, 2006), we used a different approach. The APHVIII gene cassette encoding paromomycin resistance was inserted randomly into the C. reinhardtii genome (see Methods), and an insertional mutant library of 25,000 individual colonies was generated. Genomic DNA was isolated from pools of the mutants (see Methods for details) and used to screen for strains defective in the gene encoding aCRY; specifically, the DNA was screened by PCR, using several primers situated in the aCRY gene and one primer in the APHVIII gene. Positive PCR signals were reexamined using genomic DNA subpools until a single mutant strain was identified (Gonzalez-Ballester et al., 2011); the mutant line identified was designated CRMS101. Genome regions flanking the cassette in CRMS101 were amplified by PCR and used for sequencing (see Supplemental Figures 3A to 3C online). This analysis showed that the APHVIII cassette was inserted into intron 7 of aCRY in the reverse orientation relative to the target gene along with some minor changes (see Supplemental Figure 3B online).
The transgenic mutant line CRMS101 was generated in a D66 genetic background. For functional analysis, it was backcrossed to the wild-type SAG73.72 strain (see Methods). One of the generated tetrads (SAG73.72:acry1A) was analyzed in more detail. Initially, PCR was performed to amplify the inserted cassette, which was resequenced and compared with that of the analogous sequence in the original mutant (see Supplemental Figure 3D online). The DNA from the backcrossed strain was used for DNA gel blot hybridizations to determine the copy number of the cassette in the mutant genome; only one copy of the introduced gene was detected (Figure 2C). This analysis had already been performed with the original CRMS101 mutant and yielded the same results. MluI and SgrAI digestions of genomic DNA each resulted in a single band (∼4857 and 3018 bp, respectively) that hybridized to the labeled APHVIII probe. Hybridization of the identical probe to MluI-digested wild-type genomic DNA resulted in no signal.
Finally, the aCRY protein level was analyzed in the SAG73.72:acry1A strain (abbreviated as acry mutant or acrymut from now on) relative to the wild-type strain SAG73.72 (Figure 2D). The aCRY protein is reduced in the acry mutant to ∼20% ± 4% (se) of the level measured in wild-type cells (the level in wild-type cells was assigned a value of 100%).
Blue and Red Light–Elicited Expression of Metabolic-, Cell Cycle-, and Clock-Relevant Genes
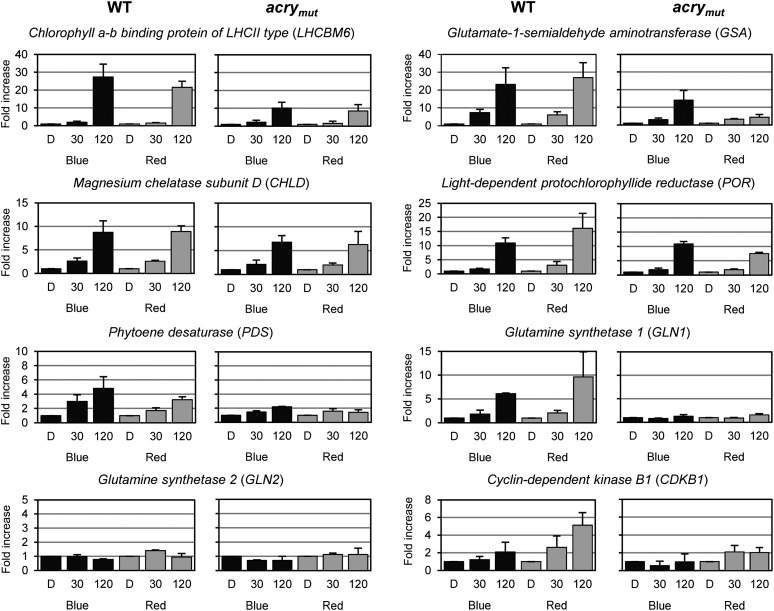
We analyzed the impact of blue light on the expression of a variety of C. reinhardtii genes, including some that were previously shown to be light induced. The selected genes encode proteins from different metabolic pathways and components associated with both the cell cycle and the circadian clock. Some of these genes, such as those encoding light-harvesting complex proteins, are regulated by phytochromes in vascular plants. Since phytochromes do not seem to be present in C. reinhardtii, it was important to verify previous results suggesting that red light could elicit elevated expression of the LHCBM6, GSA, and possibly PDS genes (Im et al., 2006). In the case of the other selected genes, blue and red light–dependent regulation had not been studied before in C. reinhardtii. For the assays, algal cells were grown in a light/dark cycle, and at the end of the third light period they were maintained for 60 h under constant darkness before being subjected to 30- and 120-min blue or red light treatments (for further details, see Methods). Genes selected for testing encode light-harvesting complex proteins (LHCBM6), proteins important for chlorophyll (GSA, CHLD, and POR) and carotenoid (PDS) biosynthesis, or proteins important for nitrogen metabolism (GLN1 and GLN2); many of these genes were significantly upregulated (approximately threefold to fivefold, PDS; greater than fivefold, GSA, CHLD, POR, LHCBM6, and GLN1), and both blue and red light could elicit changes in their expression (Figure 3). In contrast, transcripts encoding the plastidic Gln synthetase (GLN2) were not affected by either blue or red light. We also analyzed the cell cycle component gene CDKB1. It was mainly upregulated after red light treatment (fivefold; Figure 3).
Figure 3.
Blue (465 nm) and Red (635 nm) Light-Dependent Changes in Transcript Accumulation of Genes Encoding Metabolic Proteins and a Cell Cycle Component.
Transcript accumulation was quantified by RT-qPCR in the wild type (WT) and acrymut. Cells were grown in a light/dark cycle. At the end of the light period, cells were maintained for 60 h in darkness (D) before exposure to 30 and 120 min of either blue or red light (see Methods). LEDs with an energy fluence rate of 2.6 W/m2 were used. Total RNA was isolated, and equal amounts were used for RT-qPCR with RACK1 as a reference gene for normalization. The changes in transcript levels following exposure of the cells to blue or red light are presented as fold change relative to RNA from dark-grown cells. Each experiment was performed in triplicate from at least two different biological samples. Bars show means and sd.
We further examined the expression of known genes in C. reinhardtii that encode proteins associated with the circadian clock (Figure 4). A marked (∼15-fold) upregulation of the gene encoding the C3 subunit of the circadian RNA binding protein CHLAMY1 was observed 30 min after being placed in blue or red light (Figure 4). Other clock-relevant genes exhibited a relatively modest (1.5- to ∼3-fold) change in their expression following blue and/or red light exposure; these genes include those encoding the C1 subunit of CHLAMY1, ROC55, ROC66, CO, CK1, ROC114, and ROC75. Red light had little effect on the expression of ROC114 and caused a twofold decrease in the expression of ROC75. In addition to the ROC75 transcript, the transcripts of two other members of the ROC transcription factor family declined; ROC40 and ROC15 transcripts declined to about one-half and one-fifth, respectively, of their pretreatment levels following a 2-h incubation in either blue or red light (Figure 4). In sum, blue and red light impacted the accumulation of transcripts from several genes, and in nearly all cases, both lights had a similar qualitative effect (the direction of change of the transcript level was the same).
Figure 4.
Blue (465 nm) and Red (635 nm) Light-Dependent Changes in Transcript Accumulation of Genes Encoding Circadian Clock–Relevant Components.
Experiments were performed as described in Figure 3. Each experiment was performed in triplicate from at least two different biological samples. Bars show means and sd. WT, wild type.
Transcript Levels Are Significantly Altered in the acry Mutant
We examined blue and red light effects on those genes discussed above in the acry mutant to determine if aCRY is involved in modulating their expression (Figures 3 and 4). Often, the mutant exhibited a different response than wild-type cells following exposure to either red or blue light. Among the genes encoding metabolic and cell cycle proteins (Figure 3), several transcripts were significantly reduced in the acry mutant (relative to wild-type cells) following a 2-h exposure to blue or red light (LHCBM6, PDS, and GLN1), or primarily to red light (CDKB1, GSA, and POR). Our data suggest that most of the analyzed metabolism-related genes, as well as the one encoding the cell cycle component, are regulated by aCRY.
Changes in mRNA levels were also observed in the case of some genes important for clock function (Figure 4). The increase in the level of transcript encoding the C3 subunit following exposure of wild-type cells to blue or red light appeared to be suppressed in the acry mutant. In addition, the ROC55, ROC66, ROC114, CK1, and CO transcript levels were lower in the acry mutant under blue light relative to their levels in wild-type cells; this effect was either smaller or completely absent following red light exposure. Interestingly, the transcript for one of the clock-relevant genes (ROC15) was significantly elevated in the acry mutant relative to wild-type cells following exposure to red light. The mutant exhibited an ∼10-fold increase in the ROC15 mRNA level relative to wild-type cells following exposure to red light (the transcript in wild-type cells dropped to ∼25% after red light exposure). These data clearly demonstrate that aCRY is involved in regulating the levels of transcript for many genes, including those associated with circadian clock function, following exposure to blue or red light.
