The transcriptionally active chromosome from spinach chloroplasts was analyzed by two-dimensional gel electrophoresis and mass spectrometry to identify proteins involved in structuring of the nucleoid core. SWI/SNF complex B domain–containing proteins were identified that might be functional equivalents of the bacterial nucleoid-associated proteins involved in shaping of nucleoid architecture.
Abstract
A highly enriched fraction of the transcriptionally active chromosome from chloroplasts of spinach (Spinacia oleracea) was analyzed by two-dimensional gel electrophoresis and mass spectrometry to identify proteins involved in structuring of the nucleoid core. Among such plastid nucleoid-associated candidate proteins a 12-kD SWIB (SWI/SNF complex B) domain–containing protein was identified. It belongs to a subgroup of low molecular mass SWIB domain proteins, which in Arabidopsis thaliana has six members (SWIB-1 to SWIB-6) with predictions for localization in the two DNA-containing organelles. Green/red fluorescent protein fusions of four of them were shown to be targeted to chloroplasts, where they colocalize with each other as well as with the plastid envelope DNA binding protein in structures corresponding to plastid nucleoids. For SWIB-6 and SWIB-4, a second localization in mitochondria and nucleus, respectively, could be observed. SWIB-4 has a histone H1 motif next to the SWIB domain and was shown to bind to DNA. Moreover, the recombinant SWIB-4 protein was shown to induce compaction and condensation of nucleoids and to functionally complement a mutant of Escherichia coli lacking the histone-like nucleoid structuring protein H-NS.
INTRODUCTION
Chloroplast function is indispensable for plant productivity. Chloroplasts are thought to originate from ancestral cyanobacteria and hence have retained prokaryotic features in the organization of their genome and the structure of the corresponding nucleoids. Typically, chloroplasts contain hundreds of plastid genomes in the form of circular 120- to 180-kb molecules organized in aggregates that in analogy with bacterial nucleoids were named plastid nucleoids (Kuroiwa et al., 1998; Lilly et al., 2001; Sato et al., 2003). Number, position, and compactness of nucleoids are known to parallel the changes in gene expression occurring during chloroplast development. Two DNA binding proteins, the plastid envelope DNA binding protein (PEND) (Sato et al., 1998) and the matrix attachment region binding filament-like protein1 (MFP1) (Jeong et al., 2003), are known to be involved in redistribution of the plastid nucleoids from the envelope membrane to the thylakoids during the transition from proplastids to chloroplasts. However, proteins involved in regulation of nucleoid morphology and compactness and corresponding to the bacterial HU (for histone-like protein from Escherichia coli strain U93) and HU-like proteins have not been identified hitherto (Sato et al., 2003; Sakai et al., 2004).
Plastid nucleoids were prepared by different approaches. Preparations shown to preserve the morphological integrity of nucleoids involve sucrose density gradient centrifugation (Kuroiwa and Suzuki, 1981; Sato et al., 1997; Cannon et al., 1999; Majeran et al., 2012). For enrichment of transcriptional activity, nucleoids are released from thylakoids using the nonionic detergent Triton X-100 and are purified by gel filtration. These so-called transcriptionally active chromosomes (TACs) (Igloi and Kössel, 1992) contain subunits of the plastid-encoded RNA polymerase (Suck et al., 1996), which together with further DNA binding proteins are also found in transcriptionally active soluble fractions prepared from chloroplasts (Schröter et al., 2010; Steiner et al., 2011). TAC fractions have been prepared from chloroplasts of Euglena gracilis (Hallick et al., 1976), mustard (Sinapis alba; Bülow et al., 1987), barley (Hordeum vulgare; Krupinska and Falk, 1994; Suck et al., 1996), spinach (Spinacia oleracea; Krause and Krupinska, 2000), and Arabidopsis thaliana (Pfalz et al., 2006). The conventionally prepared TAC fraction of spinach chloroplasts (TAC-I) was shown to have a rather complex protein composition and was further purified by precipitation with protamine sulfate and resolubilization in the presence of heparin followed by a second round of gel filtration. The resulting fraction, enriched in the core components of the complex and showing 50-fold higher specific transcription activity rates, was termed TAC-II (Krause and Krupinska, 2000).
Apart from the proteins of the transcriptional apparatus, the TAC fraction is expected to contain a number of nucleoid-associated proteins with DNA binding properties that might be involved in replication, recombination, and repair of DNA as well as in structuring of nucleoids (Sakai et al., 2004). In bacteria, low molecular mass proteins, such as the 9-kD HU and HU-like proteins (HLPs), are responsible for formation of nucleosome-like structures (Dillon and Dorman, 2010) and affect DNA-associated processes, such as replication and transcription (Dorman and Deighan, 2003; Kamashev et al., 2008; Dillon and Dorman, 2010). However, genes encoding potential counterparts of the bacterial nucleoid-associated proteins have not been found in the genomes of higher plants (Riechmann et al., 2000; Sato et al., 2003). Among the few structural proteins of plastid nucleoids thus far identified is the bifunctional sulfite reductase, which was shown to induce compaction of plastid nucleoids coinciding with a repression of transcriptional activity (Sekine et al., 2002).
Proteome analyses performed with TAC fractions isolated from mustard and Arabidopsis did not identify DNA binding proteins with a molecular mass below 20 kD (Pfalz et al., 2006). Nevertheless, several novel TAC-associated proteins (PTACs) with DNA/RNA binding domains were identified (Pfalz et al., 2006). PTAC1 is identical with the plastid-nucleus-located Whirly1 (Grabowski et al., 2008). Its plastidial form was shown to bind to a subset of intron-containing RNA species (Prikryl et al., 2008; Melonek et al., 2010). PTAC3 belongs to the SAP (for SAF A/B, Acinus, and PIAS) domain protein family, members of which are involved in chromosomal organization (Aravind and Koonin, 2000). The PTAC11 protein (Pfalz et al., 2006) is identical with Whirly3, which recently was described to function together with Whirly1 as an antirecombination protein (Maréchal et al., 2009). PTAC14 from Arabidopsis was identified as a SET (for Suppressor of variegation 3-9 [Su(var)3-9], Enhancer of zeste [E(Z)], and Trithorax [Trx]) domain–containing protein. Nuclear proteins with a SET domain were shown to function as histone methyltransferases (Thorstensen et al., 2011). PTAC14 belongs to those plastidial proteins, which were acquired from the nucleus where they are involved in chromatin remodeling (Jacob et al., 2009).
Here, we report on identification of a low molecular mass SWIB domain–containing plastid nucleoid-associated protein (ptNAP) in the proteome of the TAC-II fraction that is highly enriched in nucleoid core proteins from spinach chloroplasts. SWIB domain–containing proteins are subunits of nuclear ATP-dependent chromatin-remodeling complexes of the SWI/SNF type that facilitate transcriptional activation (Bennett-Lovsey et al., 2002). In silico searches showed that the genome of Arabidopsis encodes six low molecular mass SWIB domain proteins. By fusion with green fluorescent protein (GFP), four of them were shown to be targeted to chloroplasts where they colocalize in speckles, indicating association with nucleoids. One of these having a histone H1-like domain (SWIB-4) was shown to bind to DNA. Recombinant SWIB-4 protein induced condensation of E. coli nucleoids in vivo and functionally complemented an E. coli mutant lacking the gene encoding the histone-like nucleoid protein H-NS. Our results indicate that the low molecular mass SWIB domain proteins in chloroplasts might be functional equivalents of the bacterial nucleoid-associated proteins (NAPs) involved in shaping of nucleoid architecture.
RESULTS
Proteomic Identification of Chloroplast Nucleoid-Associated Proteins
To identify novel proteins involved in shaping of chloroplast nucleoids, a highly enriched fraction of the TAC from spinach chloroplasts was used for proteome analysis. For our studies, we used chloroplasts isolated from spinach leaves, which provide excellent material for biochemical studies requiring highly purified and functional organelles (Gruissem et al., 1986; Babujee et al., 2010) that cannot easily be prepared from Arabidopsis (Bayer et al., 2011). To obtain a highly enriched fraction (TAC-II), a conventional TAC fraction (TAC-I) prepared from 20 kg of spinach leaves was subjected to a second round of gel filtration resulting in TAC-II (Figure 1A). The protein compositions of both fractions were analyzed by two-dimensional gel electrophoresis (2-DE). For isoelectric focusing, pH gradients from 3 to 10 were chosen. After second-dimension denaturing gel electrophoresis, proteins were visualized by staining with Coomassie blue (Figures 1B and 1C). Image analysis allowed the detection of 132 spots on the 2-DE gel of TAC-I, whereas on the 2-DE gel of TAC-II, only 85 protein spots were clearly discernible (Figure 1D). These 85 proteins were manually excised from the gel and digested with trypsin (see Supplemental Figure 1 online). By means of mass spectrometry, 53 TAC-II proteins with protein scores of 60 or more and individual peptide scores of 39 or more could be identified (see Supplemental Data Set 1 online). Thirteen of them were identified by homology with protein sequences of spinach, 15 by homologies with protein sequences from Arabidopsis, and 14 by homology with sequences from rice (Oryza sativa), and single proteins were identified by homologies with other plant species (see Supplemental Data Set 1 online). Despite precipitation with protamine sulfate and the second round of gel filtration used for preparation of TAC-II (Krause and Krupinska, 2000), the fraction contains several contaminants as it has previously been reported for other preparations enriched in nucleoid-associated proteins (Schröter et al., 2010; Majeran et al., 2012). For example, the two polygalacturonases (spots 7 and 51) are likely contaminants. These proteins are synthesized as precursors at the endoplasmic reticulum from where they are transported in vesicles to their destinations in the plasma membrane and cell wall. It is likely that these proteins in the TAC fraction are derived from the endoplasmic reticulum, which is known to be physically tightly associated with chloroplast envelope membranes (Andersson et al., 2007).
