This work finds that a maize PPR protein (MPPR6) localized to the mitochondria is directly involved in 5′ maturation and translation initiation of rps3 mRNA. This dual role supports a general principle of action for PPR proteins in RNA processing and translation.
Abstract
Pentatricopeptide repeat (PPR) proteins are members of one of the largest nucleus-encoded protein families in plants. Here, we describe the previously uncharacterized maize (Zea mays) PPR gene, MPPR6, which was isolated from a Mutator-induced collection of maize kernel mutants by a cDNA-based forward genetic approach. Identification of a second mutant allele and cosegregation analysis confirmed correlation with the mutant phenotype. Histological investigations revealed that the mutation coincides with abnormities in the transfer cell layer, retardation of embryo development, and a considerable reduction of starch level. The function of MPPR6 is conserved across a wide phylogenetic distance as revealed by heterologous complementation of the Arabidopsis thaliana mutant in the orthologous APPR6 gene. MPPR6 appeared to be exclusively present in mitochondria. RNA coimmunoprecipitation and in vitro binding studies revealed a specific physical interaction of MPPR6 with the 5′ untranslated region of ribosomal protein S3 (rps3) mRNA. Mapping of transcript termini showed specifically extended rps3 5′ ends in the mppr6 mutant. Considerable reduction of mitochondrial translation was observed, indicating loss of RPS3 function. This is consistent with the appearance of truncated RPS3 protein lacking the N terminus in mppr6. Our results suggest that MPPR6 is directly involved in 5′ maturation and translation initiation of rps3 mRNA.
INTRODUCTION
In the mature embryo sac of maize (Zea mays), double fertilization initiates the development of the embryo and endosperm. The fusion of the sperm and egg nuclei forms the zygote from which the proembryo and later on the embryo develops. The triploid primary endosperm arises from the fusion of the second sperm nucleus with the two polar nuclei. These undergo several mitotic divisions resulting in up to 512 free nuclei. Cell wall formation leads the endosperm to become cellular (Kiesselbach, 1949). Shortly thereafter, cells of the endosperm begin to differentiate, forming four distinct tissue types: aleurone, central starchy endosperm, embryo surrounding region (ESR), and basal endosperm transfer layer (BETL). The outermost epidermal single cell layer, the aleurone, provides enzymes required for hydrolysis of starch and storage proteins, which make up the main content of the central endosperm of cereals. The ESR forms the cavity in which the embryo is located, whereas the BETL constitutes the base of the endosperm and is adjacent to the maternal pedicel. Both the ESR and BETL tissue types supply the embryo with nutrients and are therefore thought to be required for seed development.
In maize, a mature dormant seed with a well-developed embryo and starchy endosperm is the normal product of fertilization. Variations from this norm with significantly reduced embryo or endosperm development are considered as defective kernel types (Lowe and Nelson, 1946). Normal grain filling and development depends on mitochondrial function (Kang et al., 2009) and sufficient nutrient supply, which in turn depends on appropriate differentiation and function of cells, such as BETL cells, specialized for the transport of solutes (Thompson et al., 2001; Royo et al., 2007).
The endosymbiotic hypothesis suggests that mitochondria are descendants of α-proteobacteria. Over time, most of the endosymbiotic genes have been either lost or transferred to the nucleus. According to estimates, the mitochondrial proteome of vascular plants is composed of 2000 to 3000 proteins (reviewed in Millar et al., 2005), the majority of which are encoded in the nucleus. Only a limited number of gene products are encoded in the plant mitochondrial genome, and most of them are necessary for building up ribosomes and the respiratory chain. Expression and regulation of these genes is subjected to various posttranscriptional steps in which many nuclear-encoded proteins are involved. A recently discovered RNA binding pentatricopeptide repeat (PPR) protein family is considered to play a central role in these processes (reviewed in Schmitz-Linneweber and Small, 2008). PPR proteins are characterized by the presence of tandem arrays of a degenerate 35–amino acid residue repeat. The PPR motif is predicted to consist of two antiparallel α-helices, which in tandem arrays form a superhelical structure enclosing a hydrophilic, positively charged groove supposed to bind hydrophilic, acidic ligands, such as RNA (Small and Peeters, 2000; Kobayashi et al., 2012). Members of the huge PPR protein family are particularly found in plant organelles. A total of 441 members are present in Arabidopsis thaliana, the majority of which is encoded by single exon genes (Lurin et al., 2004). So far characterized PPR proteins have been implicated in quite diverse posttranscriptional steps, such as RNA editing (Kotera et al., 2005; Okuda et al., 2006; Chateigner-Boutin et al., 2008; Bentolila et al., 2010; Sung et al., 2010; Uchida et al., 2011; Hammani et al., 2011), RNA splicing (Schmitz-Linneweber et al., 2006; de Longevialle et al., 2007; Hattori et al., 2007; Koprivova et al., 2010; Liu et al., 2010; Chateigner-Boutin et al., 2011), RNA stability (Beick et al., 2008; Pfalz et al., 2009; Johnson et al., 2010; Pusnik and Schneider, 2012), RNA processing (Meierhoff et al., 2003; Jonietz et al., 2010, 2011; Hölzle et al., 2011) and translation (Schmitz-Linneweber et al., 2005; Uyttewaal et al., 2008, Prikryl et al., 2011; Cai et al., 2011). Despite the obvious recent progress in understanding their key role, little is known about their exact biological function. Therefore, further studies of PPR proteins and their ligands are required to elucidate their molecular function and specific targets.