The results presented above indicate that aCRY (1) may have the ability to absorb both blue and red light, (2) may be functionally linked to a signaling pathway under the control of a C. reinhardtii red light photoreceptor that has not yet been identified, or (3) both. Indeed, the data showing a significant increase in ROC15 transcript level following exposure of the acry mutant to red light, while the level of the transcript declines in red light–exposed wild-type cells, suggest that both statements 1 and 2 may be valid.
Complementation of the acry Mutant
To determine if the phenotypes of the acry mutant are a consequence of depressed levels of aCRY and are not due to a potential second-site lesion remaining in the backcrossed line, we attempted to rescue the mutant phenotype with a wild-type copy of aCRY. The aCRY gene, with its endogenous promoter (1373 bp upstream of the translation start site), was ligated into a vector containing a hygromycin B marker gene (see Methods; see Supplemental Data Set 2 online). This aCRY-harboring vector, designated pKP39, was introduced into the acry mutant, and transgenic lines were selected on hygromycin B–containing plates and then screened for the accumulation of the aCRY protein by immunoblotting. One resistant line, acry complemented 42, accumulated aCRY to a level that was nearly identical to that of wild-type cells (104% ± 4% [n = 3]; see Supplemental Figure 4A online). This strain was used to examine blue light– and red light–elicited responses of representative genes that were altered for expression in the original acry mutant; these genes included two associated with metabolism (GLN1 and PDS) and two that are clock relevant (C3 and ROC15). In all cases, transcript accumulation patterns approached those of wild-type cells in transgenic line 42 (see Supplemental Figure 4B online). These data demonstrate that the aberrant light responses observed in the acry mutant are a consequence of reduced aCRY levels.
In Vitro Spectral Analysis Shows Photoreduction of aCRY in Response to Blue and Red Light
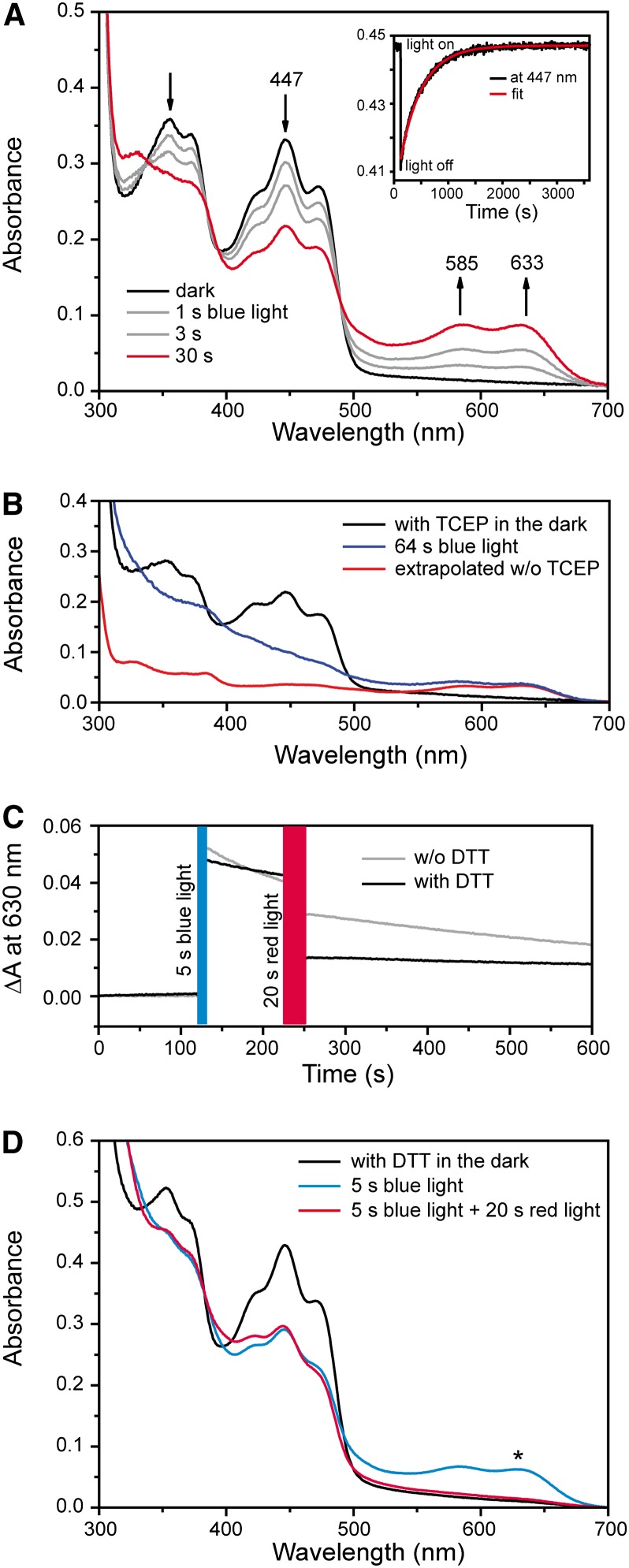
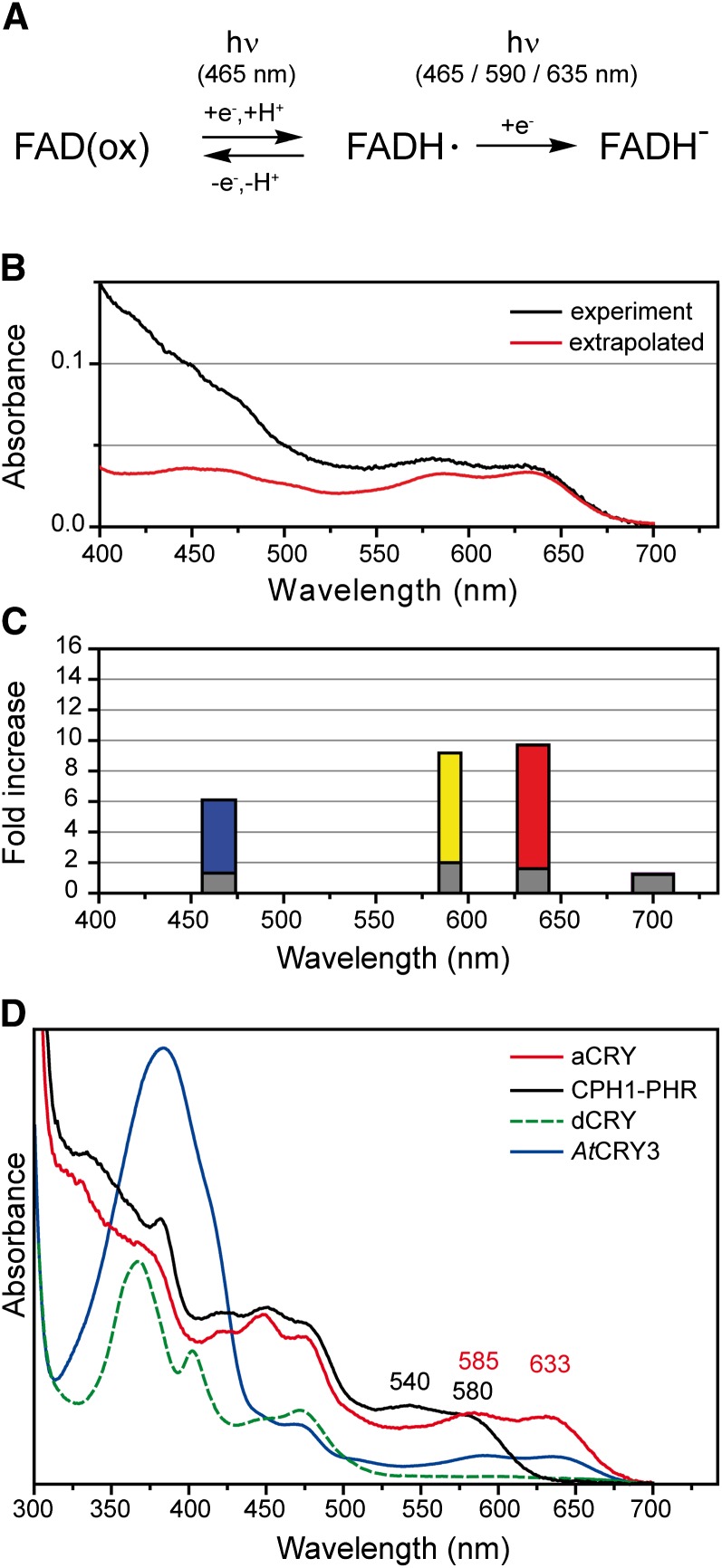
To determine if the in vivo effects can be attributed to the molecular characteristics of aCRY, we used UV/Vis spectroscopy to investigate the responses of the isolated flavoprotein, synthesized in E. coli, to blue and red illumination. The experiments were first performed without an external reductant such as DTT or Tris(2-carboxyethyl)phosphine (TCEP). The samples were exposed to blue light from a light-emitting diode (LED) with an output at 455 nm (Figure 5A). A decrease in the maximum absorbance at 447 nm was detected after an illumination of up to 30 s, indicating a loss of oxidized FAD. At the same time, a broad absorption band with peaks at 585 and 633 nm developed, demonstrating the formation of the FAD neutral radical state (Massey and Palmer, 1966). The presence of two isosbestic points at 338 and 489 nm highlights the exclusive presence of two flavin states in the sample, those of the oxidized FAD and the FAD neutral radical. In the absence of reductant, this mixture of oxidation states was obtained even after prolonged illumination. The addition of the reductant TCEP elicited further conversion of the oxidized to the radical state and facilitated the measurement of a spectrum of the neutral radical (Figure 5B). For comparison, a radical spectrum was generated by extrapolating the reaction sequence in the absence of reductant from partial to full conversion. Both spectra of the neutral radical show the same absorption maxima, albeit overlapped in the presence of TCEP by contributions at lower wavelengths from light scattering due to protein aggregation (Figure 5B). The spectral properties are similar to those of other flavoprotein neutral radicals (Massey and Palmer, 1966). In the dark, full recovery of the oxidized state was observed in the absence of external reductant. The absorbance was followed at 447 nm after a 1-s illumination with blue light (Figure 5A, inset). The analysis yielded a monoexponential kinetic with a time constant of ∼450 s.