Figure 1.
Two-Dimensional Separation of TAC-I and TAC-II Proteins.
(A) Scheme of the purification protocol of TAC-I and TAC-II fractions from spinach chloroplasts (Krause and Krupinska, 2000). After digestion with DNase I and RNase A, the proteins were separated by isoelectric focusing in a pH gradient from 3 to 10 and by 2-DE.
(B) and (C) Spot patterns obtained on gels after staining with Coomassie blue of TAC-I (B) and TAC-II (C) proteins, respectively, are shown. Image analysis of the 2-DE gels revealed 132 protein spots on the TAC-I and 85 on TAC-II gel.
(D) Relative spot intensity of proteins detected in TAC-II compared with the intensities of corresponding proteins in the TAC-I fraction as determined with Progenesis PG240 software.
Among the identified proteins in TAC-II are several putative nucleoid core components with putative roles in transcription, replication, maintenance, and structuring of DNA. As expected, subunits of the plastid-encoded RNA polymerase, namely, RpoC1 (spot 13) and RpoC2 (spot 18), were detected in TAC-II. The RpoA subunit was identified with a score under 60 and therefore was not included in Supplemental Data Set 1 online. Six of the identified TAC proteins are putative architectural ptNAPs with DNA binding properties that are able to alter the DNA compaction through bending, wrapping, or bridging (Table 1; see Supplemental Data Set 1 online, spots 10, 17, 32, 37, 39, and 53).
Table 1. Identified Putative Plastid Nucleoid-Associated Proteins in the TAC-II Fraction.
| No. (Spot No.)a | Identified ptNAP Proteins | AGI Accession Numberb | Conserved Domains/Motifs/Regions | Function | kD | pI | Lys Content |
|---|---|---|---|---|---|---|---|
| 1 (37) | SWIB complex BAF60b domain-containing protein | At2g35605 | SWIB/MDM2 domain, low complexity region | Chromatin remodeling | 12 | 10 | 9% |
| 2 (39) | Myb-related transcription factor SANT superfamily | At2g02060 | N-CoR and TFIIIB DNA binding domains | DNA binding, regulation of transcription | 29 | 7 | 6% |
| 3 (32) | Jasmonate- and ethylene-responsive factor3 (JERF3) | At3g14230 | AP2 DNA binding | Regulation of transcription | 36 | 9 | 8% |
| 4 (17) | Hypothetical protein, armadillo/β-catenin repeat family protein | At2g27430 | Armadillo-type fold, armadillo-like helical | Nucleic acid binding/protein binding | 49 | 8 | 8% |
| 5 (53) | Myb-related transcription factor SANT superfamily | At5g02320 | N-CoR and TFIIIB DNA binding domains | DNA binding, regulation of transcription | 62 | 8 | 10% |
| 6 (10) | Unknown protein, RecF/RecN/SMC N-terminal domain | At5g36780 | RecF/RecN/SMC N-terminal domain | Chromatin and DNA dynamics/DNA metabolism and recombination | 66 | 5 | 11% |
Spot number on the TAC-II 2-DE gel (see Supplemental Data Set 1 online).
AGI, Arabidopsis Genome Initiative.
Previous biochemical investigations revealed low molecular mass DNA binding proteins in nucleoid fractions, which were assigned as candidates for functional replacement of the missing HU-like proteins in plants (Briat et al., 1984). Hitherto, an identification of such basic low molecular mass proteins was not possible. Purification of the TAC fraction from spinach chloroplasts enabled the identification of a SWIB domain protein in spot 37 of the 2-DE gel (see Supplemental Figure 1 online). The peptide masses determined for the spinach protein matched with a 12-kD SWIB domain–containing protein encoded by the At2g35605 gene (Table 1). The low molecular mass SWIB domain protein was found to have a high Lys content and a high isoelectric point, thereby resembling bacterial NAP (Dillon and Dorman, 2010) (Table 1).
The smallest putative ptNAP identified in spot 37 belongs to the family of SWIB domain proteins that has 20 members in Arabidopsis as identified by The Arabidopsis Information Resource (TAIR; www.arabidopsis.org) database analyses (Figure 2A). Phylogenetic analysis with MEGA5 software (Tamura et al., 2011) revealed that based on their amino acid sequences, plant SWIB proteins can be subdivided into four major groups (Figure 2A; see Supplemental Data Set 2 online). The two group 4 proteins with a molecular mass of ∼50 kD have high similarity with the yeast SWP73 protein (Jerzmanowski, 2007), which is a component of the SWI/SNF complex with a functional role in transcriptional activation by modulation of the chromatin structure (Cairns et al., 1996). Proteins of groups 2 and 3 have, in addition to the SWIB domain, other domains, such as Really Interesting New Gene, zinc-finger, plus-3, and GYF (for Gly-Tyr-Phe) or the DEK domain. All members of these two groups are predicted to be involved in histone modification and regulation of transcription in the nucleus (TAIR) (Swarbreck et al., 2008). The 12-kD SWIB domain protein found in TAC-II belongs to group 1, consisting of so-called stand-alone SWIB domain proteins, where the SWIB domain is found as an isolated protein and is not part of a larger protein. In prokaryotes, such SWIB domain proteins were found exclusively in Chlamydia, which as parasitic bacteria probably have acquired the SWIB domain gene from a mammalian host (Bennett-Lovsey et al., 2002).
Figure 2.
Phylogenetic Analysis of 20 SWIB Domain–Containing Proteins of Arabidopsis and Characterization of the Putative Organellar SWIB Proteins.
(A) The phylogenetic tree was constructed using MEGA5 with the neighbor-joining method. Bootstrap values calculated from 1000 trials are shown at each node. The evolutionary distances were computed using the Poisson correction method and are in units of the number of amino acid substitutions per site. The multiple sequence alignment was performed with ClustalW algorithm and is available as Supplemental Data Set 2 online. The proteins could be subdivided in four groups. Members of group 1 have molecular masses ranging from 10 to 20 kD. Bar indicates number of substitutions per site.
(B) Table presenting the nomenclature and molecular properties of the six proteins of group 1. AGI, Arabidopsis gene identifier; Mw, molecular mass; H1, histone H1-like motif. TargetP localization predictions: M, mitochondria; C, chloroplasts; TP, targeting peptide length (aa, amino acids). The proteins were ordered according to their molecular mass.
The other TAC-II proteins possessing DNA binding domains have molecular masses higher than 20 kD (Table 1). The protein identified in spot 32 is a member of the APETALA2 (AP2) subfamily of the AP2/EREBP (for ethylene-responsive element binding proteins) family of transcription factors unique to plants (Okamuro et al., 1997). The AP2/EREBP domain consists of 60 to 70 amino acids and is involved in binding to DNA (Weigel, 1995). In silico analyses predicted localization for some members of the AP2/EREBP subfamily in organelles (Schwacke et al., 2007). Moreover, for the At2g44940 protein, a dual targeting to chloroplasts and nucleus could be shown (Schwacke et al., 2007). The protein identified in spot 10 represents a structural maintenance of chromosomes (SMC) protein. SMC proteins belong to a ubiquitous protein family present in almost all prokaryotic and eukaryotic organisms and have functions in chromosome condensation, segregation, cohesion, and DNA recombination and repair (Graumann and Knust, 2009). In E. coli, SMC proteins were shown to induce negative supercoiling of DNA in vivo, indicating their involvement in organization and compaction of nucleoids (Graumann and Knust, 2009). An SMC (At-SMC1) as well as an armadillo/β-catenin protein were previously identified in the proteome of chloroplasts from Arabidopsis (Kleffmann et al., 2004; Zybailov et al., 2008). The protein identified in spot 17 has similarity in sequence with the Arabidopsis armadillo/β-catenin protein. Armadillo repeat proteins form a large group of 108 proteins in Arabidopsis. Two further putative ptNAPs identified in TAC-II belong to the family of the MYB/SANT domain transcription factors having a helix-turn-helix (HTH) DNA binding domain (spots 39 and 53). Animal and fungal proteins containing SANT domains (for SWI3, ADA2, N-Cor, TFIIB) were found to be associated with histone modifying enzymes and ATP-dependent chromatin remodeling proteins (Boyer et al., 2004; Jerzmanowski, 2007).
