This work identifies ZML2 and its homolog ZML1 as key regulators of gene expression in the cry1-dependent response to excess light. ZML1/2 bind to the CryR1 cis-element in vitro and in vivo, and T-DNA insertion lines for ZML2 and ZML1 were sensitive to excess light, demonstrating misregulation of several cry1-dependent genes in response to excess light.
Abstract
Exposure of plants to light intensities that exceed the electron utilization capacity of the chloroplast has a dramatic impact on nuclear gene expression. The photoreceptor Cryptochrome 1 (cry1) is essential to the induction of genes encoding photoprotective components in Arabidopsis thaliana. Bioinformatic analysis of the cry1 regulon revealed the putative cis-element CryR1 (GnTCKAG), and here we demonstrate an interaction between CryR1 and the zinc finger GATA-type transcription factors ZINC FINGER PROTEIN EXPRESSED IN INFLORESCENCE MERISTEM LIKE1 (ZML1) and ZML2. The ZML proteins specifically bind to the CryR1 cis-element as demonstrated in vitro and in vivo, and TCTAG was shown to constitute the core sequence required for ZML2 binding. In addition, ZML2 activated transcription of the yellow fluorescent protein reporter gene driven by the CryR1 cis-element in Arabidopsis leaf protoplasts. T-DNA insertion lines for ZML2 and its homolog ZML1 demonstrated misregulation of several cry1-dependent genes in response to excess light. Furthermore, the zml1 and zml2 T-DNA insertion lines displayed a high irradiance-sensitive phenotype with significant photoinactivation of photosystem II (PSII), indicated by reduced maximum quantum efficiency of PSII, and severe photobleaching. Thus, we identified the ZML2 and ZML1 GATA transcription factors as two essential components of the cry1-mediated photoprotective response.
INTRODUCTION
Plants can detect almost all facets of light, including direction, intensity, duration, and wavelength using three major classes of photoreceptors: the red/far-red light absorbing phytochromes, the blue/UV-A light absorbing cryptochromes and phototropins, and the UV-B sensing UV-B receptors. These photoreceptors perceive light signals and initiate intracellular signaling pathways and large-scale reorganization of the transcriptional program to modulate plant growth and development (Chen et al., 2004). Arabidopsis thaliana cryptochromes cry1 and cry2 are well characterized and regulate several aspects of growth and development (Li and Yang, 2007). The more divergent member of the family, cry3, is localized to the organelles and was reported to be involved in single-stranded DNA repair (Kleine et al., 2003; Pokorny et al., 2008). Cry2 undergoes degradation following light activation via ubiquitylation and targeting to the proteasome (Lin et al., 1998; Yu et al., 2007), whereas cry1 is stable in bright light (Ahmad and Cashmore, 1993). Thus, cry1 is responsible for seedling deetiolation in high fluence rates of blue light, and cry2 mediates flowering time and deetiolation in response to lower fluence rates (Ahmad and Cashmore, 1993; Lin et al., 1998). It has also been demonstrated that cry1 plays a critical role in the photoprotective response to excess light (Kleine et al., 2007). Cry1 is present in the nucleus, and although having considerable effects on transcriptional activity, cry1 is not known to bind DNA (Cashmore et al., 1999; Lin and Shalitin, 2003). Some of the cry1 action on the transcription level has been attributed to its interaction with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1). The cry1-COP1 interaction is light dependent and suppresses the activity of COP/DEETIOLATED proteins, thereby allowing light-activated transcription (Wang et al., 2001; Yang et al., 2001). Cry1 has also been shown to be present in the cytosol (Yang et al., 2000). It has been suggested that cry1 may be involved in both nuclear and cytosolic events, and it is possible that there are separate functions for nuclear and cytoplasmic cry1 (Wu and Spalding, 2007).
Genetic screens have revealed several regulators in the cryptochrome-mediated light signaling pathway; a blue light–specific Ser/Thr protein phosphatase (PP7) has been isolated (Møller et al., 2003), and SHORT UNDER BLUE1 is a cytoplasmic calcium binding protein with a major function in cryptochrome signaling but that also modulates phyA-mediated signaling (Guo et al., 2001). Cry1 inhibits COP1-mediated degradation of a MYB transcription factor, BIT1, to activate blue light–dependent gene expression (Duek and Fankhauser, 2003; Hong et al., 2008). Furthermore, HFR1, a basic helix-loop-helix protein identified as a component in the far-red signaling has been demonstrated to be crucial for regulating global gene expression in response to cry1-mediated early blue light response (Duek and Fankhauser, 2003). In addition, HYPERSENSITIVE TO RED AND BLUE1, SHORT HYPOCOTYL UNDER BLUE1, and OBF BINDING PROTEIN3 have been shown to act both in red and blue signaling pathways (Kang et al., 2005; Ward et al., 2005; Kang and Ni, 2006). Photoexcited cry2 was found to interact with CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX1 to regulate transcription and initiation of flowering (Liu et al., 2008). In addition, cry1 has been shown to interact directly with ZTL/LKP1/ADO1 in yeast two-hybrid assays and in vitro pull-down tests (Kiyosue and Wada, 2000; Somers et al., 2000; Jarillo et al., 2001). ZTL/LKP1/ADO1 is a protein that was originally identified in a study of the circadian clock in Arabidopsis, and ZTL/LKP1/ADO1 plays an important role in regulating the circadian clock and photoperiodic flowering in Arabidopsis (Somers et al., 2000). Thus, cryptochromes are involved in numerous aspects of plant growth and development.
Cry1 has also been assigned a role in plant stress responses, and analysis of the high irradiance response of the photoreceptor mutants phyA, phyB, cry1, and cry2 revealed a function of cry1 in mediating plant responses to high irradiances (Kleine et al., 2007). In Arabidopsis, cry1 regulates a large number of genes in response to excess light, and as a consequence of the misregulation of these genes the cry1 mutant displayed a high irradiance-sensitive phenotype with significant photoinactivation of photosystem II (PSII) (Kleine et al., 2007). Thus, in addition to a role in photomorphogenesis, cry1 is essential to the induction of photoprotective mechanisms against high light stress (Kleine et al., 2007; Li et al., 2009). Numerous array experiments have shown that exposure to excess light results in dramatic changes in gene expression in plants (Kimura et al., 2001; Rossel et al., 2002; Richly et al., 2003; Kleine et al., 2007). A significant number of these excess light–regulated genes depend on the action of cry1 for correct regulation and 26 of those genes were also LONG HYPOCOTYL5 (HY5) dependent (Kleine et al., 2007). The cry1-dependent genes encode components of the protective mechanisms against light stress, such as several enzymes in the phenylpropanoid pathway, EARLY LIGHT INDUCED PROTEIN1 (ELIP1) and ELIP2, GPX7 that encodes a putative glutathione peroxidase, the glutathione S-transferase ERD9, and a MYB family transcription factor (At5g49330). Bioinformatics studies performed on the HY5-independent cry1 regulon revealed that two previously undescribed cis-elements, CryR1 (GnTCkAG) and CryR2 (ACATAwCT), were enriched in the list of cry1-dependent genes responding to excess light (Kleine et al., 2007). CryR1 was significantly enriched in the promoters of genes induced by excess light in the wild type, suggesting interaction with an activator of gene expression (Kleine et al., 2007).
Although cry1 plays an essential role in mediating plant responses to high irradiances, HY5 is the only transcription factor responding to cry1 that has been identified to play a role under these conditions (Kleine et al., 2007). Thus, to discover nuclear components involved in the cry1-mediated signaling pathway in response to excess light, we used a biochemical approach and identified interaction between the GATA-type transcription factor ZINC FINGER PROTEIN EXPRESSED IN INFLORESCENCE MERISTEM LIKE2 (ZML2; also known as TIFY2a) (Vanholme et al., 2007) and the CryR1 cis-element. We confirmed binding and specificity of ZML2 and its homolog ZML1 (TIFY2b) to the CryR1 cis-element in vitro and in vivo. We also demonstrate a function of ZML2 as a trans-activation factor. T-DNA insertion lines for ZML2 and ZML1 demonstrated an excess light-sensitive phenotype, increased accumulation of reactive oxygen species (ROS), and misregulation of several cry1-dependent genes in response to excess light. Thus, we identified two key regulators, ZML2 and ZML1, of gene expression in the cry1-dependent response to excess light.
RESULTS
Identification of a Transcription Factor Binding to the Putative CryR1 cis-Element
To test whether a nuclear protein(s) recognizes the putative CryR1 cis-element, oligonucleotides corresponding to the CryR1 element (GAAAAAGTTCTAGAATTTTTT) were used in electrophoretic mobility shift assays (EMSAs). Nuclear protein extracts for EMSA were prepared from wild-type plants grown under control conditions. When nuclear protein extracts were incubated with biotin-labeled CryR1 oligonucleotides, a DNA-protein complex with slower mobility in EMSA was observed (see Supplemental Figure 1 online).
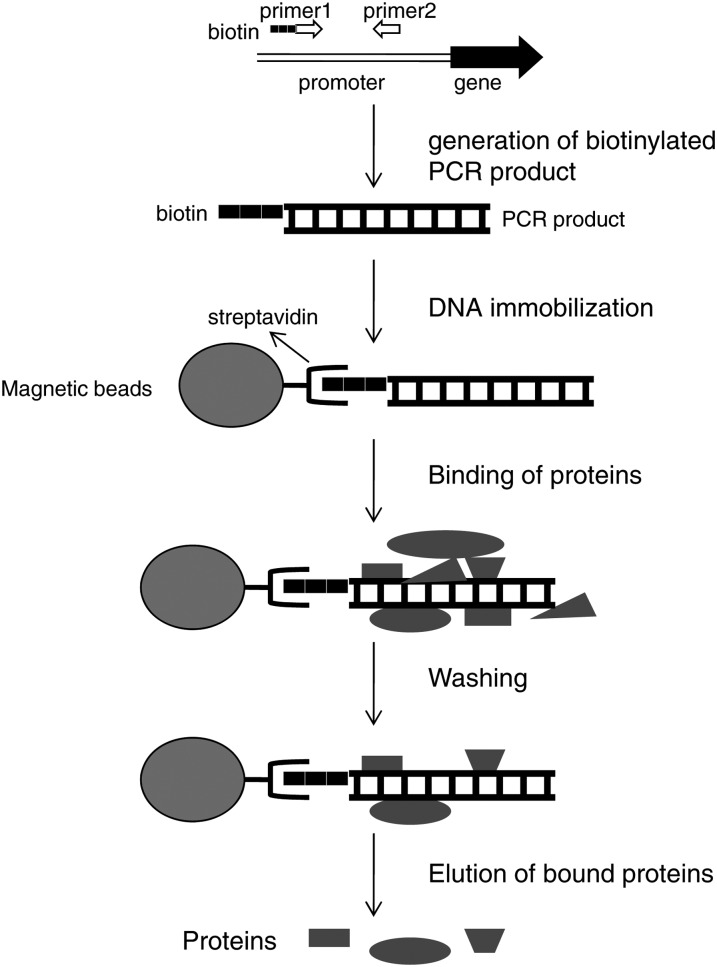
To identify the transcription factor(s) responding to the cry1-mediated signal and interacting with the putative cis-element CryR1, we used a biochemical approach where a DNA fragment containing the cis-element of interest was tagged with biotin and linked to microbeads via interaction with streptavidin (Figure 1) (Gabrielsen et al., 1989; Rey et al., 2003). The 218-bp DNA promoter fragment containing the CryR1 putative cis-element was amplified from the R2R3 transcription factor (At5g49330) (Figure 2) representing one of the genes responding to excess light in a cry1-dependent manner. MatInspector analysis for cis-elements within the 218-bp CryR1-containing promoter fragment revealed the presence of DOF, MADS box, GT-box, GAGA, L1-box, W-box, heat shock, homeobox, and AS1/AS2 elements (http://www.genomatix.de).
Figure 1.
Schematic Outline of the Method for Affinity Trapping of DNA Binding Proteins.
PCR amplification of a promoter region of interest where primer1 was modified with biotin. The biotinylated PCR product was immobilized on streptavidin-coated magnetic beads. Nuclear proteins from Arabidopsis were incubated with the DNA fragment linked to the beads to allow interaction between the protein(s) and the DNA. To eliminate binding of nonspecific proteins, several washing steps were performed. DNA binding proteins were eluted from the complex using high salt buffer.