In this work, we describe the genetic and molecular characterization of a gene encoding a PPR protein essential for kernel development in maize, which we named maize PPR6 (MPPR6). A mutation in MPPR6 caused by the insertion of a Mu1.7 transposon element creates a defective kernel phenotype associated with retarded grain development and filling, as well as morphological alterations of the BETL. The identification of one additional mppr6 allele in maize showing the same phenotype confirmed that mppr6 is causal for the mutant phenotype. We further demonstrate that the mppr6-related phenotype of the Arabidopsis ppr6 (appr6) T-DNA insertion line can be functionally complemented by heterologous expression of the MPPR6 gene, demonstrating a remarkably conserved function among land plants. Similar kernel defects in maize were described in knockouts of the mitochondria-targeted PPR protein Empty Pericarp4 (EMP4) required for accumulation of a certain set of mitochondrial transcripts. However, the precise function and targets of EMP4 remained unclear (Gutiérrez-Marcos et al., 2007). Our data show that mppr6 is specifically affected in posttranscriptional expression of the ribosomal protein S3 (rps3) gene in mitochondria. MPPR6 is associated in vivo with the 5′ untranslated region (UTR) of mitochondrial (rps3) mRNA, consistent with its proposed function in accurate rps3 5′ maturation and translation initiation.
RESULTS
Analysis of the mppr6 Mutant Phenotype
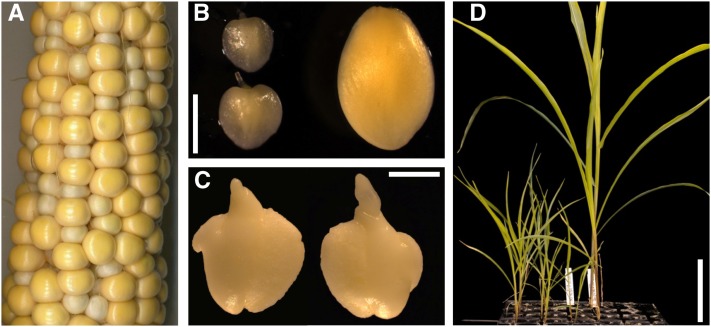
The maize mppr6 mutant originates from Biogemma’s collection of Mutator (Mu) transposon-induced defective kernel mutants. In immature heterozygous mppr6 (mppr6/+) ears, the recessive seed mutation is macroscopically visible at around 11 d after pollination (DAP). Approximately one-quarter of the kernels appeared small, pale, and distorted (Figure 1A; see Supplemental Figure 1D online). Embryos dissected from 18 DAP homozygous mppr6 (mppr6/−) kernels showed a small size and had a slightly translucent appearance (Figure 1B). Moreover, mutant embryos exhibited deformities at a later developmental stage (30 DAP), presumably caused by the pressure of adjacent wild-type kernels (Figure 1C). Since mature mppr6/− seeds did not germinate, we tried to recover embryos at 18 DAP from immature kernels and investigated growth on Murashige and Skoog (MS) medium. In contrast with wild-type plants, rescued mppr6/− individuals grew much slower but were able to grow further on soil (Figure 1D). However, alongside the pronounced dwarfism (maximal 20 cm; see Supplemental Figures 1A and 1B online) in the homozygous mppr6 mutant plants, male flowers were lacking and replaced by a female one.
Figure 1.
Phenotype of the Mu-Induced mppr6 Mutant.
(A) A 25 DAP ear segregating for mutant and wild-type kernels obtained by self-pollination of a plant heterozygous for the mppr6 mutation.
(B) Comparison between mutant (left) and wild-type (right) embryos at 18 DAP.
(C) The size of mppr6/− embryos at 30 DAP in (C) is comparable to that of the 18 DAP wild-type ones in (B).
(D) Comparison between developing mppr6 homozygous (left) and wild-type (right) rescued plants.
Bars = 1 mm in (B) and (C) and 10 cm in (D).