Subcellular Localization of the Low Molecular Mass SWIB Proteins of Group 1
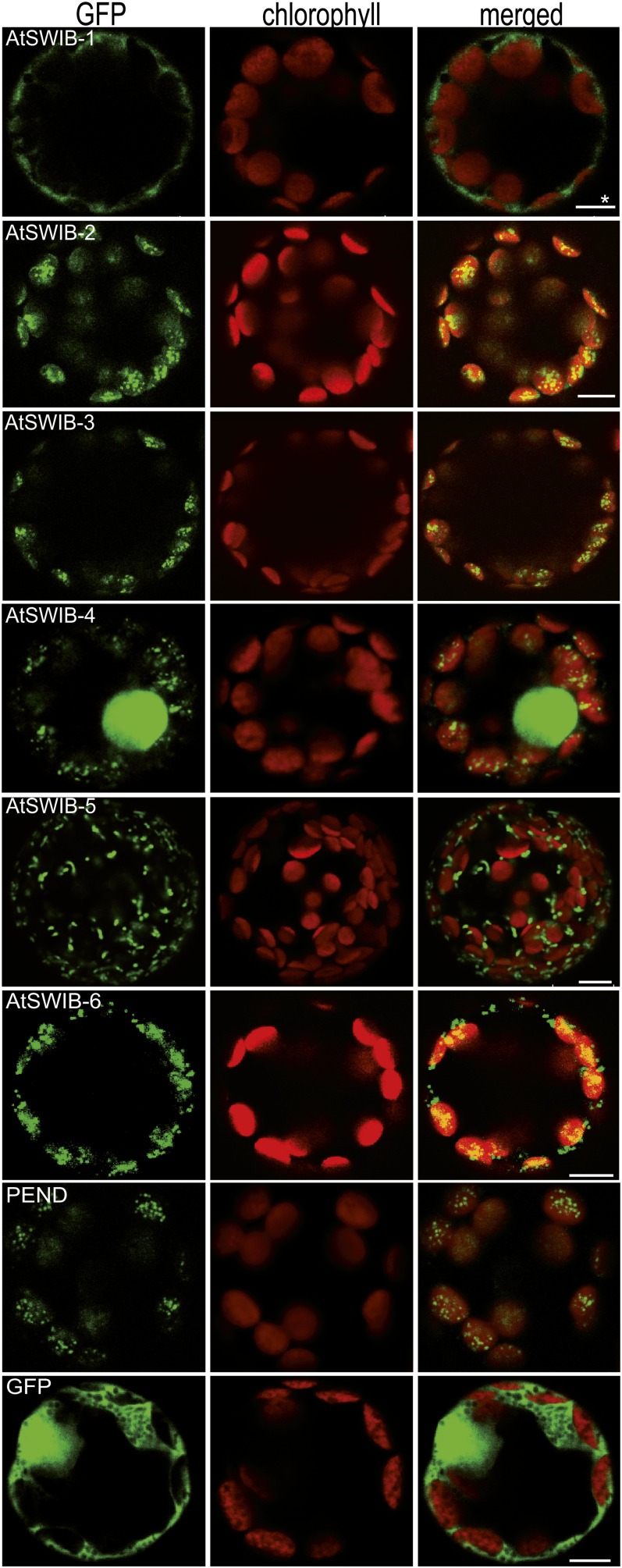
Proteome analysis of TAC-II allowed identification of a 12-kD SWIB domain protein. Masses of peptides obtained by trypsin digestion fitted best to the 12-kD SWIB-6 protein, which has the lowest molecular mass of the six members of group 1 (Figure 2). All SWIB proteins designated to this group are predicted to be targeted to either chloroplasts or mitochondria (TargetP; Figure 2B) (Emanuelsson et al., 2000). To investigate the subcellular localization of the proteins, GFP fusion constructs were prepared and employed for transient transformation assays with tobacco (Nicotiana tabacum) protoplasts. Microscopy inspections revealed that apart from SWIB-1, which was observed in the cytoplasm when fused to GFP, GFP fusion proteins of all group 1 SWIBs were targeted to chloroplasts (SWIB-2, SWIB-3, and SWIB-4), to mitochondria (SWIB-5), or to both organelles (SWIB-6) (Figure 3). All GFP chimeric proteins located in chloroplasts showed punctuate patterns resembling the distribution of nucleoids visualized by the PEND:GFP construct (Terasawa and Sato, 2005) on the red background of the chlorophyll autofluorescence (Figure 3). The mitochondrial localization of SWIB-5:GFP and SWIB-6:GFP proteins could be confirmed by labeling with MitoTracker Orange (see Supplemental Figure 2 online). As a control a construct encoding GFP alone is shown (Figure 3).
Figure 3.
Subcellular Distribution of SWIB:GFP Chimeric Proteins.
The six low molecular mass SWIB domain proteins of Arabidopsis predicted to be located in organelles were fused with GFP at their C termini and transiently expressed in tobacco protoplasts. As controls, constructs encoding PEND:GFP and GFP alone were used. Bars = 7.5 or 4 μm (as indicated by an asterisk).
Association of SWIB-2, SWIB-3, SWIB-4, and SWIB-6 with Chloroplast Nucleoids
To investigate whether the four chloroplast-located SWIB domain proteins (SWIB-2, SWIB-3, SWIB-4, and SWIB-6) might be components of the same subplastidial structures, their potential colocalization was examined by simultaneous transformation of protoplasts with GFP and red fluorescent protein (RFP) fusion constructs. Microscopy analyses showed that SWIB-2:GFP colocalized with SWIB-4:RFP, SWIB-3:GFP with SWIB-6:RFP, and SWIB-4:GFP with SWIB-6:RFP (Figure 4A). In all four GFP/RFP combinations tested, fluorescence signals frequently overlapped, indicating an association of all four SWIB proteins with the same subplastidial structures. To examine whether these structures could represent nucleoids, SWIB-4:GFP and SWIB-6:GFP constructs were transiently cotransformed with a PEND:dsRED construct in tobacco protoplasts. The PEND protein is known to bind to plastid DNA and to be suited for visualization of plastid nucleoids (Terasawa and Sato, 2005). The overlay of green and red fluorescence signals within the chloroplasts revealed that both SWIB-4 and SWIB-6 colocalize with the PEND protein (Figure 4A).
Figure 4.
Colocalization and Sequence Analysis of Plastidial SWIB Proteins.
(A) Colocalization of the plastidial SWIB proteins as well as SWIB-4:GFP and SWIB-6:GFP with the PEND:dsRED protein in chloroplast nucleoids. For better contrast, red fluorescence of chlorophyll was converted into blue. Bar = 2 µm.
(B) Sequence alignment of four chloroplast located stand-alone SWIB domain containing proteins. The SWIB domain is framed. The Lys-rich region similar to the part of histone H1 is written in red color. The alignment was performed with ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and depicted using The Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/).
Sequence Homology, Conserved Regions, and Potential DNA Binding Domains of Plastidial SWIB Proteins
The amino acid sequences of the four SWIB domain proteins targeted to chloroplast nucleoids have high similarity due to the highly conserved SWIB domain (Figure 4B). One of the masses determined by mass spectrometry belongs to a theoretical peptide derived from the SWIB domain of SWIB-6 as revealed by theoretical digestion with trypsin using the program ExPASy (Expert Protein Analysis System; http://www.expasy.ch/; Gasteiger et al., 2003). Masses of five further peptides could correspond to peptides obtained by partial digestion of SWIB-6 or to modified peptides. When the theoretical masses of peptides obtained by digestion with trypsin from the four different proteins were compared, it became obvious that SWIB-2 and SWIB-3 are very similar sharing four identical peptides with masses above 500 D. SWIB-4 and SWIB-6 were found to share two identical peptides with masses above 500 D but not to share peptides with SWIB-2 and SWIB-3. Besides the SWIB domain, SWIB-2, -3, and -4 have low compositional complexity regions (LCRs) in their sequences (Figure 4B). So far no universal function for such LCRs could be elucidated (Coletta et al., 2010). The LCR region (amino acids 31 to 59; Figure 4B) at the N terminus of the SWIB-4 protein shows high similarity to a part of the C-terminal domain of the histone H1 of tobacco (see Supplemental Figure 3A online). Moreover, in silico analysis revealed that this region of SWIB-4, which is extremely rich in Lys, is highly similar to an analogous region found at the N terminus of the CND41 (for chloroplast nucleoid DNA binding protein 41 kD) protein from tobacco (see Supplemental Figure 3A online) that was described to function as a DNA binding domain (Nakano et al., 1997; Murakami et al., 2000). This part of the SWIB-4 protein might represent a domain for binding to DNA. No such motifs could be found in the sequences of SWIB-2, -3, and -6, respectively.