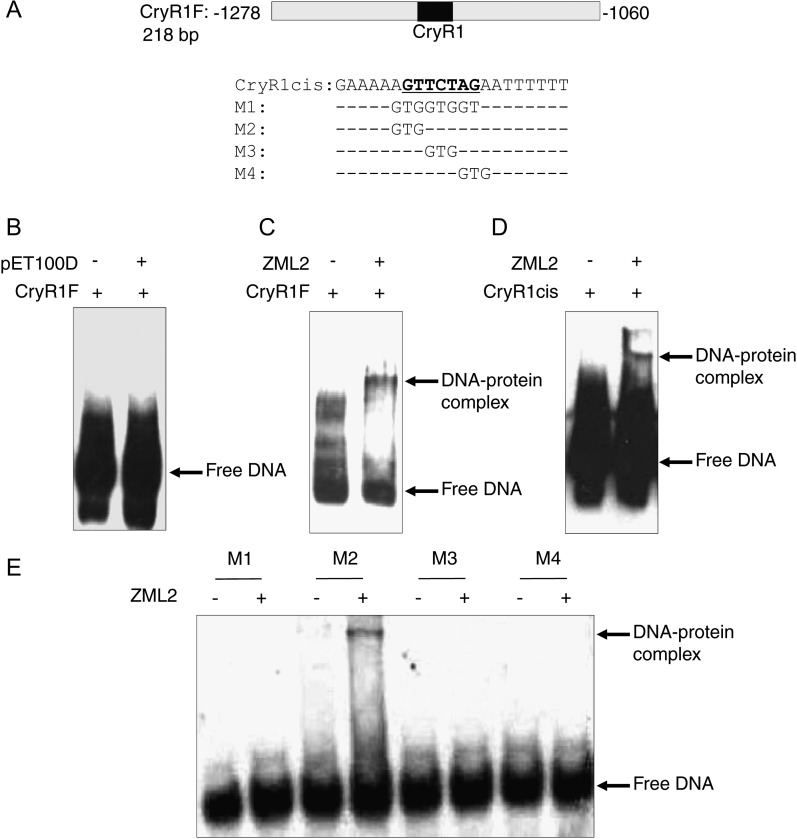
Figure 2.
Characterization of DNA Binding of Recombinant ZML2 Protein.
(A) Nucleotide sequence of the 218-bp fragment containing the CryR1 cis-element from the promoter of R2R3 family transcription factor and the upper strand sequence of the synthetic double-stranded oligonucleotide containing CryR1 cis-element or its mutagenized variants used as probes.
(B) EMSA of lysate from E. coli transformed with pET100D empty expression vector and the 218-bp CryR1 fragment.
(C) to (E) EMSA of recombinant ZML2 protein with 218-bp CryR1 fragment (C), CryR1 cis-element (D), and its mutagenized variants (E). Positions of free DNA probes and DNA-protein complexes are indicated by arrows.
Nuclear proteins were prepared from wild-type plants exposed to control conditions and to excess light. The two different sample types of nuclear proteins were incubated together with the DNA fragment containing the CryR1 cis-elements, and the proteins binding to the DNA fragment were isolated and identified using mass spectrometry. Using this approach, we identified an interaction between ZML2 (At1g51600) and the CryR1 element (Table 1). ZML2 was identified in both the control sample and the sample exposed to excess light (Table 1). The ZML2 protein belongs to the GATA-type zinc finger family, and ZIM was originally identified in Arabidopsis due to its pronounced expression in flowers and flower buds (Nishii et al., 2000). There are two ZIM homologs, ZML1 and ZML2, and these three proteins form a separate group when compared with the other typical GATA-type proteins (Teakle et al., 2002). The zinc finger domains of ZIM, ZML1, and ZML2 have 20 residues between the two Cys pairs (C-X2-C-X20-C-X2-C), while those of the other Arabidopsis GATA factors all have 18 residues (Shikata et al., 2004).
Table 1. ZML2 Was Identified to Interact with the CryR1 cis-Element.
| cis-Element | Protein | Samples in Which Peptide Was Identified | Peptide |
|---|---|---|---|
| CryR1 (GTTCTAG) | At1g51600 | Control light | YTVRKEVALR* |
| ZML2 transcription factor | Exposure to excess light |
Complete Mascot score result and ion spectra for the ZML2 identification in control sample are available in Supplemental Figure 9 online. The asterisk indicates that the same peptide was also identified in samples exposed to excess light.
EMSAs Confirmed Binding of ZML2 to the CryR1 cis-Element
EMSA was performed to confirm the interaction between the isolated protein and the DNA fragment used as bait in the biochemical approach. Full-length cDNA of ZML2 was expressed as a His-tagged protein in Escherichia coli, and the ability of the fusion protein to bind specific DNA fragments was examined (Figure 2A). Binding was not detected in the presence of control lysate from E. coli bacterial cells transformed with an empty expression vector, pET100D (Figure 2B). The ZML2 protein was demonstrated by EMSA to bind to the 218-bp DNA fragment containing the CryR1 cis-element used in the biochemical assay (Figure 2C). Furthermore, we demonstrated that ZML2 binds specifically to the CryR1 cis-element using a 21-bp DNA fragment containing the CryR1 element (Figure 2D). To determine the core sequence that is essential for ZML2 binding of the CryR1 cis-element, mutations were introduced into this element (Figure 2A) and in vitro binding to ZML2 was tested by EMSA. Changing the sequence of the CryR1 cis-element from GTTCTAG to TGGTGGT (M1) abolished DNA binding of the ZML2 protein to the CryR1 target completely. Similarly to M1, M3 in which TCT of the CryR1 element were mutated to GTG and M4 in which the nucleotides AGA were mutated to GTG resulted in loss of DNA binding (Figure 2E). By contrast, mutation of AGT to GTG (M2) did not abolish ZML2 binding (Figure 2E). Taken together, these results indicate that the CryR1 cis-element is the target motif for the ZML2 transcription factor and that the core sequence required for binding is TCTAG. Our results also validate the method used to trap DNA binding proteins. This biochemical approach was previously proven successful in bacterial and yeast systems (Gabrielsen et al., 1989; Rey et al., 2003), and our results demonstrate that it is possible to successfully apply this technique also on the more complex plant material.
Arabidopsis plants overexpressing ZIM (ZIM-ox) were shown to upregulate a significant number of genes compared with the wild type when grown under control conditions (Shikata et al., 2004). We analyzed 500 bp from the ATG site of the promoters of these genes and found that the CryR1 cis-element was overrepresented in the promoters of the genes upregulated in the ZIM-ox plants (see Supplemental Table 1 online). In 92% of the promoters, the CryR1 cis-element showed 71 to 100% identity to GTTCTAG, which is the sequence of the CryR1 cis-element in the promoter of R2R3, and the CryR1 cis-element occurred at least twice in 62% of the promoters (see Supplemental Table 1 online). This suggests that ZIM and ZMLs recognize and bind to the same cis-element.
Subcellular Localization of ZML2
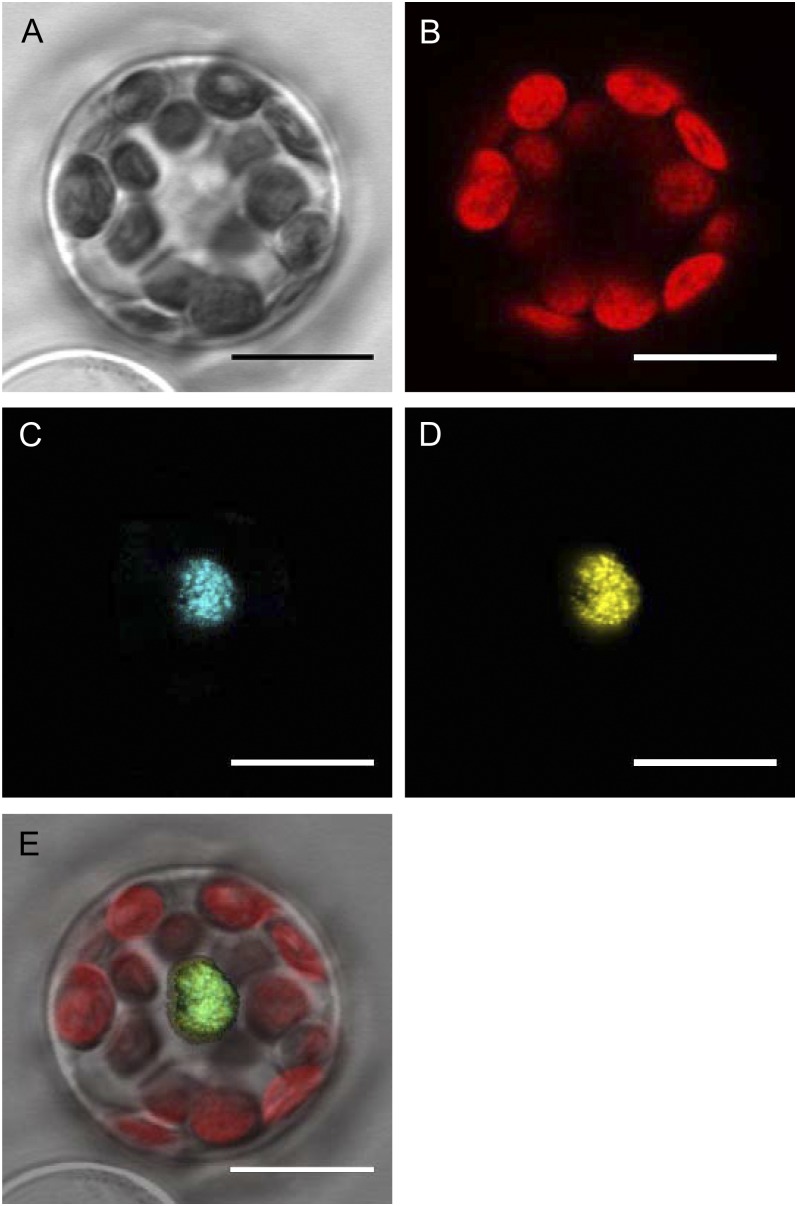
ZML2 was isolated as a nuclear protein binding to the CryR1 element, and it contains a basic residue-rich region indicating a putative NLS. The subcellular localization of ZML2 protein was investigated in transiently transfected protoplasts from Arabidopsis using confocal laser scanning microscopy. The ZML2:CFP (for cyan fluorescent protein) fusion protein was transfected into the protoplasts either separately or with the nuclear-localized bZIP transcription factor, ABI5 (Lopez-Molina et al., 2003; Shaikhali et al., 2008). Detection of ZML2:CFP and ABI5:YFP (for yellow fluorescent protein) fusion proteins demonstrated that both proteins were distributed exclusively to the nucleus (Figure 3). The fluorescence distribution of ZML2:CFP in the nucleus was consistent with expected nuclear localization of this protein and confirms that ZML2 is localized to the nucleus.
Figure 3.
Subcellular Localization of ZML2 Fusion Protein in Arabidopsis Protoplasts.
Confocal laser scanning microscope images of ZML2-CFP and ABI5-YFP coexpressed transiently in Arabidopsis protoplasts. Transmitted light image (A), chlorophyll fluorescence image (B), ZML2-CFP image (C), ABI5-YFP image (D), and merged image of (A) to (D) in (E). CFP signal is shown in blue, YFP signal is shown in yellow, and the chloroplast (chlorophyll) autofluorescence is shown in red. Bars = 10 μm.
Chromatin Immunoprecipitation Analysis Reveals in Vivo Binding of ZML2 to the CryR1 Element
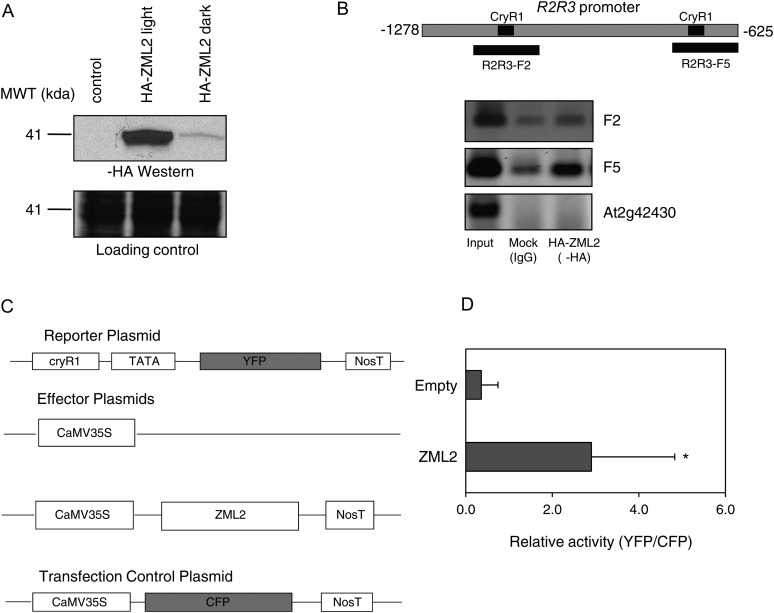
To investigate if ZML2 is also able to bind the CryR1 element in vivo, chromatin immunoprecipitation (ChIP) assays were performed using Arabidopsis protoplasts (Lee et al., 2007). The fusion protein HA-ZML2 was expressed in Arabidopsis protoplasts under dark and light conditions (Figure 4A). Interestingly, the level of ZML2 was much higher in light compared with the dark-grown samples. For ChIP analysis, Arabidopsis protoplasts were transfected with HA-ZML2 fusion protein or a mock control and incubated in light for 16 to 24 h. At the end of the incubation time, chromatin complexes were cross-linked with formaldehyde and the chromatin was fragmented by sonication and incubated with anti-HA monoclonal antibody. The immunoprecipitated complexes were captured with protein G-coated magnetic beads. PCR analysis performed on the DNA eluted from the beads revealed that R2R3 promoter fragments containing CryR1 element, F2 and F5, were enriched in the ChIP samples prepared with the anti-HA compared with the mock control (Figure 4B). By contrast, no PCR product was amplified using primers designed for the lateral root-specific promoter (At2g42430), indicating that these sequences were absent from the immunoprecipitated chromatin (Figure 4B). Taken together, the ChIP assay indicates that ZML2 specifically binds the R2R3 promoter in vivo and most likely to the CryR1 cis-element.