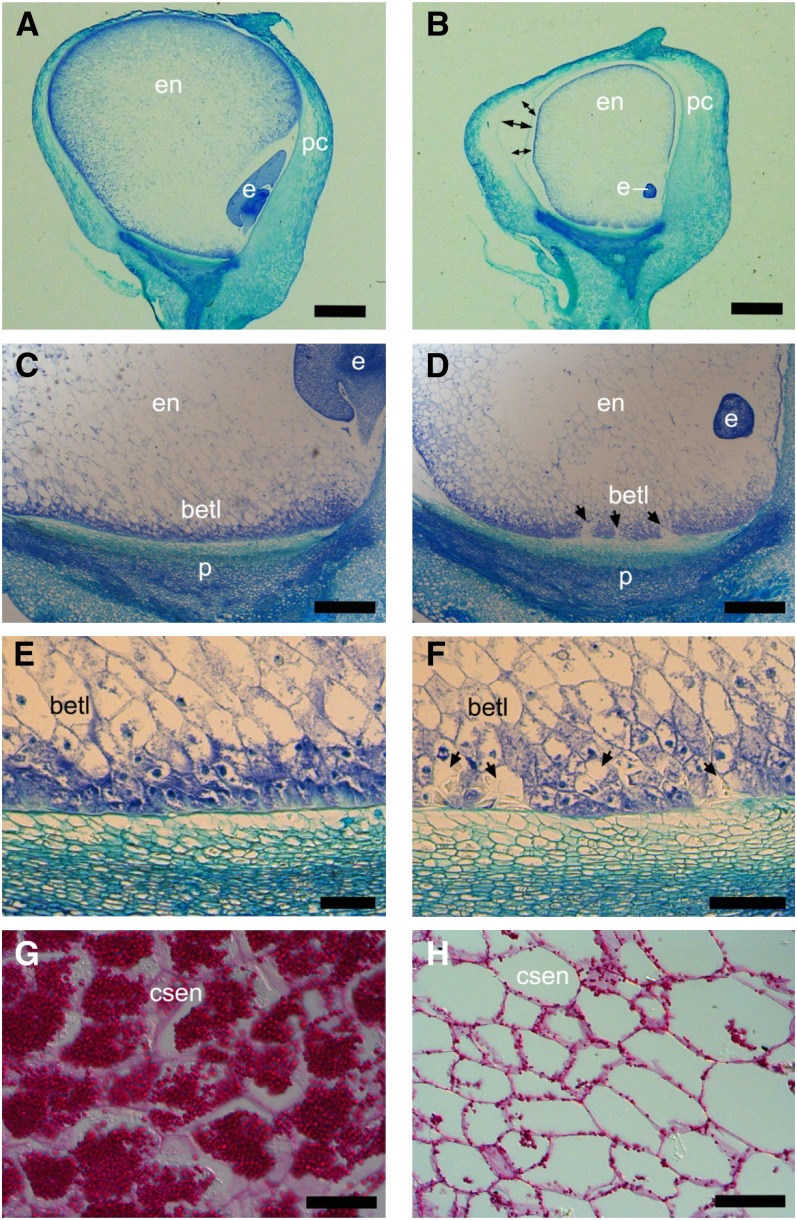
To investigate the mutant phenotype closely, wax longitudinal sections of the mppr6/− and wild-type kernels at 12 DAP were stained with Azure B and examined microscopically. Both the embryo and endosperm were affected in mppr6 mutants. A comparison between wild-type and mutant embryos revealed a significant developmental retardation of homozygous mppr6 embryos (Figures 2A and 2B). In wild-type seeds, the endosperm entirely filled the pericarp (Figure 2A), whereas the size of the mutant endosperm was notably reduced, resulting in a gap between the pericarp and the aleurone (Figure 2B). Furthermore, the BETL was fragmented by structurally irregular cells devoid of wall ingrowths (WIGs) shown in Figures 2C and 2D. In fact, most of the mppr6/− BETL cells displayed relatively weak WIG formation (Figures 2E and 2F). A similar WIG phenotype was described in two further loss-of-function alleles of kernel mutants (Costa et al., 2003; Kang et al., 2009).
Figure 2.
Embryo and Endosperm Are Altered in mppr6 Mutant Kernels.
The 12-µm longitudinal wax sections of mutant ([B], [D], [F], and [H]) and wild-type ([A], [C], [E], and [G]) kernels at 12 DAP stained with Azure B ([A] to [F]) and periodic acid Schiff’s reagent ([G] and [H]). In (B), double arrows indicate the gap between pericarp and aleurone, whereas arrows in (D) and (F) show abnormal cells in the BETL. csen, central starchy endosperm; e, embryo; en, endosperm; p, pedicel; pc, pericarp. Bars = 1 mm in (A) and (B), 100 µm in (C) and (D), 60 µm in (E) and (F), and 40 µm in (G) and (H).
Starch levels in the endosperm were investigated by staining sections with periodic acid Schiff’s reagent. A considerable starch reduction in mutant kernels was observed at 12 DAP (Figures 2G and 2H).
Molecular Cloning, Gene Structure, and Sequence Analysis of MPPR6
Genomic sequences flanking the Mu transposon and cosegregating with the mppr6 mutant phenotype were identified using a new cDNA-based transposon display technique. In this procedure, total RNA extracted from mutant kernels of two segregating sibling ears and wild-type kernels of a single sibling wild-type ear was used for cDNA synthesis according to the rapid amplification of cDNA ends (RACE) protocol (Clontech). Reactions for 3′ and 5′ RACE were conducted using a Mu-specific primer paired with an adapter primer corresponding to the attached sequence at the end of the cDNA. All primer sequences used in this work are provided in Supplemental Table 1 online. To increase the specificity of displayed bands, a first round RACE was followed by a nested RACE reaction. A Mu flanking cDNA sequence of 1 kb was present in the 3′ RACE of both mutant samples but absent in the wild-type and therefore considered for cosegregation analysis (data not shown). Sequencing identified the 1-kb Mu-adjacent cDNA fragment as part of a PPR protein-encoding gene, named MPPR6. Further analysis revealed that the mppr6 allele contains a Mu1.7 element located in its open reading frame (ORF). The Mu element was flanked by 9-bp direct repeats, known as Mu target site duplication.