Figure 5.
Conversion of the Oxidized State of aCRY to the Radical State upon Blue Light Illumination and the Effect of Red Light on the Radical State of aCRY.
(A) UV/Vis spectra of aCRY were recorded before and after blue light illumination for the indicated time intervals. The inset shows a representative kinetic of the recovery of the oxidized state in the dark as measured at 447 nm. A time constant of 450 s was determined by a monoexponential fit.
(B) UV/Vis spectra before and after illumination for 64 s in the presence of the reductant TCEP. The oxidized state was completely converted to the neutral radical state of flavin. To illustrate the contribution of light scattering, a spectrum of the neutral radical was extrapolated from the reaction sequence in (A).
(C) The presence of the neutral radical state was monitored by UV/Vis spectroscopy at 630 nm with and without the reductant DTT. Preillumination with blue light leads to formation of the flavin radical, which decays in the dark to the oxidized state. Subsequent exposure to red light results in a significant additional loss of the flavin radical state. In the presence of DTT, the decay in the dark is slowed and the effect of red light becomes more pronounced.
(D) Corresponding UV/Vis spectra show that red light illumination leads to a decay of the neutral radical but not to a recovery of the oxidized state. The asterisk indicates the wavelength at which traces were recorded in (C).
The selective conversion of the FAD neutral radical to the reduced state by illumination with green light in vitro has been demonstrated for Arabidopsis CRY1 and CRY2 (Banerjee et al., 2007; Bouly et al., 2007). Accordingly, the isolated aCRY was preilluminated with blue light and then exposed to red light at 636 nm, close to the absorption maximum of the aCRY neutral radical (Figure 5C). The kinetic trace monitoring changes of absorbance at 630 nm showed the formation of the neutral radical state by blue light and consecutive decay of the neutral radical in the dark. The decay had a time constant similar to that of the recovery of the oxidized state (Figure 5A, inset). Furthermore, exposure to red light caused an additional decline in the neutral radical level. In the presence of DTT, this red light effect was significantly more pronounced (Figure 5C), while decay to the oxidized state was much slower. These results were confirmed by spectra taken in the dark and immediately after the sample was exposed to consecutive blue and then red illumination (Figure 5D; see Supplemental Figure 5 online). Illumination with blue light led to a loss of oxidized state (absorbance peak at 447 nm) and formation of the neutral radical. Upon red illumination, the neutral radical decayed while absorbance at 447 nm remained constant. The loss of both the oxidized and neutral radical states implies the formation of the fully reduced state of FAD upon exposure to red light, which is characterized by weak absorption at >400 nm (Massey and Palmer, 1966). These results demonstrate that both blue and red light can induce photoresponses in purified aCRY.
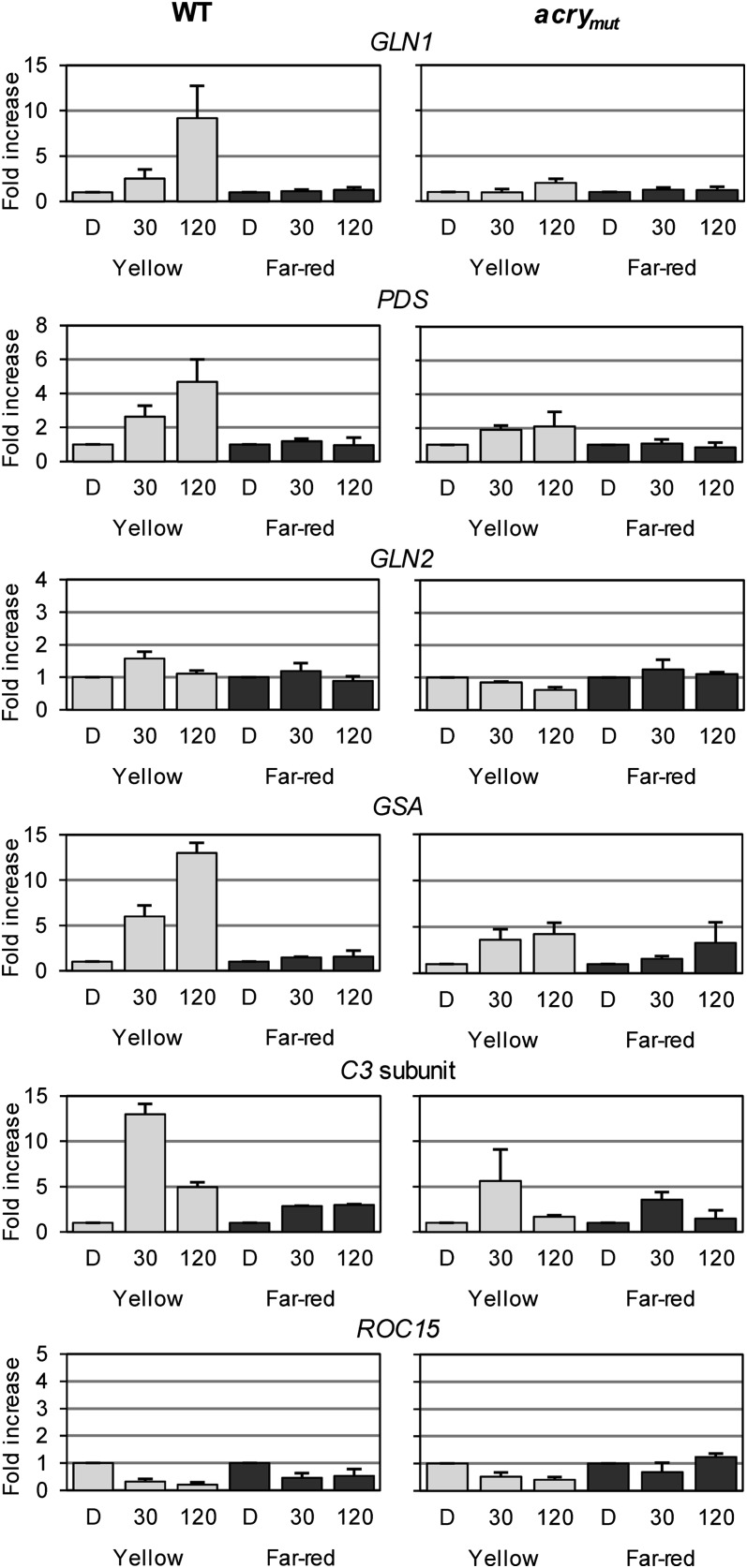
Expression of Selected Genes under Yellow and Far-Red Light in the Wild Type and the acry Mutant
Our data suggest that C. reinhardtii aCRY absorbs red light of ∼633 nm in its neutral radical state. If this hypothesis is correct, absorption of yellow light by aCRY should also impact gene expression, since the neutral radical state has absorbance peaks at both 585 and 633 nm. On the other hand, far-red light (700 nm) should not impact gene expression, since there is no absorption observed in vitro at that wavelength. To evaluate these predictions, we illuminated cells with yellow or far-red light (see Methods) and monitored the levels of transcript from selected light-regulated genes previously characterized in wild-type cells and the acry mutant (Figures 3 and 4). The transcripts analyzed included those from the blue light– and red light–regulated GLN1, GSA, PDS, C3, and ROC15 and the nonblue light– and nonred light–regulated GLN2. In all cases in which transcript levels increased following exposure to red light (GLN1, GSA, PDS, and C3), a similar effect was observed following exposure to yellow light (for 30 or 120 min); this light-elicited increase in transcript levels was significantly suppressed in the acry mutant (Figure 6). Under far-red illumination conditions, either no effect or a small effect was observed in both wild-type cells and the acry mutant (Figure 6). These data support our earlier suggestion that aCRY may act not only as a blue light but also as a yellow/red light photoreceptor in C. reinhardtii.
Figure 6.
Yellow (590 nm) and Far-Red (700 nm) Light-Dependent Expression of Genes Encoding Metabolic Proteins and Clock-Relevant Components in the Wild Type and acrymut.
Experiments were performed as described in Figure 3, but cells were exposed to either yellow or far-red light. Each experiment was performed in triplicate from at least two different biological samples. Bars show means and sd. WT, wild type.