SWIB-4 in TAC-II Binds to DNA as Revealed by Immunological Analysis
For immunological analyses, the peptide sequence KSDSPAKKTPRSTG representing the residues 50 to 63 of SWIB-4 was chosen for production of a SWIB-4–specific antibody (Figure 4B). Protein fractions derived from spinach chloroplasts and subfractions thereof were analyzed by immunoblotting. The predominant form of the protein in chloroplasts had a molecular mass of 17 kD (Figure 5), which is consistent with the molecular mass of the processed form of the in vitro translation product imported into chloroplasts (see Supplemental Figure 3B online). A second higher molecular mass form of the protein could result from posttranslational modification (Figure 5A). Two plastidial and two higher molecular mass nuclear forms of the SWIB-4 protein also could be detected in protein fractions isolated from chloroplasts and nuclei of Arabidopsis leaves (see Supplemental Figure 4 online).
Figure 5.
Immunological Detection of the SWIB-4 Protein in Chloroplast Subfractions and in TAC and DNA Binding Analysis.
(A) Immunoreactions were performed with total protein (TP), chloroplast protein (C), membrane (M), and stroma (S) fractions extracted from spinach leaves as well as TAC-I and TAC-II protein extracts (10 µg), respectively. To demonstrate the purity of the chloroplast subfractions, immunoblots decorated with antibodies directed toward cytochrome b559 apoprotein A (PsbE) and the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RbcL) are shown. In addition, the level of MFP1 in the TAC-I and TAC-II fraction was determined. For comparison, the Coomassie blue staining of the protein fractions is shown (CBB).
(B) Protein/DNA gel blot analysis with TAC-II proteins. After SDS-PAGE and blotting, TAC-II proteins were renatured and incubated with a radioactively labeled 16S rDNA probe. The sample shown in lane 1 was directly incubated with DNA, while the samples shown in lanes 2 and 3 prior to incubation with DNA were incubated with the anti-SWIB-4 antibody or with BSA, respectively. The protein band preventing binding to DNA in the presence of the SWIB-4 antibody is indicated by an arrow. For comparison, silver staining of the fraction is shown in lane 4.
We further tested the presence of SWIB-4 in TAC-I and TAC-II. When TAC-I and TAC-II fractions, having the same amount of protein, were analyzed, the level of immunoreactive protein detected by the SWIB-4 antibody was higher in TAC-II than in TAC-I, indicating an enrichment of the SWIB-4 protein during purification of TAC. In comparison, the relative level of the nucleoid-associated protein MFP1 decreased during purification of TAC-I as shown previously with corresponding fractions from barley chloroplasts (Melonek et al., 2010). SWIB-4 binds to DNA when it is not altered by posttranslational modification because TAC fractions contained only the lower molecular mass form of SWIB-4 (Figure 5A).
Next, we examined whether SWIB-4 in TAC-II binds to DNA. For this purpose, TAC-II proteins after transfer onto nitrocellulose membranes were renatured. By incubation with a radiolabeled 16S rDNA probe, three low molecular mass protein bands were detected (Figure 5B, lane 1). When the membrane prior to incubation with DNA was incubated with the SWIB-4 antibody, the intensity of the band corresponding to a DNA binding protein of ∼17 kD was diminished (Figure 5B, lane 2). This result indicates that SWIB-4 might represent one of the low molecular mass DNA binding proteins of TAC fractions detected previously (Bülow et al., 1987).
Remodeling of Nucleoid Architecture by SWIB-4
To investigate whether SWIB-4 might have an architectural function in nucleoids, we overexpressed the gene encoding SWIB-4 in E. coli. DNA staining with 4′,6-diamidino-2-phenylindole (DAPI) showed that after induction with isopropyl-β-d-thiogalactopyranoside (IPTG), cells accumulating the recombinant SWIB-4 protein had highly condensed nucleoids, while control cells showed a uniform distribution of DNA (Figure 6A). Accumulation of SWIB-4 (Figure 6B) coincided with growth arrest of the cells for several hours shortly after addition of IPTG (Figure 6C). A similar effect on growth of E. coli cells and compaction of nucleoids had been observed previously when the abundant histone-like protein H-NS was overproduced (Spassky et al., 1984; Spurio et al., 1992).
Figure 6.
Impact of Chloroplast SWIB-4 on Compactness of Bacterial Nucleoids and Complementation of the H-NS–Deficient Mutant.
(A) The M15 E. coli cells were transformed either with the pQE30 vector containing the full coding sequence of SWIB-4 or with the empty pQE30 vector. Cells were cultivated at 37°C and the IPTG was added at an OD600 ranging from 0.7 to 1. For visualization of the nucleoids, the E. coli cells were stained with DAPI. Bar = 10 µm.
(B) Immunological analysis with the SWIB-4 antibody. Proteins of bacterial cultures were resolved by denaturing gel electrophoresis on a 14% (w/v) polyacrylamide gel and either transferred onto nitrocellulose membrane or stained with Coomassie blue. The arrow indicates the protein band representing SWIB-4. Ctrl, control (uninduced E. coli cells).
(C) Effect of nucleoid condensation mediated by SWIB-4 on E. coli cell proliferation. E. coli cells expressing either the empty pQE30 vector or the pQE30 vector containing the SWIB-4 coding sequence were grown at 37°C, and after addition of IPTG, the OD600 was measured at different time points.
(D) Complementation of the H-NS–deficient mutant by SWIB-4. E. coli Δhns::FRT mutant cells grown on MacConkey agar plates supplemented with salicin, Δhns::FRT+pQE30 mutant cells transformed with the empty plasmid, Δhns::FRT+pQE30+SWIB-4 mutant cells transformed with plasmid encoding the Arabidopsis SWIB-4 gene, as well as E. coli wild-type (WT) cells are shown. Arabidopsis SWIB-4 can functionally replace the E. coli H-NS protein in vivo as demonstrated by the colorless colonies formed by the Δhns::FRT+pQE30+SWIB-4 cells.
To investigate whether SWIB-4 could be a functional analog of an abundant histone-like protein termed H-NS, we aimed at complementation of an E. coli mutant lacking this NAP (Spassky et al., 1984; Spurio et al., 1992). The H-NS protein was shown to induce compaction of nucleoids and to affect expression of ∼5% of the E. coli genes (Hommais et al., 2001). Among the genes repressed by H-NS is the cryptic β-glucoside (bgl) operon, which encodes all gene products necessary for uptake and fermentation of the aryl-β,d-glucosides, such as salicin or arbutin (Higgins et al., 1988; Schnetz, 1995; Caramel and Schnetz, 1998). Δhns mutant cells that can metabolize salicin appear as red colonies on salicin-containing indicator plates, whereas colonies unable to ferment salicin are colorless (Schnetz et al., 1987). To test whether the Arabidopsis SWIB-4 gene can functionally substitute for the E. coli hns gene, cells of the Δhns mutant strain were transformed with the pQE30 plasmid containing SWIB-4. After 2 d of growth on indicator plates, colonies of the hns mutant and the hns mutant transformed with pQE30 plasmid were red (Figure 6D), whereas colonies of the Δhns mutant strain transformed with a control pQE30+SWIB-4 plasmid stayed colorless as colonies of the wild type (Figure 6D). This result showed that SWIB-4 in vivo can functionally replace the E. coli H-NS protein.
The T-DNA insertion mutants so far available have insertions either in 5′ untranslated region (swib-4-1 and -2) or in the intron (swib-4-3) of the SWIB-4 gene (see Supplemental Figure 5A and Supplemental Table 1 online). The 20-fold to fivefold enhanced transcript levels in some of the mutants were found to coincide with retarded postgermination development of seedlings (see Supplemental Figures 5B and 5C online).
DISCUSSION
Our proteome analysis of a fraction highly enriched in transcriptionally active chromosomes prepared from spinach chloroplasts (TAC-II) revealed the presence of architectural nucleoid-associated proteins of plastids (ptNAP) possessing domains typically found in proteins with functions in chromatin organization, such as SMC, Armadillo, SANT, and SWIB. The ptNAPs identified in TAC-II have not been detected in TAC fractions from mustard and Arabidopsis (Pfalz et al., 2006), indicating that a second round of gel filtration after precipitation of conventionally prepared TAC-I with protamine sulfate (Krause and Krupinska, 2000) is suited to enrich proteins having low abundance in the conventional TAC preparations. On the other hand, due to limitations in the resolution of highly charged proteins by isoelectric focusing, several DNA binding proteins of the core nucleoids might have escaped detection. As described previously, the specific transcriptional activity of the TAC-II fraction was enhanced by more than 50-fold compared with TAC-I (Krause and Krupinska, 2000). During preparation of TAC-II from TAC-I, several proteins most probably loosely attached to DNA were lost, for example, MFP1 and PTAC1, which is identical with Whirly1 (Melonek et al., 2010). It is likely that the TAC-II fraction is enriched in proteins belonging to the nucleoid core, which has been described as central body by electron microscopy (Herrmann et al., 1974; Yoshida et al., 1978). Also for nucleoids of human mitochondria, a layered structure has been proposed (Bogenhagen et al., 2008). In this model of mitochondrial nucleoid architecture, replication and transcription were suggested to occur in the central core, whereas translation and complex assembly occur in peripheral regions (Bogenhagen et al., 2008).