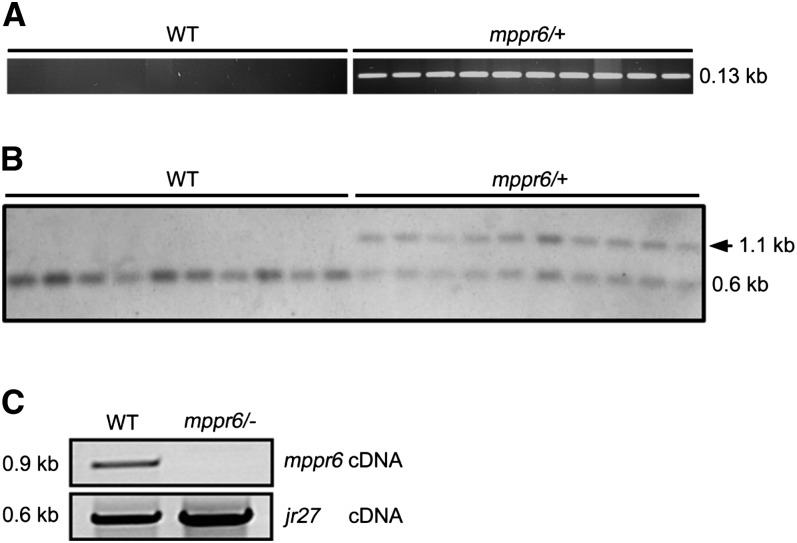
Genomic DNA extracted from leaf samples of 280 randomly selected sibling plants was sorted into segregating mppr6/+ and wild-type plants according to the ear phenotype of self-pollinated plants. A PCR-based cosegregation analysis using a Mu and an MPPR6-specific primer indicated precise cosegregation with all 200 mutant plants and no linkage with 80 wild-type sibling individuals. An example of these results is shown in Figure 3A for 10 mutant and 10 wild-type samples.
Figure 3.
The Mu Insertion within MPPR6 Cosegregates with the Mutant Phenotype.
(A) Cosegregation analysis by genomic PCR with MPPR6- and Mu-specific primers in the wild type (WT) and mppr6/− mutant.
(B) DNA gel blot hybridized with MPPR6-specific probe. The shift cosegregating with the Mu1.7 insertion in the mppr6/+ samples is indicated by an arrow (∼1.1 kb).
(C) A wild-type copy of mppr6 mRNA is absent in mppr6 mutants. RNA from wild-type and mppr6/− embryos was reverse transcribed and mppr6 mRNA was examined by RT-PCR using primer mppr6_1fw and mppr6_2rev, which flanks the Mu1.7 element. The quality of the cDNA was analyzed by amplifying the mRNA of the ubiquitous expressed gene jr27 (Lorbiecke et al., 2005) shown in the panel below.
To verify the PCR-based cosegregation, a DNA gel blot analysis was performed. Genomic DNA from 10 mppr6/+ and 10 wild-type randomly chosen plants was digested with SacI, which should result in a polymorphism when using an MPPR6-specific probe for hybridization (position of the probe is diagrammed in Figure 4A). A shift corresponding to the Mu1.7 insertion was obtained after hybridization in all mutant samples but not in the wild type (Figure 3B). Consistent with the Mu insertion, the mppr6 cDNA could not be amplified by RT-PCR in the mutants (Figure 3C).
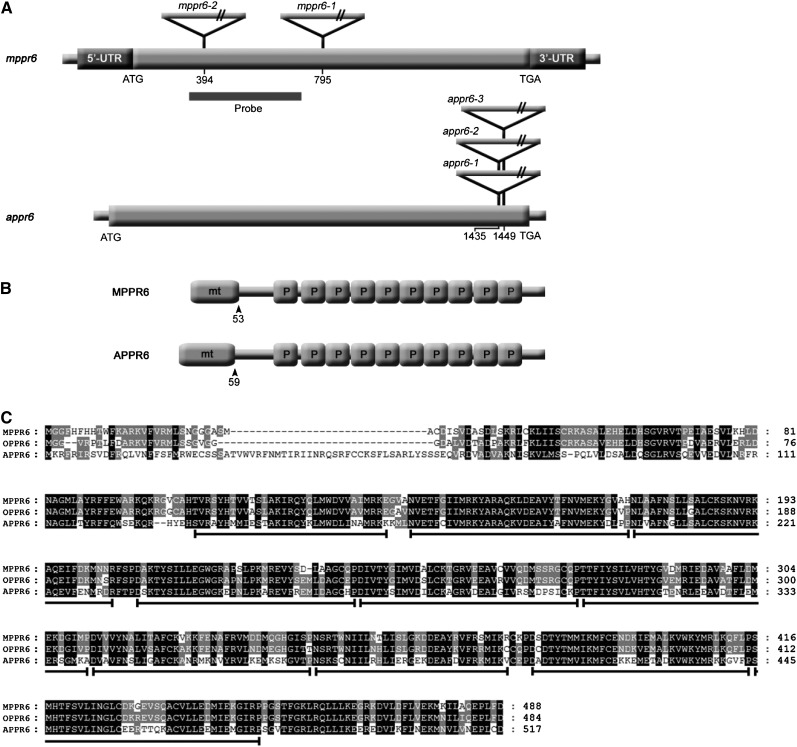
To confirm that the Mu insertion into the MPPR6 gene accounts for the phenotype, a Mu-induced mutant collection was screened for an additional mppr6 allele (see Methods). An independent insertion event was successfully recovered. The second allele, which we named mppr6-2, contains a so far uncharacterized Mu element in the ORF of MPPR6 394 bp downstream of the start codon (Figure 4A) and shows an mppr6-like kernel phenotype. Allelic crosses between mppr6-1/+ and mppr6-2/+ resulted in ears segregating for normal and defective seeds in the expected 3:1 ratio, confirming that the mutant phenotype of mppr6-1 and mppr6-2 is indeed the result of the Mu insertion in the MPPR6 gene and that both mutants are allelic (see Supplemental Figure 1C online).
Figure 4.
cDNA and Protein Structure of MPPR6 and APPR6.