Interestingly, the level of ROC15 mRNA was markedly reduced in yellow light in wild-type cells, similar to the effect of red light. However, it was also reduced following exposure of wild-type cells to far-red light, although to a lesser extent. The highly elevated ROC15 transcript levels observed in red light in the acry mutant were not elicited by yellow light. This complex regulation pattern observed for ROC15 will be discussed below.
DISCUSSION
Blue and Red Light Cause a Similar Change in Gene Expression
In this study, we measured changes in the expression of genes (mRNA levels) associated with different cellular processes in C. reinhardtii in response to blue and red light. All of the examined transcripts encoding proteins involved in chlorophyll and carotenoid biosynthesis (GSA, CHLD, POR, and PDS), as well as the LHC polypeptide, LHBM6, exhibited a gradual rise during exposure of cells to 30 and 120 min of blue and red light, with the highest levels generally being observed after 120 min. In the case of GSA, PDS, and LHCBM6, blue and red light induction was previously demonstrated (Im et al., 2006), although in that study there was little effect of red light on the level of PDS mRNA. This lack of effect on PDS transcript levels could reflect the difference in the wavelengths of light used (i.e., 650 nm in the previous study and 635 nm in this study); the wavelength used in our study better matches the local absorption maximum of the neutral radical form of aCRY (Figure 5A). GLN1 and GLN2, which encode the cytosolic and plastidic Gln synthetases, respectively, were previously shown to be induced by white light within 3 h (GLN1) or after 48 h (GLN2; Chen and Silflow, 1996). This is in agreement with our data showing a gradual increase in GLN1 mRNA during a 2-h exposure to either blue or red light, with no effect on GLN2 transcript levels within the same time frame. Also, the transcript encoding the cell cycle–dependent kinase CDKB1 gradually increases over a 2-h red light exposure.
We also examined the impact of blue and red light on transcripts encoding 10 circadian clock–relevant C. reinhardtii proteins. Both light conditions altered the levels of these transcripts, but in several cases the changes in transcript abundance were modest (1.5- to 3-fold). An exception was noted for the C3 subunit of the RNA binding protein CHLAMY1; altered expression of this protein causes phase shifts in circadian responses (Iliev et al., 2006). We observed an ∼15-fold increase in the C3 transcript after the cells were exposed to 30 min of either blue or red light, demonstrating that the gene encoding C3 belongs to a group of rapidly induced genes such as VVD in Neurospora; a disruption in VVD also causes phase shifts of circadian rhythms (Heintzen et al., 2001; Lewis et al., 2002). Notably, the level of C3 mRNA is strongly influenced by both light and temperature (Voytsekh et al., 2008; Seitz et al., 2010), suggesting that it links light- and temperature-responsive entrainment pathway(s) with the oscillatory system. Moreover, this protein can influence circadian output as part of the CHLAMY1 complex (Kiaulehn et al., 2007). Rapid blue light– or red light–triggered induction (RNA accumulation after 30 min) of other clock-relevant genes (the C1 subunit of CHLAMY1, ROC55, CO, CK1, ROC114, and ROC75) was also observed, which contrasts with the slower induction of the metabolism- and cell cycle–relevant genes. Notably, the levels of transcripts from two clock genes showed a decline after blue or red light treatment (ROC15 and ROC40), while the ROC75 mRNA appeared to specifically decline after red light exposure.
The Absorption Spectrum of the aCRY Radical State in Vitro Is Congruent with Light-Elicited in Vivo Responses
Blue, red, and yellow light can trigger elevated levels of transcripts of several C. reinhardtii genes. This effect is significantly reduced in the acry knockdown mutant, which suggests that aCRY is the primary photoreceptor mediating these effects. However, if it is mediating these effects, then the absorption spectra (Figures 1C, 5A, 5B, and 5D), which reflect the redox states of flavin in aCRY in vitro (Figure 7A), should define those wavelengths that would effectively elicit output responses. Based on the spectra of the oxidized and fully reduced states of aCRY (Figures 1C and 5D), they cannot be responsible for the yellow and red light effects. In contrast, the spectrum of the neutral radical obtained under reductive conditions (Figure 5B) has the appropriate spectral characteristics (Figure 7B). A more detailed analysis was performed to further explore whether the absorption spectrum of the aCRY neutral radical species fits with the GLN1 transcript responses (Figures 3 and 6). This comparison (Figure 7C) demonstrated that blue, yellow, and red light, but not far-red light, are absorbed by the aCRY neutral radical in vitro and that only the light qualities absorbed by this neutral radical were able to elicit GLN1 transcript accumulation in vivo. Moreover, it is not only light quality but also the quantity of light absorbed that specifies the potential photoreceptor involved in a response. Exposure to similar quanta of yellow (590 nm) and red (635 nm) light led to a similar fold increase in GLN1 mRNA (Figure 7C), as expected from the absorption coefficients of the aCRY radical at 585 and 633 nm (Figure 7B). The in vivo effect of blue light (465 nm) is weaker than expected from the absorption at this wavelength in vitro. However, full conversion of flavoproteins to their neutral radical form is difficult to achieve. Principal component analysis describing the conversion of phot-LOV domain states indicated that the neutral radical might absorb significantly less in the blue than in the red spectral region (Lanzl et al., 2010), which is in agreement with the in vivo responses that we measured (Figure 7C). Therefore, the neutral radical state of aCRY has the spectral characteristics that could account for all of the transcript abundance responses quantified in this study.
Figure 7.
Reaction Sequence of aCRY and Comparison of the Absorption Spectrum of the Neutral Radical with in Vivo Responses to Light of Different Spectral Qualities and with Absorption Spectra of FAD Radical States of Other CRYs.
(A) The three redox states associated with aCRY in vitro (i.e., oxidized, neutral radical, and fully reduced FAD) are depicted. At the specified wavelengths of light, a conversion of oxidized to neutral radical or of neutral radical to the fully reduced state of FAD takes place.
(B) The absorption spectrum of the flavin neutral radical of aCRY is shown for comparison according to Figure 5B.
(C) Relative increase in the accumulation of GLN1 mRNA after exposure to different light qualities (i.e., blue, yellow, red, and far-red light) for 120 min. Responses in both the wild type (colored bars) and the acry mutant (gray bars) are shown. The width of the bars corresponds to the FWHM of the respective LED emission spectrum.
(D) For comparison, C. reinhardtii CPH1 was selected as a representative plant CRY. It shows the blue shift of the local absorption maxima of the neutral radical to 540 and 580 nm, which is characteristic for a plant CRY. dCRY, an animal type I CRY from Drosophila, forms the anion radical with an absorption maximum at 367 nm and weak absorption at >510 nm. Arabidopsis CRY3 is a member of DASH CRYs and contains an antenna chromophore with pronounced absorption at 384 nm. All spectra contain some additional contributions from the oxidized state at <500 nm.
If all of the data obtained in this study are considered, there are some complications with respect to the picture presented above. Some genes are more strongly upregulated in blue than in red light and vice versa. This result might be explained by a regulatory contribution of the oxidized state of flavin in aCRY. The presence of a mixture of oxidized and radical states might lead to variability in output effects. In support of this proposal, our in vitro studies have shown that conversion of aCRY from the oxidized to the neutral radical is achieved without the need of excessive light or a reductive environment. On the other hand, some of the genes that we have studied (LHCBM6, GSA, and PDS) are also influenced by phot, another blue light photoreceptor (Im et al., 2006). Thus, different photoreceptors may act, at least in some cases, in concert to regulate blue light–related gene expression. Moreover, some aCRY-dependent, clock-relevant genes, including ROC15, appeared to be downregulated upon illumination, which suggests an indirect involvement of aCRY in their regulation.
A further question relating to the comparison of in vivo and in vitro data concerns the redox state of aCRY in the dark in vivo. The finding that aCRY isolated from E. coli was exclusively in its oxidized form suggests that the dark form of the photoreceptor is oxidized and that the radical state is generated by photoreduction. On the other hand, C. reinhardtii does exhibit red light responses without a blue light pretreatment, which indicates that the neutral radical is the dominant form in the dark. Photoreduction might not be required if the cellular redox potential is below the midpoint potential of the FADH·/FAD(ox) redox couple (Liu et al., 2010). Under such conditions, cellular reductants might reduce aCRY in the dark, as in the case of E. coli photolyase (Sancar, 2003). To resolve this issue, the redox midpoint potential of aCRY must be determined. However, midpoint potentials are of limited value if the redox potential in the cell is not known. While vascular plant CRYs do have known midpoint potentials, there is still no agreement on the specific redox state taken by the photoreceptor in its dark, resting state (Balland et al., 2009; Liu et al., 2010), primarily because the redox potential in the nucleus has not been determined.