The smallest ptNAP identified in the TAC-II proteome has a SWIB domain. SWIB domain–containing proteins were also found in the proteome of TAC-II fractions isolated from barley plastids (data not shown) and therefore seem to be essential components of plastid nucleoids. In a proteome study on rice etioplasts, a low molecular mass homolog of Arabidopsis SWIB proteins was identified (von Zychlinski et al., 2005). SWIB-3 was recently identified in a comprehensive proteome analysis of maize (Zea mays) chloroplast nucleoids prepared by sucrose gradient centrifugation (Majeran et al., 2012). In the TAC-II fraction, only SWIB-6 was detectable and none of the other three putative plastidial SWIB proteins. This could be due to unusual high isoelectric points of the spinach SWIB proteins whose sequences are not known so far, likely preventing their resolution by isoelectric focusing.
Database analyses showed that the SWIB domain protein identified by our study of a nucleoid subproteome belongs to a group of low molecular mass SWIB domain proteins predicted to be targeted to either chloroplasts or mitochondria. By fusion with GFP, four members of this group could be observed to be targeted to chloroplasts, where they were shown to have a speckled distribution resembling the distribution of the nucleoids. Colocalization with GFP and RFP fusion proteins showed that all four members of the group colocalize in the nucleoids. For SWIB-4 and SWIB-6, the nucleoid localization was furthermore shown by cotransformation with a PEND:dsRED construct. In addition, SWIB-6:GFP also was found to be targeted to mitochondria, whereas SWIB-4:GFP also was shown to be located in the nucleus. The nuclear localization could be confirmed by immunological analysis of isolated nuclei. Several nuclear regulatory DNA binding proteins are predicted to be dually targeted to both organelles or one organelle and the nucleus (Small et al., 1998; Schwacke et al., 2007; Krause and Krupinska, 2009). Such proteins might have roles in coordination of the nuclear genome and the plastid nucleoids, which together with the chondriome constitute the integrated genetic system of the plant cell (Herrmann et al., 2003). Most of the already described proteins targeted to two or even three of the DNA containing compartments have functions in the maintenance of DNA, telomere structuring, or in regulation of gene expression (Krause and Krupinska, 2009; Hammani et al., 2011).
Immunological analyses showed that SWIB-4 is indeed a component of TAC-II. When the same amounts of protein were loaded onto an SDS gel, the level of SWIB-4 detected in TAC-II was higher than that in the TAC-I, indicating that during enrichment of the TAC core components, the SWIB-4 protein also got enriched. SWIB-4 was shown to colocalize with the other three low molecular SWIB proteins targeted to chloroplasts. Its enrichment in TAC-II suggests that all plastidial SWIB proteins are core components of the nucleoids.
Among the four SWIB proteins shown to be targeted to chloroplast nucleoids when fused to GFP, SWIB-4 is the only one having in addition to the SWIB domain a histone-H1-like domain, making it an excellent candidate for functional replacement of the original bacterial HLPs in the plastid nucleoid. If SWIB-4 is indeed functionally related to bacterial architectural proteins, it should influence the shape of chloroplast nucleoids. The structure of plastid nucleoids and DNA topology were proposed to have important consequences for gene expression (Bogorad, 1991; Salvador et al., 1998), which is known to undergo dramatic changes during chloroplast development (Nemoto et al., 1988, 1989; Baumgartner et al., 1989; Krupinska and Falk, 1994).
We propose that the SWIB-4 protein, which has a low molecular mass, high isoelectric point, and a Lys-rich region, is a plastid-located NAP. Bacterial NAPs (e.g., the 9-kD HU, 11-kD Fis [for factor for inversion stimulation] and 15 kD H-NS proteins) have in common a low molecular mass, a high Lys content, and nonspecific binding to DNA (Luijsterburg et al., 2006; Dillon and Dorman, 2010) and to other nucleic acids (Balandina et al., 2002; Brescia et al., 2004; Kamashev et al., 2008). SWIB-4 has a histone-like domain that is likely to be involved in binding to DNA as well as RNA and single-stranded DNA as also shown for histones (Spelsberg et al., 1970).
Overexpression in E. coli revealed the architectural activity of chloroplast SWIB-4. This result confirmed that bacteria and eukaryotes use functionally conserved basic low molecular mass proteins for shaping and remodeling of their genomes (Luijsterburg et al., 2006). Complementation of an E. coli mutant lacking H-NS suggests that SWIB-4 is functionally equivalent to this most abundant NAP that when overproduced was shown to condense nucleoids resulting in death of the E. coli cells (Spassky et al., 1984; Spurio et al., 1992). For mutants having up to 20-fold enhanced level of the SWIB-4 transcript, a delayed postgermination development could be observed (see Supplemental Figure 5 online). Whether induced overexpression of the gene in plants would lead to a change in plastid nucleoid architecture and an arrest of growth remains to be demonstrated.
When we compared the expression of the genes encoding the four SWIB proteins of Arabidopsis at different developmental stages using the data of Genevestigator (Zimmermann et al., 2004), it became obvious that the gene encoding SWIB-4 has the lowest and almost constitutive level of expression, while the expression profiles of genes encoding SWIB-2, -3, and -6 show fluctuations (see Supplemental Figure 6 online). Expression of the gene encoding SWIB-4 is enhanced in plants at the time of bolting and in siliques. This pattern of expression is in accordance with a function of SWIB-4 in meristematic tissue where it might silence transcriptional activity in plastids by compaction of nucleoids when it is overexpressed.
The Lys-rich N-terminal sequence of the mature chloroplast SWIB-4 has high similarity with the sequence of the histone-H1-like protein (Hc1) from Chlamydophila felis, a chlamydial bacterium possessing the only two SWIB domain proteins described for prokaryotes. One is a low molecular stand-alone SWIB protein, and in the second the SWIB domain is fused to the COOH terminus of topoisomerase I (Barry et al., 1992). It has been demonstrated that this H1-like protein together with the SWIB domain protein functions in remodeling of the nucleoid during the chlamydial life cycle (Hackstadt et al., 1991; Barry et al., 1992). In eukaryotes, histone H1 is acting as a DNA bridging protein (Hendzel et al., 2004). Dinoflagellates, despite being eukaryotes, are known to lack histones and corresponding nucleosome structures. Three histone-like proteins, HCc1, HCc2, and HCc3, found in dinoflagellates were shown to share homology with both eukaryotic histone H1 and prokaryotic HU (Wong et al., 2003). While HCc1 and HCc2 were shown to complement mutants lacking HU, HCc3 did not complement the phenotype of HU-deficient cells and is thus not functionally equivalent to HU (Chang et al., 1999). Similarly to HCc3, SWIB-4 could not reconstitute mutants lacking HU protein (data not shown).
In accordance with the presence of histone-like proteins in plastids, in recent years, evidence has accumulated that counterparts of proteins known to be involved in histone modifications in the nucleus do also occur in plastids. One example is the Arabidopsis SET domain–containing Trithorax-related protein5 (ATXR5) acting as a monomethyltransferase in the nucleus (Raynaud et al., 2006; Jacob et al., 2009), which was found by fusion with GFP to be dually targeted to plastids and the nucleus (Raynaud et al., 2006). Other examples are two rice histone deacetylases (Os HDAC10 and Os HDAC6) (Chung et al., 2009).
Overproduction of SWIB-4 of Arabidopsis as well as HCc3 from dinoflagellates in E. coli resulted in increased compactness of nucleoids. The condensing effect of these proteins on nucleoid structure is in accordance with a function as DNA bridger. The major DNA-bridging protein in bacterial nucleoids is H-NS (Bertin et al., 2001; Dame et al., 2005; Luijsterburg et al., 2008). Complementation of a mutant lacking H-NS by chloroplast SWIB-4 indicates that the two proteins are functionally equivalent (Bertin et al., 2001; Dame et al., 2005). Recently, it has been reported that the architectural activity of H-NS can switch between bridging and stiffening and that the switch depends on pH and divalent cations (Barber, 1986; Liu et al., 2010). In chloroplasts, divalent cations are counterions of protons forming gradients above the thylakoid membrane (Izawa and Good, 1966; Barber, 1986). It will be an exciting question for future research whether SWIB-4 is involved in the adjustment of the plastid nucleoid architecture and function in response to photosynthetic performance of chloroplasts.
METHODS
Plant Materials
Spinach (Spinacia oleracea cv Deutscher Frisee) leaves used for chloroplast preparation were purchased from the local market. Tobacco (Nicotiana tabacum cv Xanthi) plants used for protoplast transformation were grown for 5 to 6 weeks on sterilized Murashige and Skoog medium at 24°C with a 16-h photoperiod.
Isolation of TAC Fractions
For isolation of TAC fractions, chloroplasts were prepared from spinach leaves by an established procedure (Douce et al., 1973). TAC extracts from spinach chloroplasts were prepared as described previously (Krause and Krupinska, 2000). To examine the transcriptional activity, aliquots of TAC fractions were incubated at 30°C with radioactively labeled α-32-UTP (Hartmann Analytic) as described (Krupinska and Falk, 1994).