Figure 4.
Binding of ZML2 to CryR1 in Vivo.
(A) Transient expression of HA-ZML2 fusion protein in protoplasts from Arabidopsis cell culture under light (lane 2) and dark (lane 3) conditions. MWT, molecular weight.
(B) ChIP assay using Arabidopsis protoplasts expressing HA-ZML2 fusion protein was performed with anti-HA antibody. The positions of the CryR1 elements identified in the R2R3 promoter and the fragments analyzed by PCR are represented in the scheme. Genomic DNA obtained from ChIP was analyzed by PCR. Immunoprecipitation with IgG antibody was used as a control. Primers for F2 and F5 of the R2R3 promoter and primers for lateral root-specific promoter (At2g42430) were used for PCR amplification. Input, total input chromatin DNA; HA, DNA precipitated using HA antibody; IgG, DNA precipitated using IgG antibody.
(C) Schematic diagram of the effector and reporter constructs used in the transactivation assay. The effector plasmid carried the ZML2 cDNA driven by CaMV 35S promoter. NosT represents the polyadenylation site of nopaline synthetase gene. In the reporter plasmid, the YFP gene was linked to the 218-bp CryR1 fragment and a 35S minimal TATA promoter. As a negative control, an empty effector plasmid was used, and as a control for transfection efficiency, CaMV35S:CFP was cotransfected in each experiment.
(D) CryR1 reporter construct was transfected into protoplasts from Arabidopsis leaves with ZML2 effector construct or 35S empty control vector. To normalize the transfection efficiency, 35S:CFP:NosT was cotransfected in each experiment. Bars represent means of three independent experiments (±sd) with five protoplasts in each experiment. The asterisk indicates significant difference according to the Student’s t test (P < 0.05).
Transactivation Assays Demonstrated That ZML2 Activates Gene Expression via the CryR1 cis-Element
We demonstrated that ZML2 interacts with the CryR1 element in vitro and in vivo by EMSA and ChIP, respectively. CryR1 was significantly enriched in the promoters of genes induced by excess light in the wild type, suggesting interaction with an activator of gene expression (Kleine et al., 2007). To determine whether the ZML2 protein could transactivate transcription, we performed a transactivation assay using protoplasts prepared from Arabidopsis leaves. The protoplasts were cotransfected with an effector plasmid and a reporter plasmid (Figure 4C). The effector plasmid carried the ZML2 cDNA driven by cauliflower mosaic virus (CaMV) 35S promoter, and in the reporter plasmid, the YFP gene was linked to the 218-bp fragment of the R2R3 promoter containing CryR1 (Figure 4C). An empty effector plasmid was used as a negative control. A vector containing the CaMV35S:CFP was cotransfected in each experiment as a control for transfection efficiency (Figure 4C). The ZML2 protein transactivated expression of the YFP reporter gene in Arabidopsis protoplasts. The YFP expression was eightfold higher compared with the empty vector control (Figure 4D). Thus, the putative CryR1 cis-element identified using bioinformatics (Kleine et al., 2007) was demonstrated to be biologically active. Furthermore, ZML2 functions as a transcriptional activator in the cry1-mediated response to excess light, which is in agreement with the bioinformatic analysis that demonstrated that CryR1 was significantly enriched in the promoters of genes induced by excess light in a cry1-dependent manner.
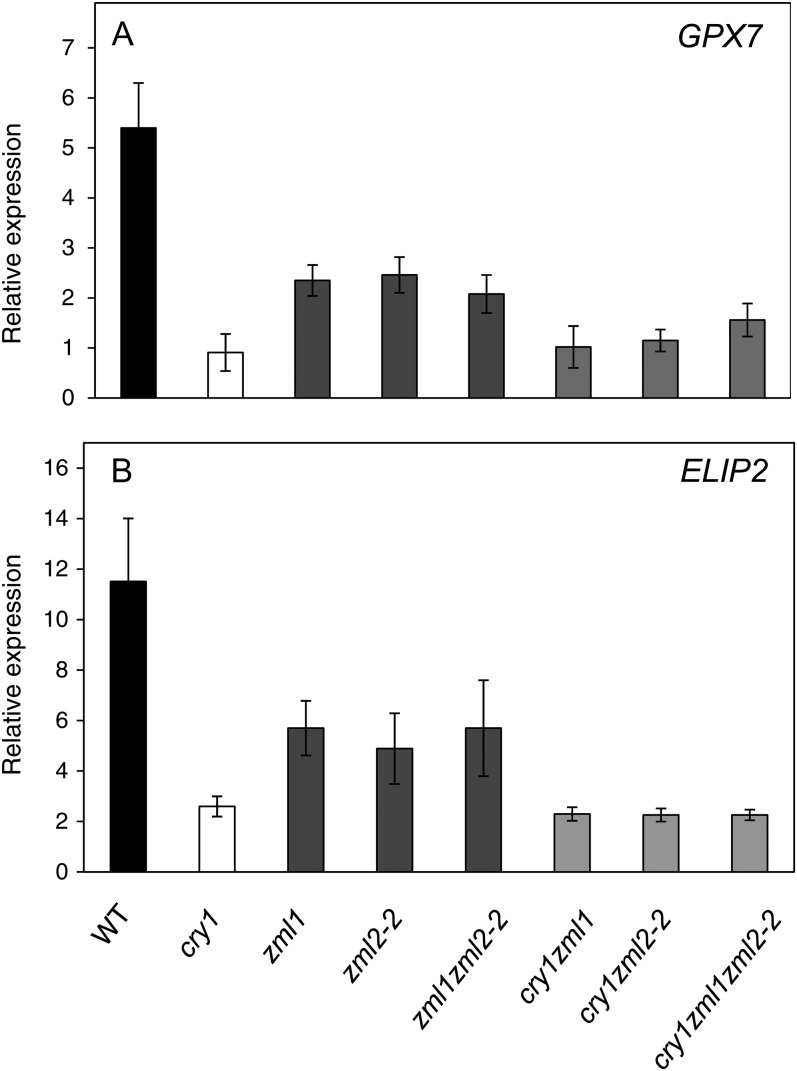
The zml Mutants Showed Impaired Regulation of Genes in the cry1 Regulon
To demonstrate whether the identified interaction between ZML2 and the CryR1 cis-element was biologically significant, we obtained T-DNA insertion lines for ZML2; zml2-1 and zml2-2, and ZML1; zml1 and ZIM; zim. The presence of the T-DNA insertions was confirmed by PCR (see Supplemental Figure 2 online). RNA was isolated from homozygous plants, and quantitative RT-PCR analysis confirmed the absence of ZML2, ZML1, and ZIM transcripts in the T-DNA insertion lines (see Supplemental Figure 2 online). Under normal growth conditions, no obvious phenotype could be observed in the zml2, zml1, and zim mutants. The expression of ZML1 and ZIM in the zml2 mutant and vice versa was not altered compared with the wild type or the cry1 mutant (see Supplemental Figure 3 online). Furthermore, no significant change in expression of the ZML1/2 or ZIM could be detected following exposure to excess light in the wild type or in the mutant lines (see Supplemental Figure 3 online).
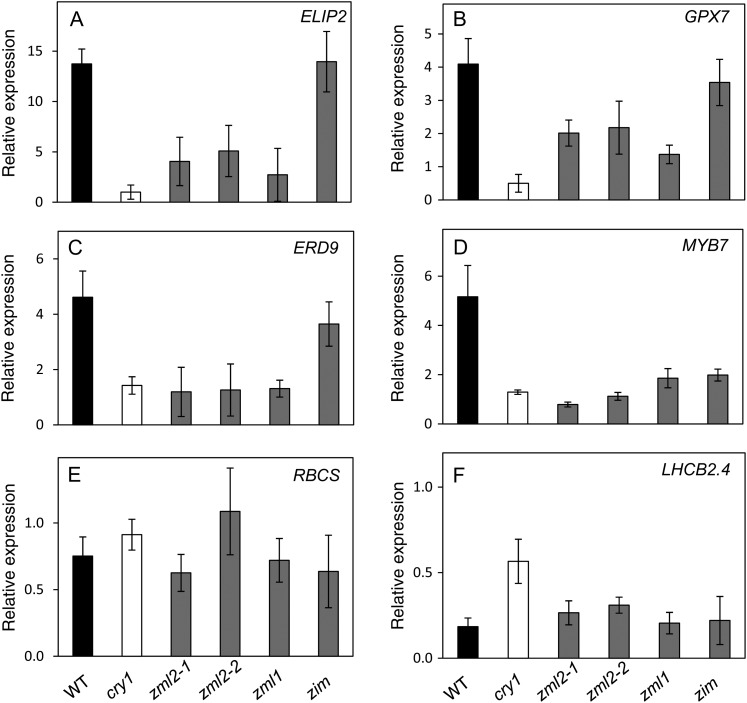
Expression of four marker genes in the cry1-dependent regulon containing the CryR1 cis-element in their promoters and two marker genes in the cry1-independent regulon (Kleine et al., 2007) was investigated in the zml2, zml1, and zim mutants and compared with the wild type and the cry1 mutant (Figure 5). Under normal growth conditions, the expression of the cry1-dependent genes ELIP2, GPX7, and ERD9 was not significantly altered in any of the mutants compared with the wild type (see Supplemental Figure 4 online). Expression of a gene encoding a MYB family transcription factor, MYB7, was significantly repressed in the zim mutant but demonstrated wild-type levels in the other mutants when grown at control conditions (see Supplemental Figure 4 online).
Figure 5.
Expression of Genes in the cry1 Regulon in the cry1, zml2-1, zml2-2, zml1, and zim Mutant Lines after Exposure to High Light.
Wild-type and the different mutant seedlings were grown for 10 d in continuous white light (100 μmol quanta m−2 s−1) at 23°C and shifted to excess light (3 h 1000 μmol quanta m−2 s−1). Expression of genes from the cry1-dependent regulon ELIP2 (At4g14690) (A), GPX7 (At4g31870) (B), ERD9 (At1g10370) (C), and MYB7 (At1g56650) (D) as well as the cry1-independent regulon RBCS (At1g67090) (E) and LHCB2.4 (At3g27690) (F) were analyzed using real-time PCR. The expression following exposure to high light was related to the control of each genotype. The results were normalized to the expression level of At4g36800 encoding a ubiquitin-protein ligase-like protein. The mean (±sd) of at least three biological replicates is shown. WT, the wild type.
When exposed to excess white light (1000 µmol m−2 s−1) for 3 h, the expression of ELIP2 was induced 15-fold compared with control conditions in the wild type and the zim mutant. The cry1, zml2, and zml1 mutants demonstrated significantly impaired induction of ELIP2 expression in response to excess light (Figure 5A). Expression of GPX7 and ERD9 was induced severalfold in response to excess light in the wild type and the zim mutant, whereas the zml2 and zml1 mutant lines demonstrated strongly attenuated induction of GPX7 and ERD9 similar to the cry1 mutant (Figures 5B and 5C). Expression of MYB7 was induced fivefold in the wild type in response to excess light (Figure 5D), whereas all of the mutants demonstrated inability to induce expression of MYB7 in response to excess light (Figure 5D). Expression of the cry1-independent genes encoding ribulose bisphosphate carboxylase small chain (RBCS) and the light-harvesting protein (LHCB2.4) in response to excess light revealed similar expression patterns in the wild type and the cry1, zml2, zml1, and zim mutants (Figures 5E and 5F). Under control conditions, the expression of RBCS and LHCB2.4 was not significantly altered in any of the mutants compared with the wild type (see Supplemental Figure 4 online). The misregulation of cry1-dependent marker genes in both the zml2 and zml1 mutants suggests that the ZML1 and ZML2 transcription factors act in concert and are significant components of the cry1-mediated response to excess light.