Spectral Properties of the aCRY Radical State Differ from Those of Other CRYs
aCRY exhibits a distinct difference from other CRYs in the absorption of the neutral radical, with band maxima at 585 and 633 nm. In comparison, plant CRYs show a strong blue shift of the lowest energy maxima to 580 and 595 nm for CPH1 and AtCRY1, respectively (Kottke et al., 2006; Immeln et al., 2010). Therefore, the lowest energy absorption bands of the CPH1 and aCRY neutral radicals are almost spectrally separable (Figure 7D). As a direct consequence, the plant CRY, CPH1, cannot be responsible for the red light responses observed in C. reinhardtii following illumination at ∼635 nm. Similarly, it was assumed that impaired red light responses in an Arabidopsis double mutant that is defective in both CRY1 and CRY2 are based on the physical interaction between CRYs and phytochromes, because Arabidopsis CRYs are not known to absorb red light (Yang et al., 2008). The same situation exists for C. reinhardtii phot, which in both the light and dark forms does not absorb light above 500 nm (Kottke et al., 2003; Guo et al., 2005).
Drosophila CRY (dCRY), a typical animal type I CRY (Stanewsky et al., 1998), shows the formation of an anion radical upon blue light illumination, which is characterized by an absorption spectrum with negligible contributions at >510 nm (Berndt et al., 2007) (Figure 7D). A similar spectrum to dCRY was recently observed for the dark form of human CRY1 (HsCRY1), an animal type II CRY (Vieira et al., 2012). The spectra of these two representatives of animal CRYs, therefore, show little similarity to the aCRY spectrum of C. reinhardtii. On the other hand, millisecond transient spectra of the bird Sylvia borin (garden warbler) CRY1a reveal the formation of a neutral radical (Liedvogel et al., 2007) as in aCRY, despite the 88% identity in sequence between SbCRY1a and HsCRY1 (Figure 1A). Furthermore, a similar radical can be generated by reoxidation of chicken CRY4 (Ozturk et al., 2009). Finally, DASH CRYs, such as Arabidopsis CRY3, display a radical spectrum that is weakly but significantly red-shifted to ∼640 nm (Pokorny et al., 2005, Zikihara et al., 2008). Additionally, the spectrum of DASH CRYs is overlapped by strong absorbance of the antenna chromophore methenyltetrahydrofolate at 380 nm (Figure 7D); this antenna chromophore is lacking in aCRY. The position of the radical band maxima of aCRY best matches those of nonsensory flavoproteins, such as Glc oxidase (Massey and Palmer, 1966) and (6-4) photolyase (Hitomi et al., 1997) or the multifunctional CPF1 protein (Usman et al., 2009). In conclusion, aCRY shows properties that are distinct from those of animal type I, DASH, and plant CRYs and also from some but not all of the animal type II CRYs.
Identification of a Flavoprotein Mediating a Red Light Response
Flavoproteins are known to act as blue light sensors in bacteria, fungi, plants, and insects but are in principle capable of acting also as red light sensors, since the absorption of the neutral radical state of flavoproteins, in most cases, extends well beyond 600 nm (Müller et al., 1972). Previous studies suggested a role of the neutral radical in photoreceptors, but only as the signaling state, which is formed from the oxidized state by the absorption of blue light (Galland and Tölle, 2003). Accordingly, illumination of the radical would then lead to depletion of the signaling state. This scenario has been suggested in a current model describing the action of plant CRYs (Chaves et al., 2011) and is based on the finding that the blue light response that is mediated by Arabidopsis CRY1 and CRY2 can be suppressed by green light and linked to the formation and depletion of the radical state of flavin in vivo (Banerjee et al., 2007; Bouly et al., 2007). Whereas this green light effect is a matter of a current controversy based on recent conflicting results with Arabidopsis CRY2 in vivo (Li et al., 2011), the neutral radical is not considered to be a resting state in plant CRYs (Liu et al., 2010; Chaves et al., 2011). In contrast, the in vivo response of aCRY from C. reinhardtii is generated by illumination of the neutral radical state, which might suggest that the fully reduced state is the signaling state. A similar, active role of the radical has been proposed for the AtCRY1- and AtCRY2-mediated blockage of two shade-inducible transcripts by green light (Zhang et al., 2011). Moreover, for aCRY, the functional relevance of red light exposure to a flavoprotein is demonstrated here in vivo.
We have shown that the regulation of several genes by blue and red light depends on aCRY (Figures 3 and 4). The absorption spectrum of the neutral radical state of aCRY extends over nearly the entire visible spectrum with a small transparent window in the far red (>680 nm). Accordingly, aCRY shows absorption and mediates responses in the blue, yellow, and red regions of the spectrum. Therefore, aCRY represents a long sought-after photoreceptor in C. reinhardtii that is responsive to red light. It is tempting to speculate that aCRY represents the only or the major red light photoreceptor in C. reinhardtii, since no other clear candidates (e.g., phytochromes) have emerged from the genome sequence (Mittag et al., 2005; Merchant et al., 2007). However, in the case of ROC15, a complex pattern of regulation was observed. ROC15 transcripts sharply decline in yellow and red light in wild-type cells, whereas transcript levels are increased in the acry mutant in red but not yellow light. Additionally, these transcripts also decline to some extent following exposure of wild-type cells to far-red light. Previous reports have demonstrated some far-red (720 and 740 nm) effects on C. reinhardtii transcript accumulation (Im et al., 2006) that cannot be attributed to aCRY. These results suggest that there is another yet unidentified red light photoreceptor in C. reinhardtii and that the future might reveal additional photoreceptors that create a complex network of light responses, similar to what is observed in vascular plants with the interconnection of phytochromes, phots, and CRYs (Más et al., 2000; Chen et al., 2004; Lariguet et al., 2006).
This study also reveals that sensory flavoproteins may not only be categorized with respect to a potential role as blue light receptor. Yellow and red light activation of flavoproteins needs to be considered as well in the design of future experiments, since responses can be directly elicited by absorption of the neutral radical state of flavin.
METHODS
Strains and Cultures of C. reinhardtii and Light Treatment
The following strains of Chlamydomonas reinhardtii were used: SAG73.72 (wild type, mt +), D66 (cw15, nit2, mt +; Pootakham et al., 2010), and CC-124 (nit1, nit2, agg1, mt−). Strain D66 was used for insertional mutagenesis followed by backcrossing of the mutant strain to CC-124 and SAG73.72. SAG73.72 was routinely used as the wild-type control and was grown under a LD12:12 light/dark cycle, unless otherwise indicated. Cells were harvested at the indicated light/dark time points, where LD0 represents the beginning of the day and LD12 represents the beginning of the night. For light treatments prior to RT-quantitative PCR (qPCR), cells were transferred at the end of the light period (LD12) to 60 h of darkness (dark control) and then exposed to the indicated light quality. Light treatments were performed using LEDs of specific wavelengths each with an energy fluence rate of 2.6 W/m2. The following equipment and settings were used: for blue light, SuperFlux LED, peak at 465 nm (with a full width at half-maximum [FWHM] of 18.5 nm) and a photon fluence rate of 10 µmol·m−2·s−1; for red light, SuperFlux LED (both Lumitronix LED-Technik), peak at 635 nm (FWHM of 17 nm) and a photon fluence rate of 13.6 µmol·m−2·s−1; for yellow light, Solarox LED (Winger Electronics), peak at 590 nm (FWHM of 12.5 nm) and a photon fluence rate of 12.7 µmol·m−2·s−1; for far-red light, TO-66 high-power array (Roithner Lasertechnik), peak at 700 nm (FWHM of 22 nm) and a photon fluence rate of 15.2 µmol·m−2·s−1.
Phylogenetics
Phylogenetic analyses were conducted using the MEGA software package version 4 (Tamura et al., 2007). Alignments of CRY and photolyase protein sequences were performed with the ClustalW algorithm (see Supplemental Data Set 1 online) using the following parameters: for pairwise alignments, a gap opening penalty of 10 and a gap extension penalty of 0.1; for multiple alignments, a gap opening penalty of 10 and a gap extension penalty of 0.2. A Gonnet weight matrix was used with residue-specific penalties ON, hydrophilic penalties ON, a gap separation distance of 4, end gap separation OFF, negative matrix OFF, and a delay divergent cutoff of 30%. The phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei, 1987), and a bootstrap consensus tree was obtained after 1000 replications. Parameters were set as follows: Poisson correction model; complete deletion of gaps/missing data; and uniform rates among sites.
Heterologous Expression, Purification, and Antibody Production
aCRY was expressed in Escherichia coli BL21(DE 3) pLysE (Invitrogen One Shot) using DYT medium (16 g/L tryptone, 10 g/L yeast extract, and 5 g/L NaCl) with 50 mg/L kanamycin. At an OD600 of 0.5, the temperature was lowered from 37°C to 18°C. The induction was initiated with 10 µM isopropyl-β-d-1-thiogalactoside at an OD600 of 0.8. After 20 h, cells were harvested by centrifugation (5000g, 20 min, 4°C) and resuspended in 50 mM sodium phosphate, pH 7.8, containing 100 mM sodium chloride and 20% (v/v) glycerol. Per 10 g of pellet, one tablet of protease inhibitor (complete EDTA-free; Roche) and ∼3 mg of DNase were added. Cells were disrupted in a French Press (SLM Aminco; 3 × 1000 p.s.i.), the debris was removed by centrifugation at 100,000g (4°C for 1 h), and the supernatant was supplemented with 60 mM imidazole and applied to a His affinity column (Novagen His.Bind Resin; Merck). The column was washed with 2 column volumes of buffer containing 60 mM imidazole, followed by elution with 300 mM imidazole. Finally, the protein was dialyzed twice (MWCO 5000, ZelluTrans; Roth) against 50 mM phosphate, pH 7.8, 100 mM sodium chloride, and 20% (v/v) glycerol, flash frozen in liquid nitrogen, and stored at –80°C.