(A) Schematic representation of mppr6 and appr6 cDNAs (diagrams not to scale). Mu (maize) and T-DNA insertions (Arabidopsis) are indicated by triangles. Positions of insertions with respect to the start codon (ATG) are shown in both cDNAs. ORFs are represented by wide gray bars, whereas dark-gray bars indicate the 5′ and 3′ UTRs in mppr6 cDNA.
(B) Protein motif organization of MPPR6 and APPR6. Mitochondrial targeting signals (mt) and PPR motifs (P) are indicated. Amino acid positions of transit peptide cleavage sites are shown by arrowheads.
(C) Alignment of MPPR6 (maize), OPPR6 (rice), and APPR6 (Arabidopsis) orthologs. The homologous areas are highlighted. PPR motifs in the sequences are indicated below by black lines.
A full-length mppr6 cDNA was generated by RACE using gene-specific primers. The cDNA consists of 1976 bp and a 5′ UTR of 201 bp followed by a 1470-bp-long ORF. The poly(A) tail is located 305 bp downstream of the stop codon (Figure 4A). Alignment of the cDNA to the corresponding genomic DNA sequence (MAGIv4_67802) revealed that the MPPR6 gene contains a single exon. The MPPR6 gene is located at the bottom of chromosome 1 and corresponds to the filter gene set GRMZM2G389645 that maps at position 279,177,101…279178807 on RefGen_V2. The deduced protein with a molecular mass of 55 to kD is composed of 489 amino acid residues and belongs to the P subclass of the PPR protein family. The 10 PPR domains stretch from amino acid residue 105 to 451. BLAST comparisons of protein sequences detected the putative orthologs in Arabidopsis and rice (Oryza sativa), At1g77360 (APPR6) and Os03g55340.1 (OPPR6), respectively. MPPR6 shares 97 and 80% similarity with OPPR6 and APPR6, respectively (Figure 4C). The proteins carry a predicted N-terminal mitochondrial transit peptide and share a highly conserved PPR motif structure (Figures 4B and 4C).
Genetic Complementation of the appr6 Mutation with MPPR6
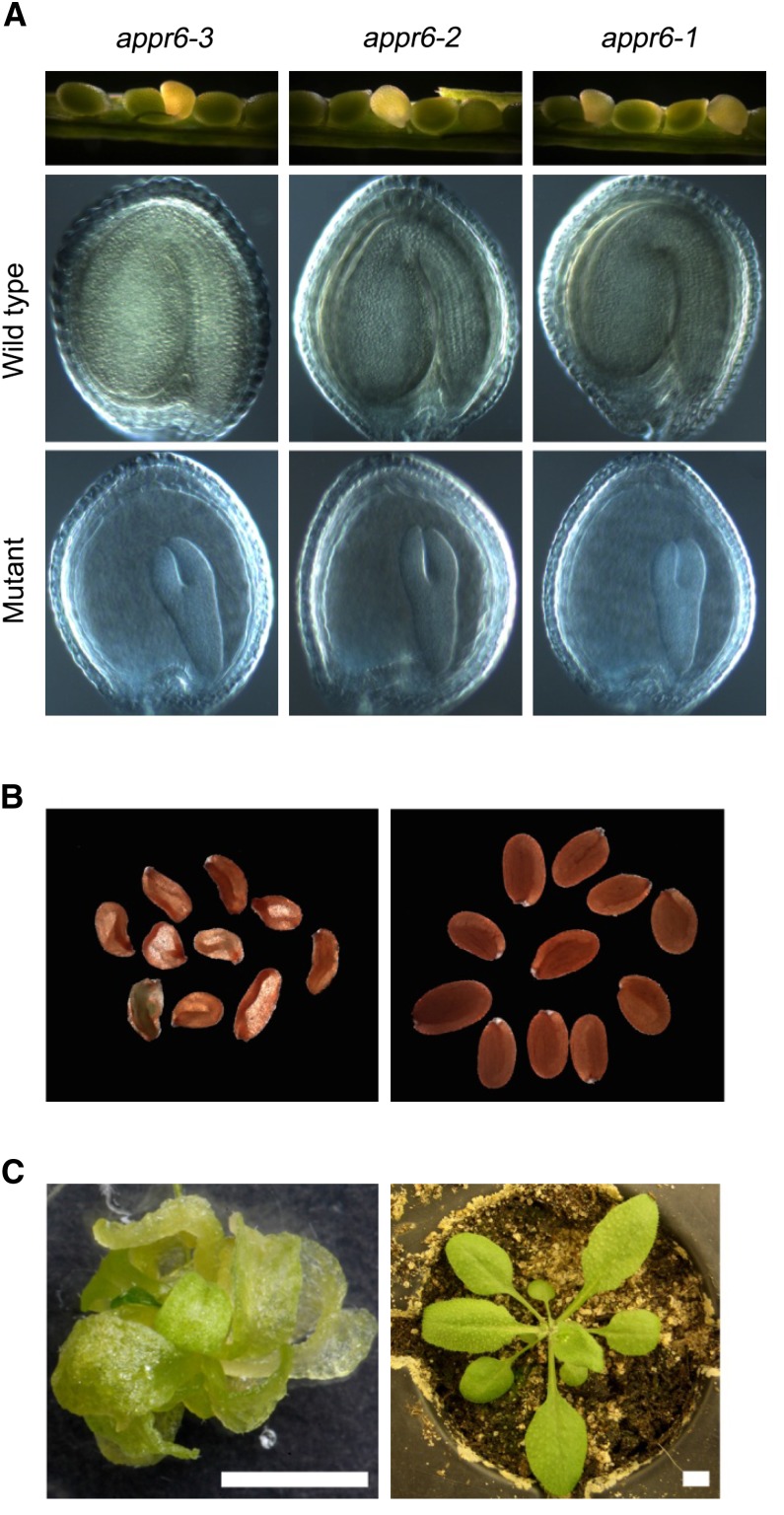
The functional relationship between the maize and Arabidopsis orthologs was investigated in a reverse genetics approach in Arabidopsis. Three transgenic Arabidopsis lines, SALK_045714 (appr6-1), SALK_091917 (appr6-2), and SALK_061950 (appr6-3), containing a T-DNA insertion in the APPR6 gene were obtained from the Salk Institute (http://signal.salk.edu/cgi-bin/tdnaexpress). Sequence analysis revealed that the T-DNAs in all three lines are inserted into the APPR6 gene and that the insertion in appr6-1 and appr6-2 is located at the same position (1435 bp downstream of the start codon; Figure 4A). Seeds from each line were sown on soil and seedlings were genotyped. For all lines, only three-quarters of the seeds were able to germinate and no homozygous mutant plants were found. One-quarter of the seeds within developing siliques of plants heterozygous for the mutations appeared translucent and exhibited a significantly delayed embryo growth (Figure 5A). Moreover, ∼25% of the