The zml Mutants Displayed Impaired Response to Excess Light
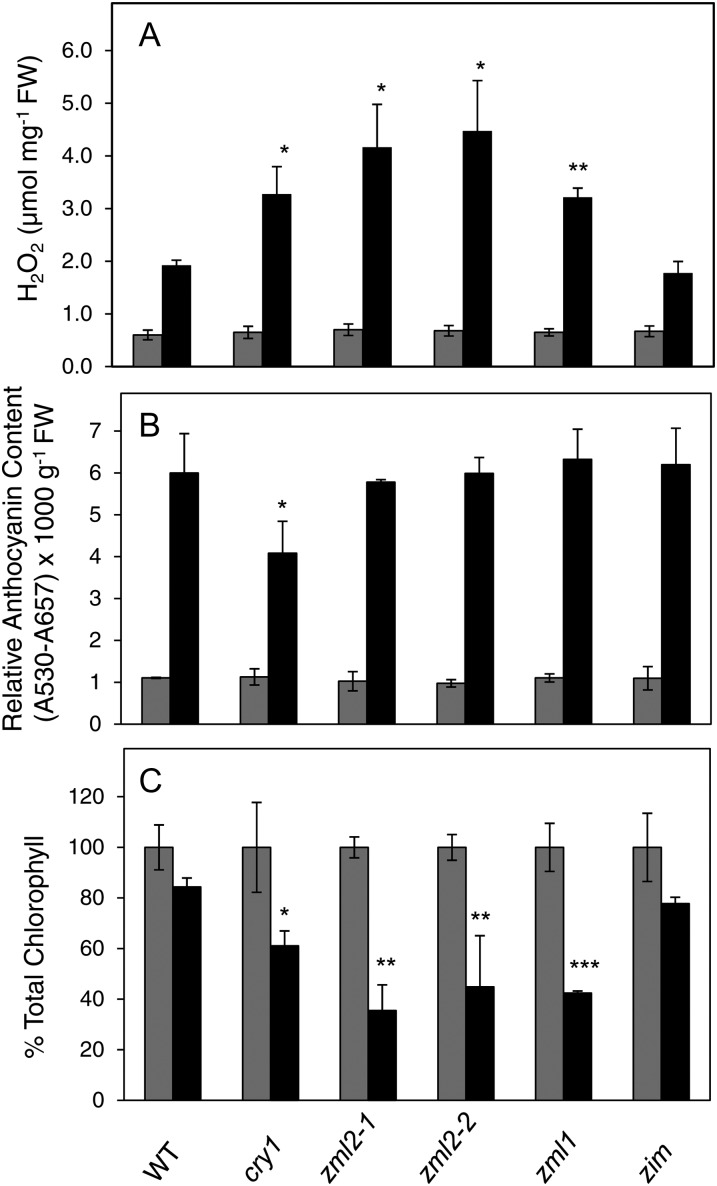
Several ROS, such as hydrogen peroxide (H2O2), singlet oxygen, superoxide, and hydroxyl radicals, are generated in chloroplasts under exposure to excess light (Asada, 2006). Superoxide is often the first reduced form of oxygen to be generated in plant tissues leading to the subsequent formation of H2O2 and hydroxyl radical. We examined the distribution of superoxide with nitro blue tetrazolium (NBT) staining in 2-week-old Arabidopsis plants of the wild type, cry1, zml2-1, zml2-2, zml1, and zim mutants exposed for 24 h to excess light (1000 µmol m−2 s−1). A clear NBT staining was observed following exposure to excess light, but no difference in superoxide accumulation and NBT staining could be detected between the wild type and the different mutants (see Supplemental Figure 5 online). In addition, we determined H2O2 accumulation in 2-week-old Arabidopsis plants of the wild type, cry1, zml2-1, zml2-2, zml1, and zim mutants following 24 h exposure to excess light (1000 µmol m−2 s−1). When exposed to excess light, a two- to threefold increase in H2O2 content was observed in the wild type and the zim mutant (Figure 6A). The cry1, zml2, and zml1 mutants accumulated significantly more H2O2 compared with the wild type following exposure to excess light (Figure 6A).
Figure 6.
Physiological Characterization of zml2, zml1, and zim T-DNA Insertion Lines in Response to Excess Light.
(A) H2O2 accumulation in control and 24 h high light–exposed 2-week-old seedlings of the wild type, cry1, zml2-1, zml2-2, zml1, and zim mutants. The mean (±sd) of three biological replicates is shown. FW, fresh weight.
(B) Relative anthocyanin content in control and 48 h high light–exposed 10-d-old seedlings of the wild type, cry1, zml2-1, zml2-2, zml1, and zim mutants. The mean (±sd) of three biological replicates is shown.
(C) Chlorophyll content relative to the control of 48 h high light–exposed 10-d-old seedlings of the wild type, cry1, zml2-1, zml2-2, zml1, and zim mutants. The mean (±sd) of three biological replicates is shown. Significant differences compared with the wild type were assessed according to Student’s t test: *P < 0.05, **P < 0.01, and ***P < 0.001. Gray bars and black bars represent control and high light samples, respectively. WT, the wild type.
In the wild type, anthocyanin accumulation increased sixfold after 48 h exposure to 1000 µmol m−2 s−1 excess light (Figure 6B). In the cry1 mutant, the anthocyanin accumulation was less compared with the wild type (Figure 6B) (Kleine et al., 2007). Reduced anthocyanin levels in cry1-deficient Arabidopsis seedlings have been shown in continuous blue light (Ahmad et al., 1995; Lin et al., 1996) and in continuous white light (Neff and Chory, 1998). In contrast with the cry1 mutant, the zml mutants accumulated wild-type levels of anthocyanins in response to excess light (Figure 6B). The anthocyanin accumulation pattern in response to excess light in mature plants (see Supplemental Figure 6 online) was similar to that observed in seedlings (Figure 6B). The cry1-mediated accumulation of anthocyanin was shown to be mediated via HY5 (Kleine et al., 2007) and is independent of ZML1 and ZML2. The CryR1 element was identified in the HY5-independent regulon of the cry1-regulated genes.
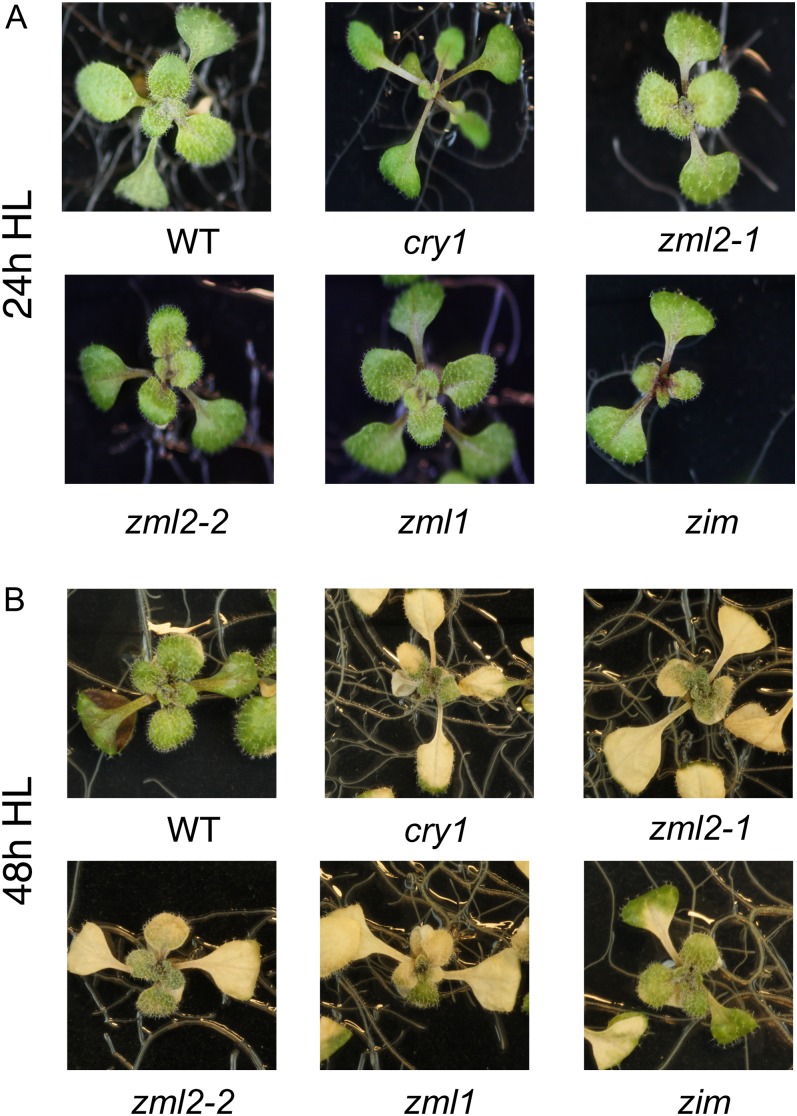
Exposure to excess light resulted in photoinactivation of PSII as demonstrated by a drop in maximum quantum efficiency of PSII (Fv/Fm) from 0.836 to 0.639 after 12 h in the wild type (Table 2). The cry1 mutant was almost twice as sensitive to excess light exposure as the wild type, shown by a drop in Fv/Fm from 0.837 to 0.374 (Table 2). The zml2 and zml1 mutant lines also demonstrated a significant increased sensitivity to excess light, measured as lower Fv/Fm compared with the wild type and the zim mutant (Table 2). Exposure to excess light for 48 h resulted in lower chlorophyll content in wild-type and zim seedlings (Figure 6C). However, the chlorophyll content was significantly lower in cry1, zml1, and zml2 mutants compared with the wild type (Figure 6C). Furthermore, 10-d-old seedlings of the cry1, zml2, and zml1 mutants demonstrated severe photobleaching following exposure to excess light (Figure 7). A similar effect was observed with mature plants (see Supplemental Figure 6 online). Thus, the high light–sensitive phenotype in the zml mutants demonstrated by the drop in Fv/Fm, the enhanced photobleaching, and the higher accumulation of ROS coincides with the misregulation of GPX7 and ERD9, encoding components involved in the photoprotective response (Kleine et al., 2007). In summary, these results support the conclusion that the ZML transcription factors are significant components of the cry1-mediated photoprotective response.
Table 2. Maximum Quantum Efficiency of PSII (Fv/Fm).
| Genotype | Control Fv/Fm | 12-h High-Light Fv/Fm |
|---|---|---|
| Wild type | 0.836 ± 0.002 | 0.693 ± 0.083 |
| cry1 | 0.837 ± 0.005 | 0.374 ± 0.065* |
| zml2-1 | 0.835 ± 0.003 | 0.569 ± 0.025* |
| zml2-2 | 0.821 ± 0.002 | 0.556 ± 0.033* |
| zml1 | 0.836 ± 0.004 | 0.397 ± 0.109* |
| zim | 0.837 ± 0.001 | 0.657 ± 0.027 |
Fv/Fm was estimated before and after exposure to 12 h of white light at 1000 μmol of photons m−2 s−1 in 5-week-old plants of the wild type and cry1, zml2-1, zml2-2, zml1, and zim mutants. Measurements were made in humidified air. Each point is the mean ± sd of nine leaves from at least three individual plants after 30 min of dark acclimation. The Fv/Fm was significantly different from the wild type in the cry1, zml2, and zml1 mutants following high light exposure (asterisk) according to the Student’s t test (P < 0.05).
Figure 7.
The zml2, zml1, and zim T-DNA Insertion Lines Exposed to Excess Light.
Ten-day-old seedlings of the wild type, cry1 mutant, and the zml2-1, zml2-2, zml1, and zim T-DNA insertion lines following 24 h (A) and 48 h (B) exposure to high light (HL; 1000 μmol quanta m−2 s−1). The severe photobleached phenotype observed in the zml2-1, zml2-2, and zml1 was reproduced in a second independent experiment. WT, the wild type.