Two-Dimensional Separation of TAC Proteins and Mass Spectrometry
Prior to 2-DE, TAC protein fractions were treated with DNase I and RNase A (MBI Fermentas) and precipitated with chloroform/methanol following the protocol of Wessel and Flügge (1984). Protein concentration was estimated with the Plus One 2D-Quant Kit (GE Healthcare). 2-DE and protein staining were performed according to Witzel et al. (2009), with the following modifications: 50 µg of TAC proteins were loaded on immobilized pH gradient dry gel strips (7 cm, pH gradient 3 to 10; GE Healthcare). The second dimension was performed on 11.25% (w/v) polyacrylamide gels containing 0.1% (w/v) of SDS. Separated proteins were stained with colloidal Coomassie Brilliant Blue (Gel Code Blue; Thermo Fisher Scientific). Protein spots were manually excised and trypsin digested (Porcine Sequencing Grade; Promega) as described (Witzel et al., 2009). Acquisition of peptide mass fingerprint data and corresponding LIFT spectra was performed using an ultrafleXtreme matrix-assisted laser desorption/ionization time-of-flight device (Bruker Daltonics) equipped with a Smartbeam-II laser with a repetition rate of 1000 Hz. The spectra were calibrated using external calibration and subsequent internal mass correction. For databank searching, Biotools 3.2 software (Bruker Daltonics) with the implemented MASCOT search engine (Matrix Science) was used, searching for Viridiplantae in the nonredundant National Center for Biotechnology Information database (26/07/2010, Green Plants 284943 sequences). Search parameters were as follows: monoisotopic mass accuracy; 50 to 100 ppm tolerance; fragment tolerance of 0.3 D; missed cleavages 1; and the allowed variable modifications were oxidation (Met), propionamide (Cys), and carbamidomethyl (Cys).
Phylogenetic Analysis
The amino acid sequences of 20 Arabidopsis SWIB domain–containing proteins were obtained from TAIR (www.arabidopsis.org). The multiple sequence alignment was performed with ClustalW (Larkin et al., 2007) and is available as Supplemental Data Set 1 online. The phylogenetic tree was constructed using MEGA5 software (Tamura et al., 2011) based on the neighbor-joining method (Saitou and Nei, 1987) with parameters of Poisson correction model, complete deletion of the gaps, and 1000 bootstrap replicates. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Cloning of GFP Fusion Constructs
For preparation of GFP constructs, the complete coding sequences of the At2g14880, At4g34290, At3g03590, and At2g356050 genes were amplified from Arabidopsis total cDNA by iProof High-Fidelity DNA polymerase (Bio-Rad) using the following specific forward and reverse primers: At3g03590for, 5′-caccatgtcttccgttgca-3′; At3g03590rev, 5′-agcagtcttcacaaagtgc-3′; At2g14880for, 5′-caccatggcggtttcttct-3′; At2g14880rev, 5′-gaggaagtgaggaccgat-3′; At4g34290for, 5′-caccatggctctttcttctg-3′; At4g34290rev, 5′-aaagtgaggaccaatgag-3′; At2g35605for, 5′-caccatgtcgagggttttc-3′; and At2g35605rev, 5′-agcagacttaggaaaatgttg-3′. The forward primers contained a CACC overhang for cloning into pENTR/D/TOPO vector (Invitrogen). pENTR plasmids encoding the particular SWIB genes were used for site-specific recombination into binary Gateway expression vector pB7FWG2,0 or pB7RWG2 (Invitrogen). The PEND:GFP and PEND:dsRED constructs were prepared as described (Terasawa and Sato, 2005; Terasawa et al., 2005). As a control, the coding sequence for the green fluorescent protein was cloned into the pBluescript vector. All constructs were under control of the 35S cauliflower mosaic virus promoter.
Transient Transformation Assays
Tobacco protoplasts used for transient transformation assay were prepared as described (Nagy and Maliga, 1976). Polyethylene glycol–mediated transformation was performed as described (Negrutiu et al., 1987). Briefly, 50 µg of plasmid DNA was gently mixed with 2 × 106 protoplasts and incubated in 40% (w/v) polyethylene glycol 1500 (Merck). In colocalization experiments, protoplasts were transformed with two plasmids simultaneously. After 24 h incubation, protoplasts were inspected by fluorescence microscopy (Leica TCS SP5; Leica Microsystems). For detection of GFP fluorescence and chlorophyll autofluorescence, the samples have been excited by the 488- and 458-nm lines of an argon laser, respectively.
Protein Isolation and Immunoblot Analysis
For extraction of total protein fractions, spinach leaves were ground in liquid nitrogen. Protein concentrations were determined using Roti-Nanoquant according to the manufacturer’s instructions (Roth) based on the method of Bradford (1976). For SDS-PAGE, the buffer system of Laemmli (1970) was used. For an efficient separation of proteins having low molecular masses, 16% (w/v) polyacrylamide gels were used and proteins were blotted onto nitrocellulose (Schleicher and Schuell). For immunoblot analysis, the polyclonal anti SWIB-4 antiserum was diluted 1:1000 with buffer consisting of 50 mM Tris/HCl, pH 7.4, and 150 mM NaCl. The antibody was directed toward the peptide KSDSPKKTPRSTG (residues 50 to 63 of the protein) and was raised in rabbits by Biogenes. Further primary antibodies used in our study were directed toward cytochrome b559 apoprotein A (PsbE; Vallon et al., 1987), large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (RbcL; Agrisera), and MFP1 (Jeong et al., 2003).
DNA Binding Blot Analysis of TAC Proteins
DNA binding assays with TAC proteins were performed as described (Bülow et al., 1987) with minor modifications. About 5 µg of total TAC proteins from spinach were separated under denaturing conditions in 14% (w/v) polyacrylamide gels (Laemmli, 1970) and subsequently transferred onto a nitrocellulose membrane employing a transfer buffer consisting of 50 mM NaCl, 2 mM EDTA, 0.1 mM DTT, and 10 mM Tris/HCl, pH 7.0. Renaturation of the proteins took place overnight in renaturation buffer consisting of 2 mM EDTA, 10 mM Tris/HCl, pH 7.0, 0.02% BSA, 1% (w/v) low-fat milk powder, and 0.02% (w/v) polyvinylpyrrolidone. The membrane was cut into two halves, and one half was incubated with the SWIB-4–specific antibody diluted 1:50 with buffer consisting of 50 mM Tris/HCl, pH 7.4, and 150 mM NaCl. The other half of the membrane was incubated with the buffer only. After washing in transfer buffer, the membrane was incubated with an [α-32P]-labeled 16S rDNA probe in renaturation buffer. After hybridization, the membranes were washed three times for 15 min with renaturation buffer.
DNA Condensation Assays in Escherichia coli Cells
The full coding sequence of the Arabidopsis SWIB-4 gene was cloned into the pQE30 vector (Qiagen) for the transformation in E. coli M15 cells. The cultures were grown in Luria-Bertani medium at 37°C for several hours and IPTG was added at OD600 0.7-1 to a final concentration of 1 mM. After staining with DAPI, the cells were examined by differential interference contrast and fluorescence microscopy with a Zeiss Axiophot microscope (Carl Zeiss).
Complementation of the E. coli hns Mutant
The E. coli strain hns::FRT, which carries a precise deletion of hns (Venkatesh et al., 2010), was transformed with the pQE30 plasmid encoding the Arabidopsis SWIB-4 gene and with the empty pQE30 plasmid as a control. The bacteria were streaked on MacConkey-salicin indicator plates, which were prepared from BD Difco MacConkey Agar Base (BD Diagnostics) and salicin (Sigma-Aldrich) as described (Schnetz et al., 1987). When required, the medium was supplemented with ampicillin (100 μg/ml) and IPTG (1 mM).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g48600 (SWIB-1), At2g14880 (SWIB-2), At4g34290 (SWIB-3), At3g03590 (SWIB-4), At1g34760 (SWIB-5), and At2g35605 (SWIB-6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Two-Dimensional Separation of the Highly Enriched Transcriptionally Active Chromosome (TAC-II) Isolated from Spinach Chloroplasts.
Supplemental Figure 2. MitoTracker Orange Labeling of Onion Epidermal Cells Transiently Expressing SWIB-5:GFP and SWIB-6:GFP Constructs.
Supplemental Figure 3. Sequence Alignment of the Histone H1 Motif of the SWIB-4 Protein and in Vitro Import Assays with Pea Chloroplasts.
Supplemental Figure 4. Immunological Detection of SWIB-4 in Chloroplasts and Nuclei Prepared from Leaves of Arabidopsis.
Supplemental Figure 5. Molecular Characterization of SWIB-4 T-DNA Insertion Mutants.
Supplemental Figure 6. Development-Dependent Expression of Genes Encoding SWIB-2 to -6 Proteins from Group 1 of the SWIB Domain Family of Arabidopsis.
Supplemental Table 1. List of Primers Used for Molecular Characterization of swib-4 Mutants.
Supplemental Data Set 1. Proteins Identified in the Spots of the TAC-II 2-DE Gel Shown in Supplemental Figure 1.
Supplemental Data Set 2. Amino Acid Alignment Used to Generate Figure 2A.