For antibody production, aCRY was further purified by heparin affinity chromatography (HiTrap Heparin; GE Healthcare). One milligram of native protein was used for antibody production in rabbit. The antibodies were generated by Pineda Antikörper Service.
Characterization of Isolated aCRY by Mass Spectrometry and UV/Vis and Fluorescence Spectroscopy
For the identification of aCRY by LC-ESI-MS/MS, 10 µg of the heterologously expressed protein was digested with 2.5 µg of sequencing-grade modified trypsin (Promega) at 37°C overnight. The digested protein was desalted with a laboratory-made Tip-column using Poros R2 10 µm column media (Applied Biosystems). Dried peptides were resuspended in an aqueous solution containing 5% (v/v) DMSO and 5% (v/v) formic acid and analyzed by LC-ESI-MS/MS according to Veith et al. (2009). A false discovery rate of ≤1% was set for data analysis.
The abundance of heterologously expressed protein was determined by SDS-PAGE. UV/Vis spectroscopy was used (UV-2450; Shimadzu) to determine both the concentration of chromophore and protein. The extinction coefficient of the protein at 280 nm was calculated with EXTCOEFF (www.expasy.org) as 107,530 L·mol−1·cm−1. For FAD, an extinction coefficient of 11,300 L·mol−1·cm−1 at 450 nm was used (Siegel, 1978). The chromophore content was calculated from the ratio of A450 to A280.
Fluorescence emission spectra were recorded for both protein and FAD (Sigma-Aldrich) at the same OD450 of 0.5. Spectra were generated by excitation at 447 nm using an F-4010 fluorescence spectrometer (Hitachi).
Illumination with Blue Light for UV/Vis Spectroscopy
UV/Vis spectra were recorded on a UV-2450 spectrometer (Shimadzu) before and after illumination of protein samples with a blue LED (455 nm, 16 mW/cm2 at the sample, FWHM of 20 nm [Luxeon Star; Lumileds]). Between each illumination, a recording time of 160 s in the dark was required. The UV/Vis spectrometer was equipped with a thermostated integration sphere (Shimadzu) to eliminate scattering and keep the sample at 20°C. For kinetic experiments, the absorbance at 447 nm was recorded. After 120 s in the dark, the sample was illuminated with blue light for exactly 1 s. The recovery of the oxidized FAD was monitored. For full conversion to the neutral radical state, the reductant TCEP was added to a final concentration of 2 mM at pH 7.4. Samples were illuminated with blue light for 64 s at 10°C.
Consecutive Illumination with Blue and Red Light for UV/Vis Spectroscopy
The kinetics of the formation and decay of the FAD radical state was monitored at 630 nm in the presence and absence of 10 mM DTT using a UV-2450 spectrometer (Shimadzu). The temperature was maintained at 20°C. The sample was first exposed to 5 s of blue light (455 nm, 10 mW/cm2 at the sample, FWHM of 20 nm [Luxeon Star; Lumileds]) and was then kept in the dark for 100 s. The sample was then exposed to 20 s of 636-nm red light (34 mW/cm2 at the sample, FWHM of 25 nm [Luxeon Star; Lumileds]).
Crude Extracts and Immunoblots
The preparation of C. reinhardtii crude protein extracts and the immunoblots to detect aCRY was performed according to Iliev et al. (2006) with the following exceptions. Proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride (PVDF) membranes and blocked in a 5% suspension of powdered milk in Tris-buffered saline (pH 8.0) with 0.1% Tween 20. The membranes were incubated with primary antibodies against aCRY at a dilution of 1:5000 for 1 h. Horseradish peroxidase–conjugated anti-rabbit IgG was used as a secondary antibody. Peroxidase activity was detected by a chemiluminescence assay. To control for differences in sample loading and protein transfer, the PVDF membranes were stained with Coomassie Brilliant Blue R 250. Quantifications of aCRY protein levels were performed using ImageMaster 2D Elite version 4.01 (GE Healthcare). The highest expression level was set to 1 (n = 3).
RT-qPCR
Total RNA was isolated from cells using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer’s instructions and also included an RNase-free DNase I treatment. The concentration of RNA was determined photometrically. Four hundred nanograms of total RNA was used in each reaction for one-step RT-qPCR; the reagents used to perform these reactions were commercially purchased (QuantiTect SYBR Green RT-PCR; Qiagen). The RT-qPCR was performed using an Mx3005P instrument (Agilent Technologies), and the relative transcript abundances of target genes were calculated based on the cycle threshold (CT) 2−ΔΔCT method (Livak and Schmittgen, 2001) using RACK1 (Mus et al., 2007) as a constitutive reference transcript for normalization. All primers used in the RT-qPCR are listed in Supplemental Table 2 online. These primers were designed to amplify a 100- to 250-bp region that spans at least one intron in the genomic DNA (based on available gene models). Cycling conditions included an initial incubation at 95°C for 15 min, followed by 40 cycles of 95°C for 20 s, 56°C for 25 s, and 72°C for 35 s. Specificity of the primers was determined by melting curve analysis; a single, sharp peak in the melting curve ensures that a single, specific DNA species was amplified. All RT-qPCR experiments were performed in triplicate with RNAs isolated from at least two different cultures.
Insertional Mutagenesis
Approximately 25,000 insertional mutants were generated using an APHVIII gene cassette, and the mutants were screened by PCR using cassette- and gene-specific primers, essentially as described previously (Pootakham et al., 2010; Gonzalez-Ballester et al., 2011). Target gene-specific primers were selected with primer design software (Clone Manager 9; Scientific and Educational Software) using the default setting for hairpin formation and dimer duplexing. Strain CRMS101 was identified with the PCR primer pair 1a/1b situated in aCRY (1a) and the cassette (1b) (see Supplemental Figures 3A and 3B online). After identification of a transgenic line with the desired lesion, a single colony was cultured, the genomic DNA was isolated, and the regions covering the borders of the insertion cassette in CRMS101 were amplified with primer pairs 1a/1b and 2a/2b and then sequenced. Primer sequences are listed in Supplemental Figure 3C online.
DNA Gel Blot Analysis
Genomic DNA was isolated from the wild type and the acry mutant as described (Lee et al., 1993). The DNA was restricted and the fragments were separated on an agarose gel and then transferred to a nylon membrane. A digoxigenin-labeled DNA probe was generated from a 304-bp fragment of the APHVIII cassette digested with BsgI and RsrII. The labeled fragment was used for hybridization and detection with anti-digoxigenin antibodies. All steps were performed using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) according to the manufacturer’s protocol.
Backcrossing
The mutant line CRMS101 was generated in a D66 (cw15, nit2, mt +) genetic background. It was backcrossed to SAG73.72 (mt +), which has a cell wall (CW) and an active nitrate reductase gene (NIT1) along with its transcriptional regulator (NIT2). Therefore, CRMS101 was first crossed with the mt− strain CC-124 (nit1, nit2, agg1). One of the mt− progeny harboring the APHVIII resistance marker gene and lacking the phenotypically detectable (Harris, 1989) agg1 mutation of CC-124 was afterward crossed with SAG73.72 (mt +). Selection of tetrads was performed on nitrate- and paromomycin-containing media. Growth on nitrate as the sole nitrogen source is based on intact NIT1 and NIT2 genes.
Gametogenesis, mating, zygote germination, and tetrad separation were performed according to the procedures of Jiang and Stern (2009).
Complementation of the acry1A Mutant by Transformation with pKP39
The pKP39 vector was used to complement the acry mutant. This vector carries the complete aCRY genomic DNA sequence, including its potential promoter region of 1076 bp, as well as the 5′ and 3′ untranslated region [chromosome 13, positions 5,252,780 to 5,259,855(+) in Joint Genome Institute version 4.0]. This vector also carries the ampicillin resistance gene for selection in E. coli and the hygromycin B resistance gene from the pHyg3 vector (kindly provided by W. Mages, University of Regensburg; Berthold et al., 2002) for selection in C. reinhardtii. The complete pKP39 sequence is presented in Supplemental Data Set 2 online. The mutant strain SAG73.72:acry1A was transformed with the AhdI-linearized pKP39 using the autolysin method as described earlier (Iliev et al., 2006). Cells were grown on Tris-acetate-phosphate agar medium (Harris, 1989) containing 20 µg/mL hygromycin B.