mature dried seeds from heterozygous plants were small, shrunken (Figure 5B; see Supplemental Figure 1D online), and failed to germinate on soil. When grown on Suc-supplemented medium, homozygous appr6 plants showed delayed growth (Figure 5C) and were able to grow further on soil although severely retarded. Nevertheless, appr6 plants were able to reach reproductive maturity although still showing reduced size of inflorescences and siliques compared with those of the wild type.
Figure 5.
appr6 Mutant Lines.
(A) Mutant phenotype of appr6 immature seeds: from the top down, immature siliques segregating for mutant and wild-type seeds and a close-up of Hoyer’s solution-cleared wild-type and mutant seeds from the corresponding siliques.
(B) Mutant (left) and wild-type (right) mature dry seeds from a plant heterozygous for appr6-3 mutation.
(C) appr6-3 homozygous (left) and wild-type (right) seedlings grown on MS medium and soil, respectively. Bars = 0.5 cm.
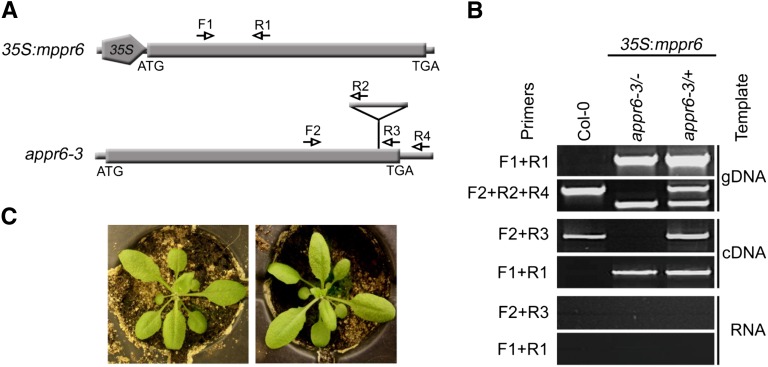
Owing to sequence similarity and the related phenotype in maize and Arabidopsis, we performed a genetic complementation of the appr6 mutation using the MPPR6 gene. For that purpose, the PCR-amplified ORF of maize MPPR6 was inserted into the binary vector pCHF5 downstream of the cauliflower mosaic virus 35S promoter (35S:mppr6; Figure 6A) and introduced into appr6-3/+ plants using the Agrobacterium tumefaciens–mediated floral dip transformation method (Clough and Bent, 1998). In the T1 generation, three independent homozygous appr6-3/− plants harboring the MPPR6 transgene were obtained (the results of one of these complemented plants is shown in Figures 6B and 6C). The phenotype and growth of these plants were indistinguishable from that of the wild type (Figure 6C). Genetic complementation proved that the appr6 mutant phenotype was indeed caused by the T-DNA insertion in At1g77360. Most importantly, heterologous expression provides genetically proven evidence of cross-species conservation of PPR function among monocot and dicot plants.
Figure 6.
MPPR6 Expression in the appr6-3 Mutant Background.
(A) Diagram of the 35S:mppr6 transgene and appr6-3 mutant locus. The positions of gene-specific and T-DNA primers used for genotyping and RT-PCR are shown by arrows. The T-DNA insertion is indicated by a triangle.
(B) Representative genomic and RT-PCR analysis of 35S:mppr6 harboring appr6-3/+ and appr6-3/− plants and a Columbia-0 plant. Genomic PCR with MPPR6-specific primers confirms the success of transformation. A PCR reaction with three primers was used to distinguish between wild-type, appr6/+, and appr6/− plants. mppr6 and appr6 transcripts were detected by RT-PCR in leaf tissue. Genomic DNA (gDNA) contaminations in the RNA used for RT-PCR were excluded by a control PCR with RNA as template.
(C) Phenotype of 3-week-old wild-type (left) and 3-week-old appr6-3/− plants (right) genetically complemented by MPPR6 expression.