ZML2 and ZML1 Physically Interact in Vivo in the Yeast Two-Hybrid System and in a Coimmunoprecipitation Assay
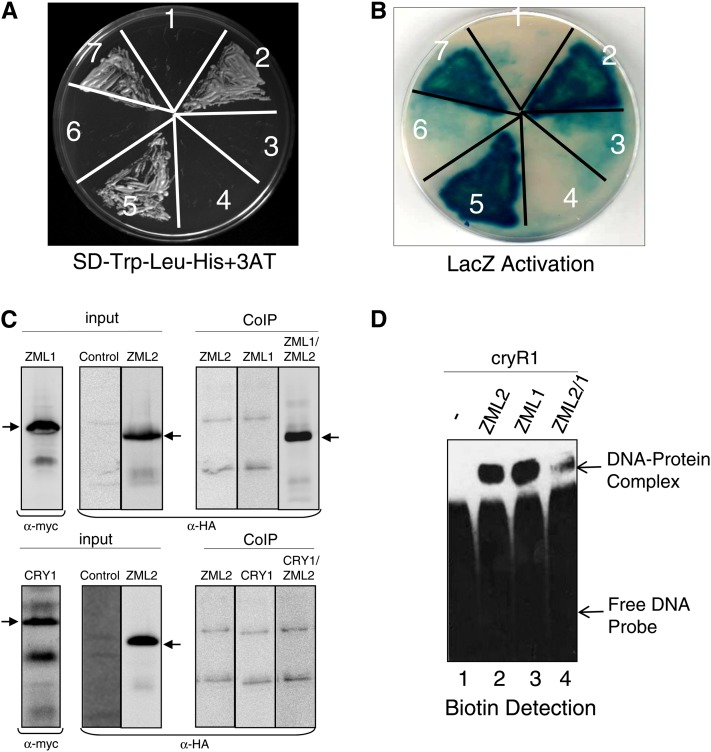
The misregulation of cry1-dependent genes in both the zml2 and zml1 mutants suggests that there is no redundancy and that ZML1 and ZML2 act in concert in the cry1-mediated response to excess light. To determine if there is a direct physical interaction between ZML2 and ZML1, a yeast two-hybrid assay was performed. A fusion protein of the GAL4 DNA binding domain and ZML2 (ZML2 bait) activated the transcription of the HIS3 reporter gene in the presence of GAL4 activation domain and ZML1 (ZML1-prey) (Figure 8A) but not with GAL4 activation domain only (empty prey) (Figure 8A), suggesting a direct protein–protein interaction between ZML2 and ZML1 in yeast cells. ZML1 not only heterodimerizes with ZML2 but also homodimerizes with itself when ZML1 bait was cotransformed with ZML1 prey (Figure 8A) but not with an empty prey. The HIS3 reporter gene activation assay was performed in the presence of 7.5 mM 3-aminotriazole (3-AT), a competitive inhibitor of the HIS3 gene. To confirm the observed interactions further, the interaction was also monitored through LacZ reporter activation, and strong activation was observed for the combinations ZML2 bait and ZML1 prey, and ZML1 bait and ZML1 prey (Figure 8B). Whereas it was clear that ZML2 interacted with ZML1, the C-terminal domain of CRY1 (CRY1-CT) was unable to interact with ZML2 or ZML1 (Figures 8A and 8B). Thus, no direct physical interaction between ZML1/2 and the C terminus of CRY1 could be detected in yeast. We also confirmed the interaction between ZML1 and ZML2 using an in vivo coimmunoprecipitation (Co-IP) assay. HA-ZML2, cMYC-ZML1, and cMYC-CRY1 fusion proteins were expressed in Arabidopsis protoplasts, and Co-IP was performed using anti-cMYC monoclonal antibody bound to protein G–coated magnetic beads. Input and Co-IP fractions were detected by immunoblot analysis with anti-cMYC and horseradish peroxidase (HRP)–conjugated anti-HA monoclonal antibodies, respectively (Figure 8C). From the in vivo Co-IP, it is clear that ZML1 and ZML2 can form heterodimers but that no direct interaction can be detected between ZML2 and CRY1 (Figure 8C).
Figure 8.
Heterodimerization of ZML2 and ZML1.
(A) and (B) The C-terminal domain of CRY1 protein does not interact with ZML2 and ZML1 proteins (3 and 6), whereas ZML2 and ZML1 homo- and heterodimerize in the yeast two-hybrid system as indicated by the activation of HIS3 reporter gene (A) and by the LacZ activation (B). Growth due to the activation of HIS3 reporter gene was examined in the presence of 3-AT. NMY51 yeast strain was cotransformed with ZML2 bait and empty prey vector (negative control) (1), ZML2 bait and ZML1 prey (2), ZML2 bait and CRY1-CT prey (3), ZML1 bait and empty prey vector (negative control) (4), ZML1 bait and ZML1 prey (5), ZML1 bait and CRY1-CT prey (6), and p53 bait and Large T prey (positive control) (7).
(C) Interaction was observed between ZML2 and ZML1, but not ZML2 and CRY1, using an in vivo Co-IP assay. HA-ZML2, cMYC-ZML1, and cMYC-CRY1 fusion proteins were expressed in Arabidopsis protoplasts, and Co-IP was performed using anti-cMYC monoclonal antibody bound to protein G–coated magnetic beads. Input and Co-IP fractions were detected by immunoblot analysis with anti-cMYC and HRP-conjugated anti-HA monoclonal antibodies, respectively. The arrows indicate the position of the respective fusion proteins.
(D) EMSA testing DNA binding activity of ZML1 and heterodimerization with ZML2. Signals were detected using a chemiluminescent nucleic acid detection module. Lane 1, free DNA probe; lane 2, ZML2 protein incubated with DNA probe; lane 3, ZML1 protein incubated with DNA probe; lane 4, ZML2 and ZML1 proteins with DNA probe. The migration positions of the free DNA probe (bottom band) and the DNA-protein complexes (top band) are marked by arrows.
[See online article for color version of this figure.]
The strong similarity between the DNA binding domains of ZML2 and ZML1 suggests that both proteins could recognize and interact with the CryR1 binding site. To confirm this hypothesis, DNA binding was tested by EMSA (Figure 8D). Similarly to recombinant ZML2 (Figure 8D), recombinant ZML1 protein was able to bind the CryR1 element (Figure 8D). It is not surprising that the position of the protein-DNA complexes was the same since the size of ZML2 and ZML1 recombinant proteins is almost the same (∼37 kD); therefore, the size of the protein-DNA complexes formed should be similar for both proteins. To confirm the formation of a heterodimer complex in vitro, ZML2 and ZML1 recombinant proteins were incubated together at room temperature prior to the binding reaction containing the CryR1 DNA probe. A shifted band was detected at the same position as that for the DNA-protein complexes formed with the single proteins ZML2 and ZML1, respectively. However, the intensity of the shifted band was significantly reduced, suggesting formation of second protein-DNA complex of high molecular weight that was unable to enter the native PAGE due to size restrictions (Figure 8D, lane 4). Thus, formation of a ZML2/ZML1 heterodimer complex is most likely occurring in vitro and the heterodimer also binds to the CryR1 element.
Analysis of Double and Triple Mutants
To test the interaction between cry1 and ZML1/2 genetically, double and triple mutants were generated for the different genotypes. Expression of two marker genes, ELIP2 and GPX7, in the cry1-dependent regulon was investigated in the zml2-2 zml1, cry1 zml1, and cry1 zml2-2 double mutants and cry1 zml1 zml2-2 triple mutant and compared with the wild type and the single mutants (Figure 9). The ELIP2 and GPX7 expression following exposure to excess light was similar in the zml2-2 zml1 double mutant compared with the respective zml2-2 and zml1 single mutants (Figure 9). Thus, no enhanced suppression of expression could be found in the double mutant compared with the single mutants, supporting the suggestion that ZML1 and ZML2 are genetically linked and act in concert in the cry1-mediated response to excess light. In addition, expression of ELIP2 and GPX7 following exposure to excess light in the cry1 zml1 and cry1 zml2-2 double mutants and the cry1 zml1 zml2-2 triple mutant was similar to the expression observed in the cry1 single mutant (Figure 9). Thus, no significant enhanced suppression of expression could be found in the double and triple mutants compared with the cry1 single mutants, demonstrating that ZML1/2 and cry1 are components in the same signaling pathway. Furthermore, it is clear that induction of ELIP2 and GPX7 expression in response to excess light is controlled by the cry1-ZML1/2 pathway.
Figure 9.
Double and Triple Mutant Analysis in Response to Excess Light.
Wild-type (WT) and the different mutant seedlings were grown for 10 d in continuous white light (100 μmol quanta m−2 s−1) at 23°C and shifted to excess light (3 h 1000 μmol quanta m−2 s−1). Expression of genes from the cry1-dependent regulon GPX7 (A) and ELIP2 (B) was analyzed using real-time PCR. The expression following exposure to high light was related to the control of each genotype. The results were normalized to the expression level of At4g36800 encoding a ubiquitin-protein ligase-like protein. The mean (±sd) of nine biological replicates is shown.
DISCUSSION
The CryR1 cis-element was identified through bioinformatic analysis and was suggested to be a putative cis-element involved in the cry1-dependent response to excess light (Kleine et al., 2007). We identified an interaction between the CryR1 cis-element and the GATA-type zinc finger protein, ZML2 (Table 1), using a biochemical approach previously proven successful in bacterial and yeast systems (Gabrielsen et al., 1989; Rey et al., 2003). ZML2 was demonstrated to be localized exclusively to the nucleus (Figure 3) and to act as a transcriptional activator of the YFP reporter gene driven by the CryR1 cis-element in Arabidopsis leaf protoplasts (Figure 4). Thus, the putative cis-element CryR1 was demonstrated to be biologically active, and an interplay between cry1, the CryR1 binding transcription factor ZML2, and its homolog ZML1 is essential for the induction of genes encoding photoprotective components in response to excess light in Arabidopsis.
ZIM, ZML2, and ZML1 belong to a group of atypical plant-specific GATA factors. The plant GATA zinc finger type transcription factors have been divided into subfamilies I, II, III, and IV, where group III is also known as the atypical GATA zinc finger type (Shikata et al., 2004; Manfield et al., 2007). The GATA factors were first identified as proteins that interacted with GATA motif (WGATAR) in vertebrates (Martin et al., 1989). In fungi, the GATA (GATAAGG) motif has been identified as the binding site for AREA (Kudla et al., 1990), and the sequence motif ATGATAAGG was found to be present in the promoter of many LHC and RBCS genes from different plant species (Dean et al., 1985; Grob and Stüber, 1987). In vitro binding studies with GATA1, GATA2, GATA3, and GATA4 from subfamily I, which represents the majority the plant GATA factors, demonstrated specificity of these proteins for the GATA motif (Teakle et al., 2002; Jeong and Shih, 2003). However, to the best of our knowledge, no previous report is available on the binding site of the atypical GATA transcription factors. Here, we demonstrated that ZML1 and ZML2 specifically bind to the CryR1 cis-element in vitro (Figures 2 and 8). In addition, the bases TCTAG of the CryR1 cis-element were demonstrated to be the core sequence required for ZML2 binding (Figure 2E). In a ChIP assay, we demonstrated that ZML2 pulled down two fragments from the R2R3 promoter both containing the CryR1 element (Figure 4B). Furthermore, using transient expression in Arabidopsis leaf protoplasts, we demonstrated that ZML2 transactivated the R2R3 promoter fragment containing CryR1 cis-element (Figure 4D). Thus, in agreement with the indications from the bioinformatic analysis where the CryR1 cis-element was enriched in the promoters of genes induced by excess light in the wild type, we could demonstrate that the ZML2 protein acts as transcriptional activator. An acidic region in the N terminus of the ZIM protein has been demonstrated to play a role as a transcriptional activation domain in transactivation experiments using cultured tobacco (Nicotiana tabacum) Bright Yellow-2 cells (Shikata et al., 2003). The entire amino terminal region (76 amino acids) of ZML2 shares 44% similarity with ZIM. Although the sequence itself is not highly conserved, the ZML2 amino terminal region is rich in acidic amino acids, suggesting a similar role for this region. ZIM was overexpressed using the CaMV 35S promoter in Arabidopsis, and the ZIM-ox plants demonstrated elongated hypocotyls and petioles and an upward positioning of the leaves (Shikata et al., 2004). The phenotype was observed under all wavelengths of light and in the presence of inhibitors of brassinosteroid and gibberellin biosynthesis (Shikata et al., 2004). A significant number of genes were shown to be upregulated in the ZIM-ox plants, and in 92% of the promoters of those genes, one or two CryR1 cis-elements were found either in exact copies as it occurs in the promoter of R2R3 or in slight variants (see Supplemental Table 1 online). This suggests that ZIM, ZML2, and ZML1 recognize and bind to the same cis-element. In addition to the GATA domain, the ZIM and ZML proteins are characterized by a 36–amino acid domain containing the conserved ZIM/TIFY motif (TIFF/YXG) (Vanholme et al., 2007). The ZIM/TIFY domain has been found in other plant proteins, and the corresponding genes have been grouped into the plant-specific TIFY family with 18 members in Arabidopsis. According to a suggested nomenclature, the ZIM, ZML2, and ZML1 were named TIFY1, TIFY2a, and TIFY2b, respectively (Vanholme et al., 2007). The TIFY family is very diverse, and ZIM/TIFY1, ZML2/TIFY2a, and ZML1/TIFY2b are the only proteins with a GATA zinc finger (Vanholme et al., 2007). The TIFY domain has been demonstrated to be involved in protein–protein interactions (Chung and Howe, 2009).