Supplementary Material
Acknowledgments
We thank Karin Schnetz (University of Cologne, Germany) for expert advice and for providing E. coli K12 and Δhns::FRT strains for complementation studies. We thank Central Microscopy (Biology Center, Christian-Albrechts-University of Kiel) for providing the confocal microscope and Christine Desel (Christian-Albrechts-University of Kiel) for expert assistance and advice. We also thank Iris Meier (The Ohio State University, Columbus, OH) for providing the antibody directed towards MFP1 and René Lorbiecke (Hamburg University, Germany) for providing the PEND:GFP and PEND:dsRED plasmids. We thank Annegret Wolf (IPK Gatersleben, Germany) and Andreas Prescher (Christian-Albrechts-University of Kiel) for technical assistance and Kirsten Krause (University of Tromsø, Norway) for stimulating discussions. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Kr1350/8).
AUTHOR CONTRIBUTIONS
K.K. and H.-P.M. designed the research. J.M., A.M., and M.T. performed research. J.M. and A.M. analyzed data. J.M. and K.K. wrote the article.
Glossary
- TAC
transcriptionally active chromosome
- GFP
green fluorescent protein
- NAP
nucleoid-associated protein
- 2-DE
two-dimensional gel electrophoresis
- ptNAP
to be defined
- TAIR
The Arabidopsis Information Resource
- RFP
red fluorescent protein
- LCR
low compositional complexity regions
- IPTG
to be defined
References
- Andersson M.X., Goksör M., Sandelius A.S. (2007). Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J. Biol. Chem. 282: 1170–1174 [DOI] [PubMed] [Google Scholar]
- Aravind L., Koonin E.V. (2000). SAP–A putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25: 112–114 [DOI] [PubMed] [Google Scholar]
- Babujee L., Wurtz V., Ma C., Lueder F., Soni P., van Dorsselaer A., Reumann S. (2010). The proteome map of spinach leaf peroxisomes indicates partial compartmentalization of phylloquinone (vitamin K1) biosynthesis in plant peroxisomes. J. Exp. Bot. 61: 1441–1453 [DOI] [PubMed] [Google Scholar]
- Balandina A., Kamashev D., Rouviere-Yaniv J. (2002). The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 277: 27622–27628 [DOI] [PubMed] [Google Scholar]
- Barber J. (1986). Regulation of energy transfer by cations and protein phosphorylation in relation to thylakoid membrane organisation. Photosynth. Res. 10: 243–253 [DOI] [PubMed] [Google Scholar]
- Barry C.E., III, Hayes S.F., Hackstadt T. (1992). Nucleoid condensation in Escherichia coli that express a chlamydial histone homolog. Science 256: 377–379 [DOI] [PubMed] [Google Scholar]
- Baumgartner B.J., Rapp J.C., Mullet J.E. (1989). Plastid transcription activity and DNA copy number increase early in barley chloroplast development. Plant Physiol. 89: 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer R.G., Stael S., Csaszar E., Teige M. (2011). Mining the soluble chloroplast proteome by affinity chromatography. Proteomics 11: 1287–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey R., Hart S.E., Shirai H., Mizuguchi K. (2002). The SWIB and the MDM2 domains are homologous and share a common fold. Bioinformatics 18: 626–630 [DOI] [PubMed] [Google Scholar]
- Bertin P., Hommais F., Krin E., Soutourina O., Tendeng C., Derzelle S., Danchin A. (2001). H-NS and H-NS-like proteins in Gram-negative bacteria and their multiple role in the regulation of bacterial metabolism. Biochimie 83: 235–241 [DOI] [PubMed] [Google Scholar]
- Bogenhagen D.F., Rousseau D., Burke S. (2008). The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283: 3665–3675 [DOI] [PubMed] [Google Scholar]
- Bogorad L. (1991). Replication and transcription of plastid DNA. In Cell Culture and Somatic Cell Genetics of Plants, Vol. 7A, L. Bogorad and I.K. Vasil, eds (San Diego, CA: Academic Press), pp. 93–124 [Google Scholar]
- Boyer L.A., Latek R.R., Peterson C.L. (2004). The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5: 158–163 [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brescia C.C., Kaw M.K., Sledjeski D.D. (2004). The DNA binding protein H-NS binds to and alters the stability of RNA in vitro and in vivo. J. Mol. Biol. 339: 505–514 [DOI] [PubMed] [Google Scholar]
- Briat J.-F., Letoffe S., Mache R., Rouviere-Yaniv J. (1984). Similarity between the bacterial histone-like protein HU and a protein from spinach chloroplasts. FEBS Lett. 172: 75–79 [Google Scholar]
- Bülow S., Reiss T., Link G. (1987). DNA-binding proteins of the transcriptionally active chromosome from mustard (Sinapis alba L.) chloroplasts. Curr. Genet. 12: 157–159 [Google Scholar]
- Cairns B.R., Levinson R.S., Yamamoto K.R., Kornberg R.D. (1996). Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 10: 2131–2144 [DOI] [PubMed] [Google Scholar]
- Cannon G.C., Ward L.N., Case C.I., Heinhorst S. (1999). The 68 kDa DNA compacting nucleoid protein from soybean chloroplasts inhibits DNA synthesis in vitro. Plant Mol. Biol. 39: 835–845 [DOI] [PubMed] [Google Scholar]
- Caramel A., Schnetz K. (1998). Lac and lambda repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J. Mol. Biol. 284: 875–883 [DOI] [PubMed] [Google Scholar]
- Chang C.C., Sheen J., Bligny M., Niwa Y., Lerbs-Mache S., Stern D.B. (1999). Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell 11: 911–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung P.J., Kim Y.S., Park S.H., Nahm B.H., Kim J.K. (2009). Subcellular localization of rice histone deacetylases in organelles. FEBS Lett. 583: 2249–2254 [DOI] [PubMed] [Google Scholar]
- Coletta A., Pinney J.W., Solís D.Y., Marsh J., Pettifer S.R., Attwood T.K. (2010). Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame R.T., Luijsterburg M.S., Krin E., Bertin P.N., Wagner R., Wuite G.J.L. (2005). DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 187: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S.C., Dorman C.J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8: 185–195 [DOI] [PubMed] [Google Scholar]
- Dorman C.J., Deighan P. (2003). Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13: 179–184 [DOI] [PubMed] [Google Scholar]
- Douce R., Holtz R.B., Benson A.A. (1973). Isolation and properties of the envelope of spinach chloroplasts. J. Biol. Chem. 248: 7215–7222 [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31: 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski E., Miao Y., Mulisch M., Krupinska K. (2008). Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 147: 1800–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P.L., Knust T. (2009). Dynamics of the bacterial SMC complex and SMC-like proteins involved in DNA repair. Chromosome Res. 17: 265–275 [DOI] [PubMed] [Google Scholar]
- Gruissem W., Greenberg B.M., Zurawski G., Hallick R.B. (1986). Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol. 118: 253–270 [DOI] [PubMed] [Google Scholar]
- Hackstadt T., Baehr W., Ying Y. (1991). Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc. Natl. Acad. Sci. USA 88: 3937–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick R.B., Lipper C., Richards O.C., Rutter W.J. (1976). Isolation of a transcriptionally active chromosome from chloroplasts of Euglena gracilis. Biochemistry 15: 3039–3045 [DOI] [PubMed] [Google Scholar]
- Hammani K., Gobert A., Hleibieh K., Choulier L., Small I., Giegé P. (2011). An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23: 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Lever M.A., Crawford E., Th’ng J.P.H. (2004). The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279: 20028–20034 [DOI] [PubMed] [Google Scholar]
- Herrmann R.G., Kowallik K.V., Bohnert H.J. (1974). Structural and functional aspects of the plastome. I. The organization of the plastome. Port. Acta Biol. 14: 91–110 [Google Scholar]
- Herrmann R.G., Maier R.M., Schmitz-Linneweber C. (2003). Eukaryotic genome evolution: rearrangement and coevolution of compartmentalized genetic information. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 87–97, discussion 97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C.F., Dorman C.J., Stirling D.A., Waddell L., Booth I.R., May G., Bremer E. (1988). A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52: 569–584 [DOI] [PubMed] [Google Scholar]
- Hommais F., Krin E., Laurent-Winter C., Soutourina O., Malpertuy A., Le Caer J.P., Danchin A., Bertin P. (2001). Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40: 20–36 [DOI] [PubMed] [Google Scholar]
- Igloi G.L., Kössel H. (1992). The transcriptional apparatus of chloroplasts. Crit. Rev. Plant Sci. 10: 525–558 [Google Scholar]
- Izawa S., Good N.E. (1966). Effects of salts and electron transport on the conformation of isolated chloroplasts. II. Electron microscopy. Plant Physiol. 41: 544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y., Feng S.H., LeBlanc C.A., Bernatavichute Y.V., Stroud H., Cokus S., Johnson L.M., Pellegrini M., Jacobsen S.E., Michaels S.D. (2009). ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 16: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.Y., Rose A., Meier I. (2003). MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzmanowski A. (2007). SWI/SNF chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta 1769: 330–345 [DOI] [PubMed] [Google Scholar]
- Kamashev D., Balandina A., Mazur A.K., Arimondo P.B., Rouviere-Yaniv J. (2008). HU binds and folds single-stranded DNA. Nucleic Acids Res. 36: 1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjölander K., Gruissem W., Baginsky S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Krause K., Krupinska K. (2000). Molecular and functional properties of highly purified transcriptionally active chromosomes from spinach chloroplasts. Physiol. Plant. 109: 188–195 [Google Scholar]