Tissue-Specific Expression and Subcellular Localization of MPPR6
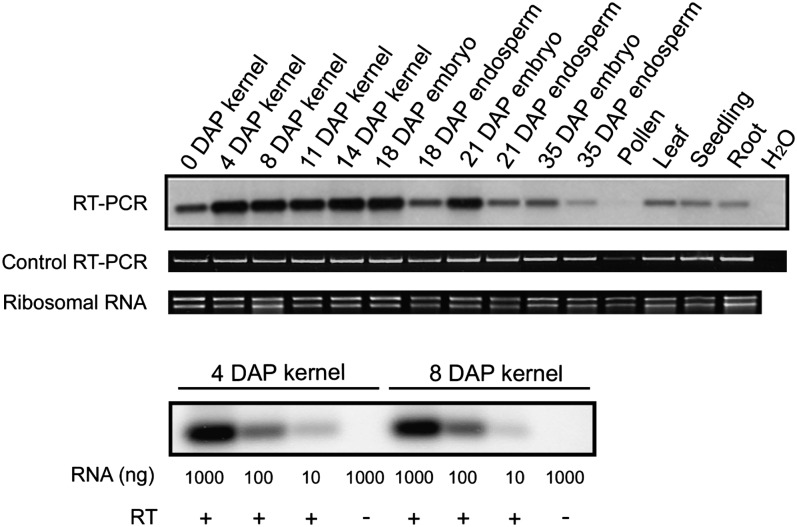
Due to the fact that the mppr6 mutation primarily affects kernel development and could partially be overcome by growth on Suc during early development, we hypothesized that MPPR6 is mainly expressed in kernel tissue. To test this, we examined the expression of MPPR6 in several maize tissues by RNA gel blot analysis. However, no hybridization signal could be detected, indicating very low gene expression. Therefore, we analyzed accumulation of mppr6 mRNA by RT-PCR (Figure 7). MPPR6 was found to be primarily expressed in kernel and embryo tissue (0 to 21 DAP), but weak signals were also obtained in endosperm, leaves, seedlings, pollen, and roots.
Figure 7.
Expression Level of MPPR6 in Several Maize Tissues Determined by RT-PCR.
Total RNA from diverse maize tissues was used in reverse transcription reactions. First-strand cDNA was applied to a low cycle RT-PCR using MPPR6-specific primers. The resulting products were electrophoretically separated, blotted to a nylon membrane, and hybridized with a DIG-labeled MPPR6-specific probe (probe is diagrammed in Figure 4A). The quality check of the cDNA by a control RT-PCR using primers specific to the housekeeping maize gene jr27 (Lorbiecke et al., 2005) is shown below. RNA quality is indicated below by staining of the rRNA with ethidium bromide. Proportional decreasing of the signal to the imputed RNA dilutions of two samples (4 and 8 DAP) is shown in the bottommost panel.
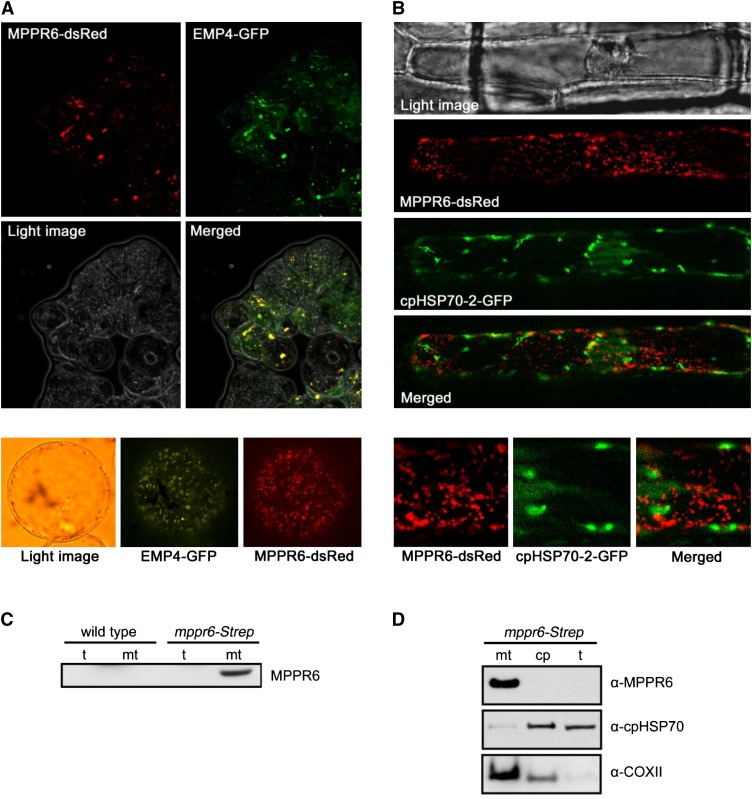
We used TargetP (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2000) and Predotar (Small et al., 2004) to predict the subcellular localization of MPPR6. Both programs detected a putative mitochondrial transit peptide. To verify experimentally the predicted mitochondrial localization, the first 80 amino acid residues of MPPR6 were fused with DsRed. As a positive control, EMP4, a mitochondrion-targeted PPR protein (Gutiérrez-Marcos et al., 2007), was fused with green fluorescent protein (GFP) and both constructs were transiently coexpressed in Black Mexican Sweet (BMS) cells. An overlap of the red and the green signals was observed using fluorescence microscopy, confirming mitochondrial localization of MPPR6 (Figure 8A). To investigate whether MPPR6 is also localized in the plastids, MPPR6-DsRed and cpHPS70-2-GFP, which is targeted to plastids, were transiently co-expressed in onion epidermis cells. Red and green fluorescence signals failed to overlap, indicating that MPPR6 targets only mitochondria (Figure 8B).
Figure 8.
MPPR6 Is Targeted to Mitochondria.