ZML1 and ZML2 are involved in the cry1-dependent induction of ELIP2, GPX7, ERD9, and MYB7 expression in response to excess light, and induction of these genes was significantly impaired in the zml2 and zml1 single and zml2-2 zml1 double mutants (Figures 5 and 9). Analysis of the cry1 zml1 and cry1 zml2-2 double mutants and the cry1 zml1 zml2-2 triple mutant confirmed that cry1 and ZML1/2 are components in the same signaling pathway since no enhanced suppression of ELIP2 and GPX7 expression could be found in the double and triple mutants compared with the cry1 single mutant (Figure 9). Similarly to the cry1 mutant, the zml mutants displayed a clear excess light–sensitive phenotype as demonstrated by the drop in Fv/Fm, the enhanced photobleaching, reduced chlorophyll content, and higher accumulation of ROS compared with the wild type (Table 2, Figures 6 and 7; see Supplemental Figure 6 online). The excess light–sensitive phenotype of the cry1, zml2, and zml1 mutants could possibly be explained by the misregulation of the stress-related genes encoding the chloroplastic glutathione peroxidase7 (GPX7) and the glutathione S-transferases (ERD9) in the mutants. Glutathione peroxidases have been shown to function as both ROS transducers and scavengers, and glutathione S-transferases have been shown to respond to various stresses such as high light, cold, and drought (Wagner et al., 2002; Seki et al., 2003; Goulas et al., 2006; Miao et al., 2006). A gpx7 T-DNA insertion mutant demonstrated similar phenotype to the cry1 and zml mutants with compromised photooxidative stress tolerance and higher H2O2 levels in the leaves following exposure to excess light, demonstrating an important role for GPX7 during photoprotection (Chang et al., 2009). In contrast with cry1, the zml2 and zml1 lines accumulated anthocyanin following exposure to excess light (Figure 6). It was demonstrated that the induction of the genes encoding components of the phenylpropanoid pathway in response to excess light is a cry1-HY5–mediated response (Kleine et al., 2007). The CryR1 binding factor ZML2 and its homolog, ZML1, represent components of the HY5-independent cry1-mediated response. Thus, no effect on the anthocyanin accumulation was expected in the zml mutants.
No functional redundancy was observed between ZML2 and ZML1. ELIP2 and GPX7 expression following exposure to excess light was similar in the zml2-2 zml1 double mutant compared with the respective single mutants, supporting the suggestion that ZML1 and ZML2 act in concert and that they bind the CryR1 element as a heterodimer in vivo. We demonstrated that ZML1 and ZML2 homo- and heterodimerize in yeast (Figures 8A and 8B) (Chung and Howe, 2009) and that ZML1 and ZML2 interact in Arabidopsis protoplasts (Figure 8C). Similarly to recombinant ZML2, recombinant ZML1 protein was also able to bind the CryR1 element (Figure 8D). When ZML2 and ZML1 recombinant proteins were incubated together, the intensity of the shifted band was significantly reduced, suggesting formation of second protein-DNA complex of high molecular weight that was unable to enter the native PAGE due to size restrictions, and formation of a ZML2/ZML1 heterodimer complex is most likely occurring in vitro (Figure 8). By analyzing deletion constructs of the JASMONATE ZIM-domain (JAZ) proteins in the TIFY family, it was demonstrated that the conserved TIFY domain mediates homo- and heteromeric interactions between several JAZ proteins in Arabidopsis, and it was suggested that combinatorial interactions between various JAZ proteins play a role in generating diverse jasmonate signal outputs (Chung and Howe, 2009). Thus, it is possible that the ZML1 and ZML2 interact via the TIFY domain. However, unlike the other GATA proteins or proteins in the TIFY family in Arabidopsis, ZML2 and ZML1 also contain a CCT domain (Shikata et al., 2004). The CCT domain is present in the flowering time protein CONSTANS (CO; Robson et al., 2001; Suárez-López et al., 2001), CO-like protein (Griffiths et al., 2003), and in TIMING OF CAB EXPRESSION1 (Strayer et al., 2000). CCT was first identified in CO, and it was suggested to mediate protein–protein interactions (Robson et al., 2001). This was supported by the observation that the CCT domain of CO interacted with the Arabidopsis transcription factor ABI3 in yeast cells (Kurup et al., 2000). Thus, it is also possible that ZML2 interacts with ZML1 through the CCT domain.
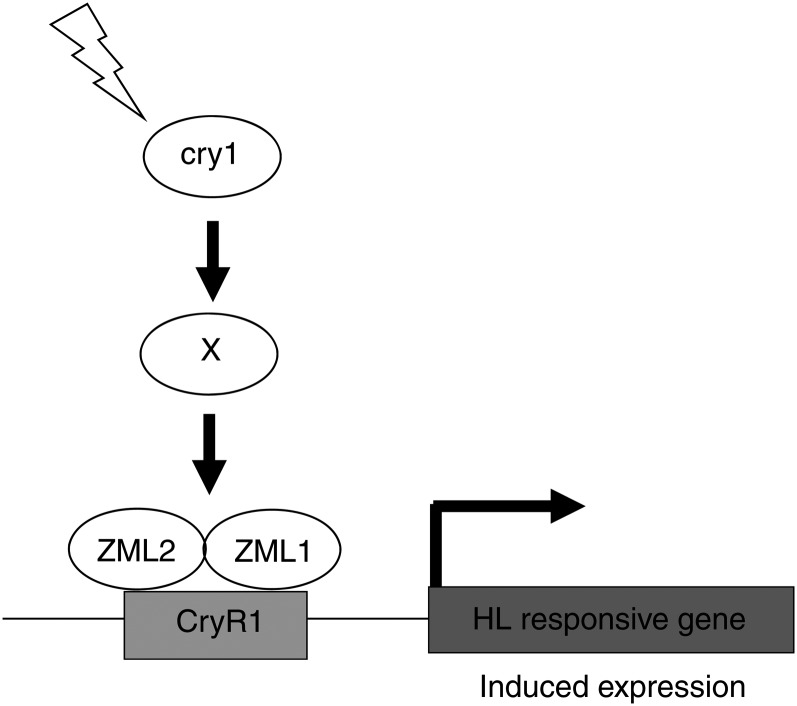
No direct interaction between the C terminus of CRY1 and ZML1 or ZML2 could be detected in yeast (Figures 8A and 8B) or between the full-length CRY1 and ZML2 in Arabidopsis protoplasts (Figure 8C). Our results suggest that cry1 and ZML1/2 are genetically linked and that cry1 action, via some unknown component, stimulates the formation and/or activation of the ZML heterodimer required for full expression of genes encoding photoprotective components (Figure 10). It is also possible that cry1 action modifies the ZML1/2 protein levels or phosphorylation status. Expression of the genes containing the CryR1 cis-element in their promoters was not impaired in zml mutant lines or in cry1 under control conditions (see Supplemental Figure 4 online), suggesting that cry1 activates the ZML1/2 complex only in response to excess light to stimulate induction of genes encoding photoprotective components (Figure 10). Furthermore, the hypocotyl growth response to blue light was not affected in seedlings of the zml2 and zml1 mutants (see Supplemental Figure 7 online), suggesting that normal blue light response is not mediated via the ZMLs. The ZIM, ZML1, and ZML2 proteins homo- and heterodimerize in all possible combinations (Chung and Howe, 2009), and it is possible that different combinations of the GATA transcription factors are triggered by different cues and regulate specific gene sets. It is also possible that heterodimers could be formed with other members of the TIFY family, such as the PEAPOD1 (PPD1) and PPD2. Similarly to ZIM, PPD1 and PPD2 have been shown to regulate leaf size and shape in Arabidopsis (Shikata et al., 2004; White, 2006). Further experiments will determine the exact interplay between the TIFY proteins and their role in gene regulation. However, it is clear that the TIFY protein ZML2 and its homolog, ZML1, are essential components of the cry1-mediated response to excess light.
Figure 10.
Working Model for the cry1-Mediated HL Response.
In response to excess light, cry1 activates the ZML complex via some unknown component, possibly by facilitating heterodimerization between ZML1 and ZML2. The ZML complex binds to the CryR1 cis-element, and expression of genes encoding photoprotective components is induced.
METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis thaliana Columbia-0 wild type, cry1-304 (Ahmad and Cashmore, 1993), zml2-1 (GK448C11), zml2-2 (GK833C07), zml1 (SALK_069271), and zim (SALK_144513) were used. The T-DNA insertion lines obtained from the European Arabidopsis Stock Center were grown on soil at 23°C (16 h light 100 μmol quanta m−2 s−1) and 18°C (8 h dark) at 60% relative humidity. For aseptic growth, seeds were sterilized for 15 min with 75% (v/v) ethanol, 0.01% Triton X-100, washed three times with 95% ethanol, and plated on 0.27% phytoagar plates containing 1× Murashige and Skoog salt mixture including vitamins (Duchefa) and 2% Suc. The plates were maintained in darkness at 4°C for 2 d for stratification and then placed for 10 d in continuous light (100 μmol quanta m−2 s−1). For high light treatment, either 10-d-old seedlings or 4-week-old plants were subjected to 3 h 1000 μmol quanta m−2 s−1 (metal halide HQI-T 400-W day light bulbs; Orsam).
Nuclear Protein Extraction
Control and 3 h high light–treated leaves of 4-week-old Arabidopsis plants were used for nuclear protein extractions using the CelLytic PN-Plant nuclei isolation/extraction kit (Sigma-Aldrich) according to the manufacturer’s instructions. The plant tissue was ground in liquid nitrogen, homogenized in nuclei extraction buffer, and filtered through nylon net, and the pellets were collected by centrifugation. For cell membrane lysis, Triton X-100 was added to 0.3% final concentration. Nuclear proteins were extracted from crude nuclei in protein extraction buffer.
DNA-Affinity Trapping of DNA Binding Proteins
DNA promoter fragments of the R2R3 transcription factor (At5g49330) containing CryR1 was used to isolate proteins binding to the cis-elements in these promoter fragments. The promoter region was −1060 to −1278 of R2R3. Biotinylated DNA promoter fragments were generated by PCR using the following primers: CryR1-F, biotin-5′-CTTCTTTAACTCGTTAAATC-3′; CryR1-R, 5′-TTTATGGTCCAGAGACCAGT-3′. Biotinylated DNA promoter fragment was immobilized on Dynabeads M-280 streptavidin (Invitrogen) according to the manufacturer’s instructions in which 2 mg beads were immobilized using 2× binding buffer (10 mM Tris HCl, pH 7.5, 1 mM EDTA, and 2 M NaCl). Protein binding to the DNA was performed as described by Gabrielsen et al. (1989) with some modifications. Incubation (15 min) at 25°C was performed after the beads were resuspended in protein binding buffer (20 mM Tris HCl, pH 8.0, 1 mM EDTA, 10% glycerol, 100 mM NaCl, 0.05% Triton X-100, and 1 mM DTT) and mixed with Arabidopsis nuclear protein extracts. The Dynabeads were washed three times with protein binding buffer before proteins were eluted in elution buffer (20 mM Tris HCl, pH 8.0, 1 mM EDTA, 10% glycerol, 1 M NaCl, 0.05% Triton X-100, and 1 mM DTT).
Protein Digestion, Mass Spectrometry, and Data Analysis
Total proteins in solution were incubated at 95°C for 15 min in presence of 0.1 M NH4HCO3 and 10 mM DTT, cooled to room temperature, mixed with 8 M urea, and incubated for 1 h. Subsequently, alkylation reaction was done at 37°C for 30 min in dark after adding 55 mM iodoacetamide. Urea concentration was reduced to 0.8 M by diluting with 0.1 M NH4HCO3. Trypsin was added at 1:50 enzyme-to-substrate ratio, and digestion was performed overnight at 37°C. The resulting peptides were lyophilized, resuspended in 1% trifluoroacetic acid, and desalted using a Poros 50 reverse-phase R2 microcolumn (PerSeptive Biosystems) as described by Yanamandra et al. (2009).
The desalted tryptic peptides were separated by reversed-phase ultraperformance liquid chromatography using a nanoACQUITY UPLC system (Waters) prior to mass spectrometry analysis. Each sample (peptides) was concentrated on a C18 trap column (Symmetry 180 µm × 20 mm 5 µm; Waters) and washed with 5% acetonitrile and 0.1% formic acid at 15 µL/min for 1 min. The samples were eluted from the trap column and separated on a C18 analytical column (75 µm × 100 mm 1.7 µm; Waters) at 350 nL/min using 0.1% formic acid as solvent A and 0.1% formic acid in acetonitrile as solvent B, in a gradient. The following gradients were used: linear from 0 to 40% B in 25 min, linear from 40 to 80% B in 1 min, isocratic at 80% B in 1 min, linear from 80 to 5% B in 1 min, and isocratic at 5% B for 7 min. The eluting peptides were sprayed into the mass spectrometer (Q-Tof Ultima; Waters) with the capillary voltage set to 2.6 kV and cone voltage to 40 V. The instrument was operated in data-dependent mode as described (Srivastava et al., 2009) without any further changes.