Accession Numbers
Sequence data from this article can be found in the Joint Genome Institute (Chlre4) or GenBank/EMBL databases under the following accession numbers: C1, jgi|182403; C3, jgi|99329; CDKB1, jgi|59842; CHLD, jgi|134594; CK1, jgi|159133; CO, jgi|159133; GLN1, jgi|133971; GLN2, jgi|129468; GSA, jgi|138524; LHCBM6, gi|184490; PDS, jgi|78128; POR, jgi|136589; RACK1, jgi|105734; ROC15, AB363964; ROC40, AB363965; ROC55, AB363966; ROC66, AB363967; ROC75, AB363968; ROC114, AB363970; aCRY, jgi|206002; CPH1, jgi|185758.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence of the Codon-Adapted acry Gene with a His Tag for Heterologous Expression in E. coli.
Supplemental Figure 2. Fluorescence Excitation and Emission of the aCRY Chromophore before and after Acidification.
Supplemental Figure 3. Sequences in acry Mutant Strains and Primers for Characterization.
Supplemental Figure 4. Complementation of the acry Mutant by Transformation with pKP39 to Generate Transgenic Line 42.
Supplemental Figure 5. UV/Vis Absorption Spectra Showing the Effect of Consecutive Blue and Red Light Illumination on aCRY in Vitro in the Absence of Reductant.
Supplemental Table 1. LC-ESI-MS/MS Analysis of Heterologously Expressed aCRY with a C-Terminal 6× His Tag.
Supplemental Table 2. Oligonucleotides Used in RT-qPCR.
Supplemental Data Set 1. Sequence Alignments for Phylogenetic Analysis.
Supplemental Data Set 2. Sequence of pKP39 Used for Complementation of the acry Mutant.
Supplementary Material
Acknowledgments
We thank Karen Roos for comparing the annotated aCRY gene with the cDNA sequence, Rainer Nötzold for cloning parts of the aCRY gene, Sandra Künzel for help with backcrossing, Wolfram Weisheit for help with LC-ESI-MS/MS, and Wolfgang Mages for pHyg3. T.K. thanks Joachim Heberle and Thomas Hellweg for generous support. The mutant library generation and screen were supported by the National Science Foundation (Grants MCB-0235878 and MCB-824469 to A.R.G.). This work was supported by the German Research Foundation within the framework of Research Group FOR 1261 (Grants Mi373/11-1 and Mi373/12-1 to M.M. and Grant Ko3580/1-1 to T.K.) as well as the Swiss National Science Foundation (Grant PA00P3_124169 to S.S.).
AUTHOR CONTRIBUTIONS
B.B., K.P., M.S., S.S., A.R.G., T.K., and M.M. designed the project, B.B., K.P., M.S., S.S., D.W., N.M., M.H., D.D., and D.I. performed experiments, and all authors contributed to analysis and interpretation of results. T.K. and M.M. wrote the article with contributions and edits from A.R.G., B.B., K.P., M.S., and S.S.
Glossary
- FAD
flavin adenine dinucleotide
- LC-ESI-MS/MS
liquid chromatography-electrospray ionization-tandem mass spectrometry
- Vis
visible
- TCEP
Tris(2-carboxyethyl)phosphine
- LED
light-emitting diode
- qPCR
quantitative PCR
- FWHM
full width at half-maximum
- PVDF
polyvinylidene difluoride
- phot
phototropin
- PHR
photolyase homology region
References
- Alizadeh D., Cohen A. (2010). Red light and calmodulin regulate the expression of the psbA binding protein genes in Chlamydomonas reinhardtii. Plant Cell Physiol. 51: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altimus C.M., Güler A.D., Villa K.L., McNeill D.S., Legates T.A., Hattar S. (2008). Rods-cones and melanopsin detect light and dark to modulate sleep independent of image formation. Proc. Natl. Acad. Sci. USA 105: 19998–20003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balland V., Byrdin M., Eker A.P., Ahmad M., Brettel K. (2009). What makes the difference between a cryptochrome and DNA photolyase? A spectroelectrochemical comparison of the flavin redox transitions. J. Am. Chem. Soc. 131: 426–427 [DOI] [PubMed] [Google Scholar]
- Banerjee R., Schleicher E., Meier S., Viana R.M., Pokorny R., Ahmad M., Bittl R., Batschauer A. (2007). The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J. Biol. Chem. 282: 14916–14922 [DOI] [PubMed] [Google Scholar]
- Berndt A., Kottke T., Breitkreuz H., Dvorsky R., Hennig S., Alexander M., Wolf E. (2007). A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J. Biol. Chem. 282: 13011–13021 [DOI] [PubMed] [Google Scholar]
- Berthold P., Schmitt R., Mages W. (2002). An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412 [DOI] [PubMed] [Google Scholar]
- Bouly J.P., Schleicher E., Dionisio-Sese M., Vandenbussche F., Van Der Straeten D., Bakrim N., Meier S., Batschauer A., Galland P., Bittl R., Ahmad M. (2007). Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 282: 9383–9391 [DOI] [PubMed] [Google Scholar]
- Bowd A., Byrom P., Hudson J.B., Turnbull J.H. (1968). Excited states of flavine coenzymes. III. Fluorescence and phosphorescence emissions. Photochem. Photobiol. 8: 1–10 [Google Scholar]
- Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L.O., van der Horst G.T., Batschauer A., Ahmad M. (2011). The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62: 335–364 [DOI] [PubMed] [Google Scholar]
- Chen C.H., Dunlap J.C., Loros J.J. (2010). Neurospora illuminates fungal photoreception. Fungal Genet. Biol. 47: 922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen Q., Silflow C.D. (1996). Isolation and characterization of glutamine synthetase genes in Chlamydomonas reinhardtii. Plant Physiol. 112: 987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J.M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58: 21–45 [DOI] [PubMed] [Google Scholar]
- Christie J.M., Reymond P., Powell G.K., Bernasconi P., Raibekas A.A., Liscum E., Briggs W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282: 1698–1701 [DOI] [PubMed] [Google Scholar]
- Coesel S., Mangogna M., Ishikawa T., Heijde M., Rogato A., Finazzi G., Todo T., Bowler C., Falciatore A. (2009). Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 10: 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S., Finazzi G., Wollman F.A. (2008). The dynamics of photosynthesis. Annu. Rev. Genet. 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Etchegaray J.P., Lee C., Wade P.A., Reppert S.M. (2003). Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421: 177–182 [DOI] [PubMed] [Google Scholar]
- Faeder E.J., Siegel L.M. (1973). A rapid micromethod for determination of FMN and FAD in mixtures. Anal. Biochem. 53: 332–336 [DOI] [PubMed] [Google Scholar]
- Galland P., Tölle N. (2003). Light-induced fluorescence changes in Phycomyces: Evidence for blue light-receptor associated flavo-semiquinones. Planta 217: 971–982 [DOI] [PubMed] [Google Scholar]
- Geisselbrecht Y., Frühwirth S., Schroeder C., Pierik A.J., Klug G., Essen L.O. (2012). CryB from Rhodobacter sphaeroides: A unique class of cryptochromes with new cofactors. EMBO Rep. 13: 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballester D., Pootakham W., Mus F., Yang W., Catalanotti C., Magneschi L., de Montaigu A., Higuera J.J., Prior M., Galván A., Fernandez E., Grossman A.R. (2011). Reverse genetics in Chlamydomonas: A platform for isolating insertional mutants. Plant Methods 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Kottke T., Hegemann P., Dick B. (2005). The phot LOV2 domain and its interaction with LOV1. Biophys. J. 89: 402–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook. (San Diego, CA: Academic Press; ). [Google Scholar]
- Hegemann P. (2008). Algal sensory photoreceptors. Annu. Rev. Plant Biol. 59: 167–189 [DOI] [PubMed] [Google Scholar]
- Heijde M., et al. (2010). Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ. 33: 1614–1626 [DOI] [PubMed] [Google Scholar]
- Heintzen C., Loros J.J., Dunlap J.C. (2001). The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104: 453–464 [DOI] [PubMed] [Google Scholar]
- Hendrischk A.K., Frühwirth S.W., Moldt J., Pokorny R., Metz S., Kaiser G., Jäger A., Batschauer A., Klug G. (2009). A cryptochrome-like protein is involved in the regulation of photosynthesis genes in Rhodobacter sphaeroides. Mol. Microbiol. 74: 990–1003 [DOI] [PubMed] [Google Scholar]
- Hitomi K., Kim S.T., Iwai S., Harima N., Otoshi E., Ikenaga M., Todo T. (1997). Binding and catalytic properties of Xenopus (6-4) photolyase. J. Biol. Chem. 272: 32591–32598 [DOI] [PubMed] [Google Scholar]
- Huang K., Beck C.F. (2003). Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 100: 6269–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D., Voytsekh O., Schmidt E.M., Fiedler M., Nykytenko A., Mittag M. (2006). A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 142: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im C.-S., Eberhard S., Huang K., Beck C.F., Grossman A.R. (2006). Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 48: 1–16 [DOI] [PubMed] [Google Scholar]
- Immeln D., Pokorny R., Herman E., Moldt J., Batschauer A., Kottke T. (2010). Photoreaction of plant and DASH cryptochromes probed by infrared spectroscopy: The neutral radical state of flavoproteins. J. Phys. Chem. B 114: 17155–17161 [DOI] [PubMed] [Google Scholar]
- Immeln D., Schlesinger R., Heberle J., Kottke T. (2007). Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J. Biol. Chem. 282: 21720–21728 [DOI] [PubMed] [Google Scholar]