- Krause K., Krupinska K. (2009). Nuclear regulators with a second home in organelles. Trends Plant Sci. 14: 194–199 [DOI] [PubMed] [Google Scholar]
- Krupinska K., Falk J. (1994). Changes in RNA-polymerase activity during biogenesis, maturation and senescence of barley chloroplasts. Comparative analysis of transcripts synthesized either in run-on assays or by transcriptionally active chromosomes. J. Plant Physiol. 143: 298–305 [Google Scholar]
- Kuroiwa T., Kuroiwa H., Sakai A., Takahashi H., Toda K., Itoh R. (1998). The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 181: 1–41 [DOI] [PubMed] [Google Scholar]
- Kuroiwa T., Suzuki T. (1981). Circular nucleoids isolated from chloroplasts in a brown alga Ectocarpus siliculosus. Exp. Cell Res. 134: 457–461 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lilly J.W., Havey M.J., Jackson S.A., Jiang J. (2001). Cytogenomic analyses reveal the structural plasticity of the chloroplast genome in higher plants. Plant Cell 13: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chen H., Kenney L.J., Yan J.J. (2010). A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 24: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg M.S., Noom M.C., Wuite G.J.L., Dame R.T. (2006). The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: molecular perspective. J. Struct. Biol. 156: 262–272 [DOI] [PubMed] [Google Scholar]
- Luijsterburg M.S., White M.F., van Driel R., Dame R.T. (2008). The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 43: 393–418 [DOI] [PubMed] [Google Scholar]
- Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: A new conceptual framework for nucleoid functions. Plant Physiol. 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A., Parent J.S., Véronneau-Lafortune F., Joyeux A., Lang B.F., Brisson N. (2009). Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 14693–14698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melonek J., Mulisch M., Schmitz-Linneweber C., Grabowski E., Hensel G., Krupinska K. (2010). Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232: 471–481 [DOI] [PubMed] [Google Scholar]
- Murakami S., Kondo Y., Nakano T., Sato F. (2000). Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Lett. 468: 15–18 [DOI] [PubMed] [Google Scholar]
- Nagy J.I., Maliga P. (1976). Callus induction and plant regeneration from mesophyll protoplasts of Nicotiana sylvestris. Z. Pflanzenphysiol. 78: 453–455 [Google Scholar]
- Nakano T., Murakami S., Shoji T., Yoshida S., Yamada Y., Sato F. (1997). A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell 9: 1673–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutiu I., Shillito R., Potrykus I., Biasini G., Sala F. (1987). Hybrid genes in the analysis of transformation conditions. I. Setting up a simple method for direct gene transfer in plant protoplasts. Plant Mol. Biol. 8: 363–373 [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Kawano S., Nakamura S., Mita T., Nagata T., Kuroiwa T. (1988). Studies on plastid-nuclei (nucleoids) in Nicotiana tabacum L. I. Isolation of proplastid-nuclei from cultured cells and identification of proplastid-nuclear proteins. Plant Cell Physiol. 29: 167–177 [Google Scholar]
- Nemoto Y., Nagata T., Kuroiwa T. (1989). Studies on plastid-nuclei (nucleoids) in Nicotiana tabacum L. II. Disassembly and reassembly of proplastid-nuclei isolated from cultured cells. Plant Cell Physiol. 30: 445–454 [Google Scholar]
- Okamuro J.K., Caster B., Villarroel R., Van Montagu M., Jofuku K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Liere K., Kandlbinder A., Dietz K.-J., Oelmüller R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Watkins K.P., Friso G., van Wijk K.J., Barkan A. (2008). A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Sozzani R., Glab N., Domenichini S., Perennes C., Cella R., Kondorosi E., Bergounioux C. (2006). Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 47: 395–407 [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sakai A., Takano H., Kuroiwa T. (2004). Organelle nuclei in higher plants: Structure, composition, function, and evolution. Int. Rev. Cytol. 238: 59–118 [DOI] [PubMed] [Google Scholar]
- Salvador M.L., Klein U., Bogorad L. (1998). Endogenous fluctuations of DNA topology in the chloroplast of Chlamydomonas reinhardtii. Mol. Cell. Biol. 18: 7235–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Misumi O., Shinada Y., Sasaki M., Yoine M. (1997). Dynamics of localization and protein composition of plastid nucleoids in light-grown pea seedlings. Protoplasma 200: 163–173 [Google Scholar]
- Sato N., Ohshima K., Watanabe A., Ohta N., Nishiyama Y., Joyard J., Douce R. (1998). Molecular characterization of the PEND protein, a novel bZIP protein present in the envelope membrane that is the site of nucleoid replication in developing plastids. Plant Cell 10: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Terasawa K., Miyajima K., Kabeya Y. (2003). Organization, developmental dynamics, and evolution of plastid nucleoids. Int. Rev. Cytol. 232: 217–262 [DOI] [PubMed] [Google Scholar]
- Schnetz K. (1995). Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 14: 2545–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Toloczyki C., Rak B. (1987). Beta-glucoside (bgl) operon of Escherichia coli K-12: Nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J. Bacteriol. 169: 2579–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter Y., Steiner S., Matthäi K., Pfannschmidt T. (2010). Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics 10: 2191–2204 [DOI] [PubMed] [Google Scholar]
- Schwacke R., Fischer K., Ketelsen B., Krupinska K., Krause K. (2007). Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Genet. Genomics 277: 631–646 [DOI] [PubMed] [Google Scholar]
- Sekine K., Hase T., Sato N. (2002). Reversible DNA compaction by sulfite reductase regulates transcriptional activity of chloroplast nucleoids. J. Biol. Chem. 277: 24399–24404 [DOI] [PubMed] [Google Scholar]
- Small I., Wintz H., Akashi K., Mireau H. (1998). Two birds with one stone: Genes that encode products targeted to two or more compartments. Plant Mol. Biol. 38: 265–277 [PubMed] [Google Scholar]
- Spassky A., Rimsky S., Garreau H., Buc H. (1984). H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 12: 5321–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelsberg T.C., Kizer P.E., Hnilica L.S. (1970). Dissociation of RNA-histone complexes by DNA. Experientia 26: 136–138 [DOI] [PubMed] [Google Scholar]
- Spurio R., Dürrenberger M., Falconi M., La Teana A., Pon C.L., Gualerzi C.O. (1992). Lethal overproduction of the Escherichia coli nucleoid protein H-NS: Ultramicroscopic and molecular autopsy. Mol. Gen. Genet. 231: 201–211 [DOI] [PubMed] [Google Scholar]
- Steiner S., Schröter Y., Pfalz J., Pfannschmidt T. (2011). Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck R., Zeltz P., Falk J., Acker A., Kössel H., Krupinska K. (1996). Transcriptionally active chromosomes (TACs) of barley chloroplasts contain the α-subunit of plastome-encoded RNA polymerase. Curr. Genet. 30: 515–521 [DOI] [PubMed] [Google Scholar]
- Swarbreck D., et al. (2008). The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 36 (Database issue): D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K., Fujiwara M., Sato N. (2005). Real-time visualization of plastids and plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol. 46 (Suppl): S68. [DOI] [PubMed] [Google Scholar]
- Terasawa K., Sato N. (2005). Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol. 46: 649–660 [DOI] [PubMed] [Google Scholar]
- Thorstensen T., Grini P.E., Aalen R.B. (2011). SET domain proteins in plant development. Biochim. Biophys. Acta 1809: 407–420 [DOI] [PubMed] [Google Scholar]
- Vallon O., Hoyer-Hansen G., Simpson D.J. (1987). Photosystem II and cytochrome b-559 in the stroma lamellae of barley chloroplasts. Carlsberg Res. Commun. 52: 405–421 [Google Scholar]
- Venkatesh G.R., Kembou Koungni F.C., Paukner A., Stratmann T., Blissenbach B., Schnetz K. (2010). BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 192: 6456–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zychlinski A., Kleffmann T., Krishnamurthy N., Sjölander K., Baginsky S., Gruissem W. (2005). Proteome analysis of the rice etioplast: Metabolic and regulatory networks and novel protein functions. Mol. Cell. Proteomics 4: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Weigel D. (1995). The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell 7: 388–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D., Flügge U.I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Witzel K., Weidner A., Surabhi G.K., Börner A., Mock H.P. (2009). Salt stress-induced alterations in the root proteome of barley genotypes with contrasting response towards salinity. J. Exp. Bot. 60: 3545–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.T.Y., New D.C., Wong J.C.W., Hung V.K.L. (2003). Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot. Cell 2: 646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Laulhere J.P., Rozier C., Mache R. (1978). Visualization of folded chloroplast DNA from spinach. Biol. Cell. 32: 187–190 [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.