ProteinLynx Global Server software (V2.2.5) was used to convert raw data to peak lists for database searching. Proteins were identified by a local version of Mascot search program (V2.1.04; Matrix Science Limited) using Arabidopsis protein database from The Arabidopsis Information Resource (version 9.0; July 19, 2009; 33,410 sequences). The following settings were used for the database search: trypsin-specific digestion with two missed cleavage allowed, carbamidomethylated Cys set as fixed modification, oxidized Met in variable mode, peptide tolerance of 80 ppm, and fragment tolerance of 0.1 D. Peptides with Mascot ion scores exceeding the threshold for statistical significance of P < 0.05 were selected and processed manually to validate their significance (see Supplemental Figure 9 online).
Expression and Purification of Recombinant Proteins
The full-length open reading frame of ZML2 and ZML1 were PCR amplified using the primers ZML2-F1 (5′-CACCATGGATGACCTACATGGAA-3′), ZML2-R (5′-TCACTGTGAGTTGCTTATGTCATT-3′), ZML1-F (5′-CACCATGGATGATCTTCATGG-3′), and ZML1-R (5′-TCACTGTGTGTTGCTAA-3′) (see Supplemental Table 2 online). The ZML2 and ZML1 PCR products were cloned into the pET100D TOPO vector according to the manufacturer’s instructions (Invitrogen). After 5 h induction with 1 mM isopropylthio-β-galactoside, Escherichia coli (BL21)–expressed proteins were affinity purified on Ni2+-NTA agarose resin (Qiagen).
EMSAs
To generate a 218-bp CryR1-containing fragment, the promoter region −1060 to −1278 of R2R3 factor gene family was used for PCR amplification using the primers (cryR1-F: 5′-CTTCTTTAACTCGTTAAATC-3′; cryR1-R: 5′-TTTATGGTCCAGAGACCAGT-3′). To generate G-box cis-element, CryR1 cis-element, and its mutant variants, forward primers cryR1cis-F (5′-TCAACTGACACGTGGCATAAC-3′), cryR1cis-F (5′-GAAAAAAGTTCTAGAATTTTTT-3′), cryR1-M1-F (5′-GAAAAGTGGTGGTAATTTTTT-3′), cryR1-M2-F (5′-GAAAAGTGTCTAGAATTTTTT-3′), cryR1-M3-F (5′-GAAAAAGTGTGAGAATTTTTT-3′), and cryR1-M4-F (5′-GAAAAAGTTCTGTGATTTTTT-3′) were annealed with their complementary oligonucleotides at room temperature after they were incubated at 70°C for 5 min.
DNA probes were labeled using the AlkPhos direct labeling and detection system (Amersham) according to the supplier’s instructions with some modifications. Reaction buffer (10 μL) was added to 10 μL DNA (ng/μL diluted DNA) after the DNA was cooled on ice for 5 min, 2 μL labeling reagent was added to the reaction followed by the addition of 10 μL of cross-linker, and the reactions were incubated for 30 min at 37°C. DNA–protein interactions were performed in 25-μL reactions that contained 2.5 μL 10× binding buffer (100 mM Tris HCl, 250 mM KCl, and 10 mM DTT), 1 μg poly dI.dC (Sigma-Aldrich), 2.5% glycerol, 0.05% Triton X-100, 5 mM MgCl2, 10 mM EDTA, 20 ng DNA, and 2.5 μg protein and were then incubated at room temperature for 30 min. The reactions were run on 6% native PAGE in 0.5× Tris-borate-EDTA at 100 V. Gels were transferred to a positively charged nylon membrane (Amersham) using a wet transfer cell (Bio-Rad), and the DNA was cross-linked to the membrane using a UV linker (Spectroline). Membranes were blocked for 1 h at room temperature in blocking buffer containing 2% blocking reagent (AlkPhos direct labeling and detection system; Amersham) and 1% milk powder in 1× TBS. Membranes were washed four times for 7 min before the DNA was detected with CDPstar (AlkPhos direct labeling and detection system).
Biotin Labeling and Detection
PCR products and double-stranded oligonucleotides were labeled at their 3′ end with biotin-14-dCTP using biotin-labeling kit (Pierce) according to the supplier’s instructions. Biotin detection was performed using the Chemiluminescent Nucleic Acid Detection Module (Pierce) according to the supplier’s instructions.
Subcellular Localization and Transactivation Assay
To study the subcellular localization of ZML2 protein, full-length coding sequence, lacking the stop codon, of ZML2 was fused upstream of CFP in the 35S:CFP vector to produce the fluorescence fusion ZML2:CFP. ZML2:CFP and ABI5:YFP (Shaikhali et al., 2008) were transfected and coexpressed transiently in protoplasts isolated from Arabidopsis leaves (see Supplemental Table 2 online). After 16 to 24 h incubation of the transfected cells, fluorescence was visualized using a SP2 confocal laser scanning system equipped with an inverted microscope (Leica) and ×63 water immersion objective (numerical aperture of 0.75). Representative images were taken at 433 and 514 nm for the specific emission of CFP and YFP, respectively. In transactivation assays, effector plasmids used were constructed using full-length cDNAs of ZML2 that were cloned into NcoI and EcoRI sites of 35S:CFP-NosT (Seidel et al., 2005), replacing the CFP gene, and the construct was designated 35S:ZML2 (see Supplemental Table 2 online). The empty vector control was constructed after 35S:CFP-NosT vector was digested with NcoI-EcoRI and religated. To construct reporter plasmids, the 35S promoter of 35S:YFP-NosT (Seidel et al., 2005) was replaced with 35S minimal TATA promoter in BamHI-NcoI sites, and then the 218-bp cryR1-containing fragments were cloned into the HindIII site upstream of the 35S minimal TATA promoter. Isolation and transfection of Arabidopsis mesophyll protoplasts was performed as described (Seidel et al., 2004). CFP and YFP fluorescence was quantified using ImageJ (http://rsbweb.nih.gov/ij/). Ten different regions displaying fluorescence signal were selected for quantification, and YFP brightness was normalized to CFP signal. 35S-CFP-NosT was transfected in all experiments to serve as an internal control to normalize the transfection efficiency.
ChIP Assays
For ChIP assays, protoplasts were prepared as described above, and ChIP assays were performed as described by Lee et al. (2007). Protoplasts were transfected with 5 μg of ZML2 cDNA fused to HA tag (HA-ZML2) construct or with water and incubated for 16 to 24 h at room temperature. The expression of HA-ZML2 fusion protein was assessed by immunoblots using protoplast protein extracts. Protoplasts were fixed with formaldehyde, and chromatin was isolated and sheared by sonication to obtain fragments of sizes between 100 and 1500 bp. Anti-HA monoclonal antibody bound to protein G–coated magnetic beads (Dynabeads Protein G Immunoprecipitation kit; Invitrogen) were used to immunoprecipitate the genomic DNA fragments. Semiquantitative PCR was performed with immunoprecipitated genomic DNA using primer pairs corresponding to fragments of R2R3 promoter (F2 and F5). Primers specific for lateral root promoter (At2g42430) were used to amplify a negative control.
Genotyping of T-DNA Insertion Lines
Two mutant alleles of zml2 (GabiKat line 448C11 and GabiKat line 833C07), one mutant allele of zml1 (Salk_069271), and one mutant allele of zim (Salk_144513) were obtained from the European Arabidopsis Stock Center. Individual plants were screened for T-DNA insertion by PCR using gene-specific primers and primers anchored in the T-DNA borders. Homozygous plants were identified by the absence of the PCR products obtained from gene-specific primers, which compose the T-DNA insertion site (see Supplemental Figure 8 online). Gene-specific primers used for genotyping were GK448C11-F (5′-GTGGTAGTGAACAACAAGGAGATC-3′) and GK448C11-R (5′-AGTATGTCAGGAACTCGCAGT-3′) for zml2-1 (448C11), GK833C07-F (5′-ACATGAGCATGGAACCTATACG-3′) and GK833C07-R (5′-CTACAGAACCTGAGGCGATTCA-3′) for zml2-2 (833C07), salk069271-F (5′-CCTCGTATCATGTGAAGAATGG-3′) and salk069271-R (5′-CATGTTCACAACCATTTGACG-3′) for zml1 (069271), and salk144513-F (5′-CAGGCTCTTTTTGTGTTCCTG-3′) and salk144513-R (5′-CCGATGGCTCGAATTACTTC-3′) for zim (144513). Primers for the T-DNA left borders were o8409 (5′-ATATTGACCATCATACTCATTGC-3′) for the Gabi T-DNA and LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) for the Salk T-DNA.
RNA Isolation, cDNA Synthesis, and Real-Time PCR
A plant RNA mini kit (EZNA) was used for total RNA isolation according to the manufacturer’s instructions. One microgram of total RNA was used for cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. The primers (see Supplemental Table 2 online) used for real-time PCR analysis were designed to flank intron sites to detect amplification of genomic DNA. Real-time PCR was performed in 20-μL reactions containing 2 μL cDNA (1:10 dilution) using iQSYBER Green Supermix (Bio-Rad). Two-step thermal cycling protocol consisted of an initial step at 95°C for 3 min followed by 40 cycles of 10 s at 95°C and 30 s at 60°C before performing a melt curve 60°C to 95°C was performed using the CFX96 Real-Time system (C1000 Thermal Cycler; Bio-Rad). All reactions were performed in triplicate, and the relative transcript abundance of each tested gene was normalized to the expression level of ubiquitin (At4g36800). The data were analyzed using LinRegPCR software (Pfaffl, 2001; Kindgren et al., 2012).
Maximal PSII Quantum Yield
In vivo chlorophyll a fluorescence of single leaves was measured using the Dual-PAM-100 measuring system (Walz) in ambient air at room temperature. Plants were dark adapted for 30 min, and minimum fluorescence (F0) excited by weak measuring light at open PSII centers was measured (settings: measuring light, 10). Then, saturating pulses (0.6 s) of white light (3000 μmol photons m−2 s−1) were used to determine the maximum fluorescence (Fm) at closed PSII centers, and the ratio maximum quantum yield of PSII (Fv/Fm = (Fm − F0)/Fm) was calculated.
Chlorophyll and Anthocyanin Measurements
The chlorophyll and anthocyanin measurements were performed as described (Porra et al., 1989; Neff and Chory, 1998). Ten-day-old seedlings or 5-week-old plants were harvested, weighed, and ground in liquid nitrogen to fine powder. For the chlorophyll measurements, total chlorophyll was extracted in 80% acetone and calculated according to Porra et al. (1989). For the anthocyanin measurement, the pigments were extracted in 1% HCl in methanol. Water was added and the chlorophyll was extracted with an equal volume of chloroform. The anthocyanin quantity was determined by spectrophotometric measurement of the aqueous phase at A530 to A657 and normalized to the total fresh weight used in each sample.
H2O2 Measurement
Two-week-old plants were used to measure H2O2 in control and high light treatment. Leaf powder (50 to 200 mg) was homogenized in 1 mL of 0.2 M HClO4, incubated on ice for 5 min, and then centrifuged at 10,000g for 10 min at 4°C. The acidic supernatant was neutralized to pH 7.0 to 8.0 with 0.2 M NH4OH, pH 9.5, and briefly centrifuged at 3000g for 2 min to sediment the insoluble material. Quantification of H2O2 in the extracts was performed using the Amplex Red Hydrogen Peroxide-Peroxidase Assay kit (Molecular Probes) according to the manufacturer’s instructions. Fluorescence was measured with a plate reader (Spectra Max Gemini; Molecular Devices) using excitation at 530 nm and fluorescence detection at 590 nm. The concentration of H2O2 was calculated using a standard curve.
ROS (Superoxide) Staining
Two-week-old plants were exposed to high light for 12 h. For superoxide staining, the plants were incubated for 1.5 h in 0.1% NBT at room temperature before they were destained in ethanol.
Yeast Two-Hybrid System
The yeast two-hybrid was performed with the DUALhybrid system (Biotech) according to the manufacturer’s instructions. Full-length coding sequence of ZML2 and ZML1 genes was cloned into pLexA-N vector to generate fusion proteins with LexA DNA binding domain (bait) (see Supplemental Table 2 online). ZML1 full-length coding sequence and CRY1 C-terminal domain coding sequence (CRY1-CT) were cloned into pGAD-HA vector to generate fusions of the prey protein with the GAL4 activation domain. NMY51 yeast strain was cotransformed with bait and prey vectors and the transformants were selected on selective media SD-Trp-Leu-HIS and containing 7.5 mM 3-AT. β-Galactosidase overlay assay to detect LacZ activation was performed with an overlay buffer containing 0.5 M potassium phosphate, pH 7.0, 6% dimethylformamide, 0.1% SDS, 50 μL/100 mL β-mercaptoethanol, 5 mg/mL low melting agarose, and 0.05% X-Gal (Fermentas).