- Jiang X., Stern D. (2009). Mating and tetrad separation of Chlamydomonas reinhardtii for genetic analysis. J. Vis. Exp. 30: pii: 1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiaulehn S., Voytsekh O., Fuhrmann M., Mittag M. (2007). The presence of UG-repeat sequences in the 3′-UTRs of reporter luciferase mRNAs mediate circadian expression and can determine acrophase in Chlamydomonas reinhardtii. J. Biol. Rhythms 22: 275–277 [DOI] [PubMed] [Google Scholar]
- Kondo T., Johnson C.H., Hastings J.W. (1991). Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas. I. Cells in darkness. Plant Physiol. 95: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaki A., Naoi M., Yagi K. (1970). Effect of proton donors on the absorption spectrum of flavin compounds in apolar media. J. Biochem. 68: 287–292 [DOI] [PubMed] [Google Scholar]
- Kottke T., Batschauer A., Ahmad M., Heberle J. (2006). Blue-light-induced changes in Arabidopsis cryptochrome 1 probed by FTIR difference spectroscopy. Biochemistry 45: 2472–2479 [DOI] [PubMed] [Google Scholar]
- Kottke T., Heberle J., Hehn D., Dick B., Hegemann P. (2003). Phot-LOV1: Photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys. J. 84: 1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbacher T., Immeln D., Dick B., Kottke T. (2009). Microsecond light-induced proton transfer to flavin in the blue light sensor plant cryptochrome. J. Am. Chem. Soc. 131: 14274–14280 [DOI] [PubMed] [Google Scholar]
- Lanzl K., Sanden-Flohe M.V., Kutta R.J., Dick B. (2010). Photoreaction of mutated LOV photoreceptor domains from Chlamydomonas reinhardtii with aliphatic mercaptans: Implications for the mechanism of wild type LOV. Phys. Chem. Chem. Phys. 12: 6594–6604 [DOI] [PubMed] [Google Scholar]
- Lariguet P., Schepens I., Hodgson D., Pedmale U.V., Trevisan M., Kami C., de Carbonnel M., Alonso J.M., Ecker J.R., Liscum E., Fankhauser C. (2006). PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. USA 103: 10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Mittag M., Sczekan S., Morse D., Hastings J.W. (1993). Molecular cloning and genomic organization of a gene for luciferin-binding protein from the dinoflagellate Gonyaulax polyedra. J. Biol. Chem. 268: 8842–8850 [PubMed] [Google Scholar]
- Lewis Z.A., Correa A., Schwerdtfeger C., Link K.L., Xie X., Gomer R.H., Thomas T., Ebbole D.J., Bell-Pedersen D. (2002). Overexpression of White Collar-1 (WC-1) activates circadian clock-associated genes, but is not sufficient to induce most light-regulated gene expression in Neurospora crassa. Mol. Microbiol. 45: 917–931 [DOI] [PubMed] [Google Scholar]
- Li X., Wang Q., Yu X., Liu H., Yang H., Zhao C., Liu X., Tan C., Klejnot J., Zhong D., Lin C. (2011). Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc. Natl. Acad. Sci. USA 108: 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedvogel M., Maeda K., Henbest K., Schleicher E., Simon T., Timmel C.R., Hore P.J., Mouritsen H. (2007). Chemical magnetoreception: Bird cryptochrome 1a is excited by blue light and forms long-lived radical-pairs. PLoS ONE 2: e1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Robertson D.E., Ahmad M., Raibekas A.A., Jorns M.S., Dutton P.L., Cashmore A.R. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269: 968–970 [DOI] [PubMed] [Google Scholar]
- Liu B., Liu H., Zhong D., Lin C. (2010). Searching for a photocycle of the cryptochrome photoreceptors. Curr. Opin. Plant Biol. 13: 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Más P., Devlin P.F., Panda S., Kay S.A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211 [DOI] [PubMed] [Google Scholar]
- Massey V., Palmer G. (1966). On the existence of spectrally distinct classes of flavoprotein semiquinones. A new method for the quantitative production of flavoprotein semiquinones. Biochemistry 5: 3181–3189 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Ishiura M. (2011). Chlamydomonas reinhardtii as a new model system for studying the molecular basis of the circadian clock. FEBS Lett. 585: 1495–1502 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Okamoto K., Onai K., Niwa Y., Shimogawara K., Ishiura M. (2008). A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 22: 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag M., Kiaulehn S., Johnson C.H. (2005). The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 137: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Brüstlein M., Hemmerich P., Massey V., Walker W.H. (1972). Light-absorption studies on neutral flavin radicals. Eur. J. Biochem. 25: 573–580 [DOI] [PubMed] [Google Scholar]
- Mus F., Dubini A., Seibert M., Posewitz M.C., Grossman A.R. (2007). Anaerobic acclimation in Chlamydomonas reinhardtii: Anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Oldenhof H., Bisová K., van den Ende H., Zachleder V. (2004). Effect of red and blue light on the timing of cyclin-dependent kinase activity and the timing of cell division in Chlamydomonas reinhardtii. Plant Physiol. Biochem. 42: 341–348 [DOI] [PubMed] [Google Scholar]
- Ozturk N., Selby C.P., Song S.H., Ye R., Tan C., Kao Y.T., Zhong D., Sancar A. (2009). Comparative photochemistry of animal type 1 and type 4 cryptochromes. Biochemistry 48: 8585–8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorny R., Klar T., Essen L.O., Batschauer A. (2005). Crystallization and preliminary X-ray analysis of cryptochrome 3 from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorny R., Klar T., Hennecke U., Carell T., Batschauer A., Essen L.O. (2008). Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc. Natl. Acad. Sci. USA 105: 21023–21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W., Gonzalez-Ballester D., Grossman A.R. (2010). Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorph N.A., Small G.D. (2004). The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol. 134: 1546–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sancar A. (2003). Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103: 2203–2237 [DOI] [PubMed] [Google Scholar]
- Schmidt M., et al. (2006). Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18: 1908–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M. (2006). RNA silencing in Chlamydomonas: Mechanisms and tools. Curr. Genet. 49: 69–84 [DOI] [PubMed] [Google Scholar]
- Schulze T., Prager K., Dathe H., Kelm J., Kiessling P., Mittag M. (2010). How the green alga Chlamydomonas reinhardtii keeps time. Protoplasma 244: 3–14 [DOI] [PubMed] [Google Scholar]
- Seitz S.B., Weisheit W., Mittag M. (2010). Multiple roles and interaction factors of an E-box element in Chlamydomonas reinhardtii. Plant Physiol. 152: 2243–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C.P., Sancar A. (2006). A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 103: 17696–17700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano G., Herrera-Palau R., Romero J.M., Serrano A., Coupland G., Valverde F. (2009). Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr. Biol. 19: 359–368 [DOI] [PubMed] [Google Scholar]
- Siegel L.M. (1978). Quantitative determination of noncovalently bound flavins: Types and methods of analysis. Methods Enzymol. 53: 419–429 [DOI] [PubMed] [Google Scholar]
- Sineshchekov O.A., Jung K.H., Spudich J.L. (2002). Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 99: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Devlin P.F., Kay S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., Kay S.A., Rosbash M., Hall J.C. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Usman A., Brazard J., Martin M.M., Plaza P., Heijde M., Zabulon G., Bowler C. (2009). Spectroscopic characterization of a (6-4) photolyase from the green alga Ostreococcus tauri. J. Photochem. Photobiol. B 96: 38–48 [DOI] [PubMed] [Google Scholar]
- Veith T., Brauns J., Weisheit W., Mittag M., Büchel C. (2009). Identification of a specific fucoxanthin-chlorophyll protein in the light harvesting complex of photosystem I in the diatom Cyclotella meneghiniana. Biochim. Biophys. Acta 1787: 905–912 [DOI] [PubMed] [Google Scholar]
- Vieira J., et al. (2012). Human cryptochrome-1 confers light independent biological activity in transgenic Drosophila correlated with flavin radical stability. PLoS ONE 7: e31867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytsekh O., Seitz S.B., Iliev D., Mittag M. (2008). Both subunits of the circadian RNA-binding protein CHLAMY1 can integrate temperature information. Plant Physiol. 147: 2179–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.J., Zuo Z.C., Zhao X.Y., Li X., Klejnot J., Li Y., Chen P., Liang S.P., Yu X.H., Liu X.M., Lin C.T. (2008). Blue-light-independent activity of Arabidopsis cryptochromes in the regulation of steady-state levels of protein and mRNA expression. Mol Plant 1: 167–177 [DOI] [PubMed] [Google Scholar]
- Zhang T., Maruhnich S.A., Folta K.M. (2011). Green light induces shade avoidance symptoms. Plant Physiol. 157: 1528–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Sauman I., Yuan Q., Casselman A., Emery-Le M., Emery P., Reppert S.M. (2008). Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 6: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikihara K., Ishikawa T., Todo T., Tokutomi S. (2008). Involvement of electron transfer in the photoreaction of zebrafish cryptochrome-DASH. Photochem. Photobiol. 84: 1016–1023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.