Co-IP
To generate expression constructs for Co-IP assays, full-length coding sequence of ZML2 was cloned into BamHI-EcoRI sites in pRT104_3HA vector, and the construct was designated HA-ZML2 (see Supplemental Table 2 online). To generate MYC-ZML1 and MYC-CRY1 constructs, full-length coding sequences of ZML1 and CRY1 were cloned into EcoRI-KpnI and SmaI-EcoRI sites in pRT104_3MYC vector, respectively. Proteins from protoplasts transformed with expression constructs were extracted in 300 μL immunoprecipitation buffer (25 mM Tris- HCl, pH 7.8, 75 mM NaCl, 10 mM MgCl2, 2 mM DTT, 5 mM EGTA, 0.2% Triton X-100, 10% glycerol, and 0.2 mM PMSF) for 40 min at 4°C. Protein extracts were incubated with 5 µg of anti-cMYC monoclonal antibody (Bio-Site) bound to protein G–coated magnetic beads (Dynabeads Protein G Immunoprecipitation kit; Invitrogen) for 1 h at 4°C. Subsequent washing steps were performed according to kit recommendations (Invitrogen), and target antigen was eluted with SDS-PAGE sample loading buffer. To detect the fusion proteins tagged with cMYC, immunoblots were detected with anti-cMYC chicken IgY fraction (Invitrogen) and rabbit HRP-anti-chicken IgY (H+L) (Invitrogen), respectively. HA-tagged proteins were detected with anti-HA peroxidase (Roche).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ZML2, At1g51600; ZML1, At3g21175; ZIM, At4g24470; ELIP2, At4g14690; GPX7, At4g31870; ERD9, At1g10370; MYB7, At1g56650; RBCS, At1g67090; and LHCB2.4, At3g27690.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Interaction of Synthetic Double-Stranded Oligonucleotide Containing CryR1 cis-Element with Nuclear Protein Extracts.
Supplemental Figure 2. Genotyping zml2, zml1, and zim T-DNA Insertion Lines.
Supplemental Figure 3. Expression Analysis of ZML2, ZML1, and ZIM in Response to HL and in the Different Genotypes.
Supplemental Figure 4. Real-Time Analysis of cry1-Dependent and -Independent Genes in the cry1, zml2-1, zml2-2, zml1, and zim Mutant Lines under Control Conditions.
Supplemental Figure 5. Superoxide Accumulation of zml2, zml1, and zim T-DNA Insertion Lines in Response to Excess Light.
Supplemental Figure 6. The zml2, zml1, and zim T-DNA Insertion Lines Exposed to Excess Light.
Supplemental Figure 7. Hypocotyl Growth Response to Blue Light.
Supplemental Figure 8. Genotyping of zml2, zml1, and cry1 Double and Triple Mutants.
Supplemental Figure 9. Mascot Score Result and Ion Spectra for the ZML2 Identification.
Supplemental Table 1. Occurrence of the CryR1 cis-Element in 500 bp of the Promoters of Genes Induced in ZIM-ox Plants.
Supplemental Table 2. Primers Used in Real-Time PCR Analysis and for Generation of Constructs.
Supplementary Material
Acknowledgments
We thank Thorsten Seidel for providing the YFP and CFP vectors. This work was supported by grants from the Swedish Research Foundation, Vetenskapsrådet, and the FFL2 grant from the Foundation for Strategic Research (Å.S.). Å.S. is a Royal Swedish Academy of Sciences Research Fellow supported by a grant from the Knut and Alice Wallenberg Foundation.
AUTHOR CONTRIBUTIONS
Å.S. and J.S. designed the research. J.S., J.B.-L., K.Ö., D.K., A.S.G., V.S., and Å.S. performed the experiments and analyzed data. G.W., K.Ö., and L.B. contributed new analytic and experimental tools and analyzed data. All authors contributed to writing the article.
Glossary
- Fv/Fm
to be defined
- PSII
photosystem II
- ROS
reactive oxygen species
- EMSA
electrophoretic mobility shift assay
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- ChIP
chromatin immunoprecipitation
- CaMV
cauliflower mosaic virus
- H2O2
hydrogen peroxide
- NBT
nitro blue tetrazolium
- Co-IP
coimmunoprecipitation
- HRP
horseradish peroxidase
- 3-AT
to be defined
References
- Ahmad M., Cashmore A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore A.R., Jarillo J.A., Wu Y.J., Liu D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284: 760–765 [DOI] [PubMed] [Google Scholar]
- Chang C.C., Slesak I., Jordá L., Sotnikov A., Melzer M., Miszalski Z., Mullineaux P.M., Parker J.E., Karpinska B., Karpinski S. (2009). Arabidopsis chloroplastic glutathione peroxidases play a role in cross talk between photooxidative stress and immune responses. Plant Physiol. 150: 670–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C., Elzen P.V., Tamaki S., Dunsmuir P., Bedbrook J. (1985). Differential expression of the eight genes of the petunia ribulose bisphosphate carboxylase small subunit multi-gene family. EMBO J. 4: 3055–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 34: 827–836 [DOI] [PubMed] [Google Scholar]
- Gabrielsen O.S., Hornes E., Korsnes L., Ruet A., Oyen T.B. (1989). Magnetic DNA affinity purification of yeast transcription factor tau—A new purification principle for the ultrarapid isolation of near homogeneous factor. Nucleic Acids Res. 17: 6253–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas E., Schubert M., Kieselbach T., Kleczkowski L.A., Gardeström P., Schröder W.P., Hurry V. (2006). The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J. 47: 720–734 [DOI] [PubMed] [Google Scholar]
- Griffiths S., Dunford R.P., Coupland G., Laurie D.A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131: 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob U., Stüber K. (1987). Discrimination of phytochrome dependent light inducible from non-light inducible plant genes. Prediction of a common light-responsive element (LRE) in phytochrome dependent light inducible plant genes. Nucleic Acids Res. 15: 9957–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Mockler T., Duong H., Lin C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291: 487–490 [DOI] [PubMed] [Google Scholar]
- Hong S.H., Kim H.J., Ryu J.S., Choi H., Jeong S., Shin J., Choi G., Nam H.G. (2008). CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J. 55: 361–371 [DOI] [PubMed] [Google Scholar]
- Jarillo J.A., Capel J., Tang R.H., Yang H.Q., Alonso J.M., Ecker J.R., Cashmore A.R. (2001). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410: 487–490 [DOI] [PubMed] [Google Scholar]
- Jeong M.J., Shih M.C. (2003). Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem. Biophys. Res. Commun. 300: 555–562 Erratum. Biochem. Biophys. Res. Commun. 333: 1385. [DOI] [PubMed] [Google Scholar]
- Kang X., Chong J., Ni M. (2005). HYPERSENSITIVE TO RED AND BLUE 1, a ZZ-type zinc finger protein, regulates phytochrome B-mediated red and cryptochrome-mediated blue light responses. Plant Cell 17: 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Ni M. (2006). Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Yoshizumi T., Manabe K., Yamamoto Y.Y., Matsui M. (2001). Arabidopsis transcriptional regulation by light stress via hydrogen peroxide-dependent and -independent pathways. Genes Cells 6: 607–617 [DOI] [PubMed] [Google Scholar]
- Kindgren P., Kremnev D., Blanco N.E., de Dios Barajas López J., Fernández A.P., Tellgren-Roth C., Small I., Strand A. (2012). The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 70: 279–291 [DOI] [PubMed] [Google Scholar]
- Kiyosue T., Wada M. (2000). LKP1 (LOV kelch protein 1): A factor involved in the regulation of flowering time in arabidopsis. Plant J. 23: 807–815 [DOI] [PubMed] [Google Scholar]
- Kleine T., Kindgren P., Benedict C., Hendrickson L., Strand A. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Lockhart P., Batschauer A. (2003). An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 35: 93–103 [DOI] [PubMed] [Google Scholar]
- Kudla B., Caddick M.X., Langdon T., Martinez-Rossi N.M., Bennett C.F., Sibley S., Davies R.W., Arst H.N., Jr (1990). The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9: 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup S., Jones H.D., Holdsworth M.J. (2000). Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21: 143–155 [DOI] [PubMed] [Google Scholar]
- Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.H., Yang H.Q. (2007). Cryptochrome signaling in plants. Photochem. Photobiol. 83: 94–101 [DOI] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Lin C., Shalitin D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Lin C., Yang H., Guo H., Mockler T., Chen J., Cashmore A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield I.W., Devlin P.F., Jen C.H., Westhead D.R., Gilmartin P.M. (2007). Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol. 143: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Lv D., Wang P., Wang X.C., Chen J., Miao C., Song C.P. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller S.G., Kim Y.S., Kunkel T., Chua N.H. (2003). PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Chory J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorny R., Klar T., Hennecke U., Carell T., Batschauer A., Essen L.O. (2008). Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc. Natl. Acad. Sci. USA 105: 21023–21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Rey D.A., Pühler A., Kalinowski J. (2003). The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J. Biotechnol. 103: 51–65 [DOI] [PubMed] [Google Scholar]
- Richly E., Dietzmann A., Biehl A., Kurth J., Laloi C., Apel K., Salamini F., Leister D. (2003). Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep. 4: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Costa M.M., Hepworth S.R., Vizir I., Piñeiro M., Reeves P.H., Putterill J., Coupland G. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28: 619–631 [DOI] [PubMed] [Google Scholar]
- Rossel J.B., Wilson I.W., Pogson B.J. (2002). Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 130: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel T., Golldack D., Dietz K.J. (2005). Mapping of C-termini of V-ATPase subunits by in vivo-FRET measurements. FEBS Lett. 579: 4374–4382 [DOI] [PubMed] [Google Scholar]
- Seidel T., Kluge C., Hanitzsch M., Ross J., Sauer M., Dietz K.J., Golldack D. (2004). Colocalization and FRET-analysis of subunits c and a of the vacuolar H+-ATPase in living plant cells. J. Biotechnol. 112: 165–175 [DOI] [PubMed] [Google Scholar]
- Seki M., Kamei A., Yamaguchi-Shinozaki K., Shinozaki K. (2003). Molecular responses to drought, salinity and frost: Common and different paths for plant protection. Curr. Opin. Biotechnol. 14: 194–199 [DOI] [PubMed] [Google Scholar]
- Shaikhali J., Heiber I., Seidel T., Ströher E., Hiltscher H., Birkmann S., Dietz K.J., Baier M. (2008). The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 8: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata M., Matsuda Y., Ando K., Nishii A., Takemura M., Yokota A., Kohchi T. (2004). Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 55: 631–639 [DOI] [PubMed] [Google Scholar]
- Shikata M., Takemura M., Yokota A., Kohchi T. (2003). Arabidopsis ZIM, a plant-specific GATA factor, can function as a transcriptional activator. Biosci. Biotechnol. Biochem. 67: 2495–2497 [DOI] [PubMed] [Google Scholar]
- Somers D.E., Schultz T.F., Milnamow M., Kay S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Srivastava V., Srivastava M.K., Chibani K., Nilsson R., Rouhier N., Melzer M., Wingsle G. (2009). Alternative splicing studies of the reactive oxygen species gene network in Populus reveal two isoforms of high-isoelectric-point superoxide dismutase. Plant Physiol. 149: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Más P., Panda S., Kreps J.A., Kay S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Teakle G.R., Manfield I.W., Graham J.F., Gilmartin P.M. (2002). Arabidopsis thaliana GATA factors: Organisation, expression and DNA-binding characteristics. Plant Mol. Biol. 50: 43–57 [DOI] [PubMed] [Google Scholar]
- Vanholme B., Grunewald W., Bateman A., Kohchi T., Gheysen G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12: 239–244 [DOI] [PubMed] [Google Scholar]
- Wagner U., Edwards R., Dixon D.P., Mauch F. (2002). Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 49: 515–532 [DOI] [PubMed] [Google Scholar]
- Wang H., Ma L.G., Li J.M., Zhao H.Y., Deng X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Ward J.M., Cufr C.A., Denzel M.A., Neff M.M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D.W. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Spalding E.P. (2007). Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 104: 18813–18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanamandra K., et al. (2009). Amyloid formation by the pro-inflammatory S100A8/A9 proteins in the ageing prostate. PLoS ONE 4: e5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Q., Tang R.H., Cashmore A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Q., Wu Y.J., Tang R.H., Liu D., Liu Y., Cashmore A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yu X., Klejnot J., Zhao X., Shalitin D., Maymon M., Yang H., Lee J., Liu X., Lopez J., Lin C. (2007). Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.