(A) Subcellular localization of MPPR6 in BMS maize suspension cells. Cells were biolistically cotransformed with Ubi:mppr6-dsRed and Ubi:emp4-GFP. Micrographs show red fluorescence signals of MPPR6-dsRed, green fluorescence signals of EMP4-GFP, light image of BMS cells, and merged fluorescence of MPPR6-dsRed and EMP4-GFP. Close-up of a single transformed cell is shown below. Due to the quick movement of mitochondria within the cell caused by cytoplasmic streaming, no precise merging of both signals was possible in the close-up.
(B) Transient coexpression of MPPR6-dsRed and cpHPS70-2-GFP in onion epidermis cells. A merged image of the nonoverlapping fluorescence is shown. Close-ups of selected regions are shown below.
(C) Immunodetection of the MPPR6 protein in total protein (t) and crude mitochondria extract (mt) obtained from wild-type and mppr6-Strep overexpressing maize seedlings.
(D) Immunoblot analyses detected the MPPR6 protein in mitochondria but not in chloroplast (cp) fractions, both obtained from the mppr6-Strep overexpressor. The same filter was reprobed with chloroplast- (α-cpHPS70-2) and mitochondria-specific (α-COXII) antibodies.
To confirm this further, a polyclonal antibody raised against recombinant MPPR6 was used in immunoblot analysis. In accordance with the low MPPR6 expression, the antibody recognizes MPPR6 only in mitochondrial protein extracts of mppr6-Strep overexpressing maize plants generated in this work (see Methods and Supplemental Figure 2 online) but not in the wild type (Figure 8C). For that reason, total protein extract and enriched fractions of mitochondria and chloroplasts were prepared from the overexpressor line mppr6-Strep. In agreement with the results of the previous subcellular localization studies, MPPR6 (∼50 kD) was detectable only in the mitochondrial fraction but was absent in the chloroplast fraction and total leaf extract (Figure 8D).
MPPR6 Binds to rps3 mRNA in Vivo and in Vitro
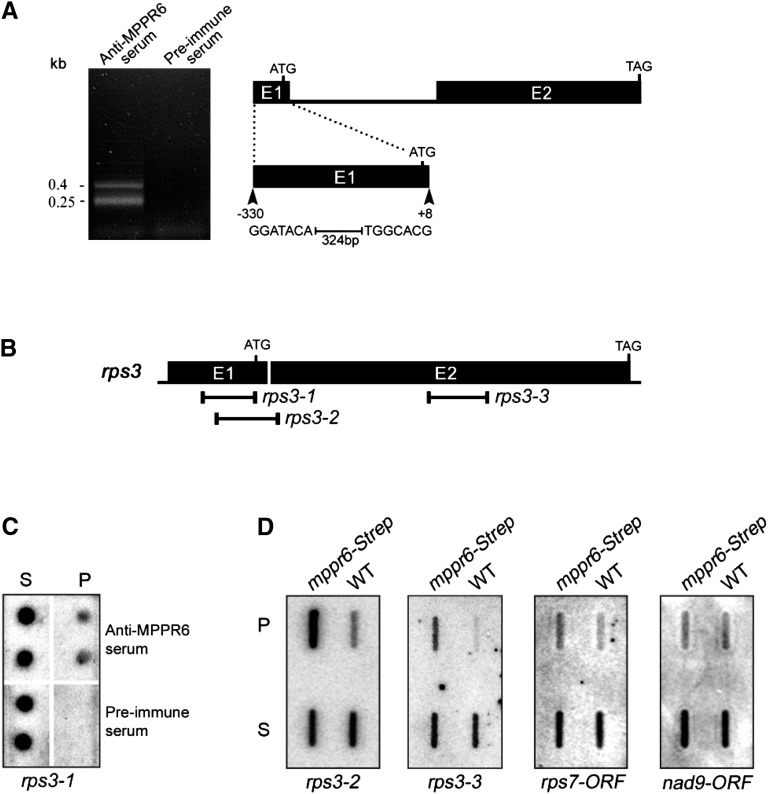
So far, characterized PPR proteins have been found to be involved in binding and processing of organellar RNAs. Therefore, we conducted RNA coimmunoprecipitation (Co-IP) analysis to detect a potential mitochondrial RNA associated with MPPR6 in vivo. Using polyclonal anti-MPPR6 rabbit serum, MPPR6 was immunoprecipitated from crude mitochondrial extract prepared from maize plants overexpressing mppr6-Strep. The RNA from the pellet was purified and polyadenylated. First-strand cDNA synthesis and subsequent cDNA amplification were performed by long-distance RT-PCR with adapter primers. As a negative control, the same Co-IP procedure was performed using preimmune serum. Two prominent RT-PCR products were obtained indicating a limited number of targets (Figure 9A). Cloning and subsequent sequencing revealed that besides the 5′ UTR of the mitochondrial rps3 mRNA, only four clones of the plastid 23S rRNA, one of the cytoplasmic 25S rRNA, and one of unknown origin were recovered. As rps3 was the single putative target of mitochondrial origin (Figure 9A), it appeared to be the only reasonable MPPR6 interaction partner; thus, it was selected for further investigations. Dot blot analysis of immunoprecipitated RNAs revealed that the 5′ UTR of rps3 was indeed enriched in the pellet obtained by Co-IP with the MPPR6 antibody but absent in the pellet fraction obtained with the preimmune serum (Figure 9C).
Figure 9.
MPPR6 Is Associated with rps3 in Vivo.
(A) RT-PCR products obtained from the Co-IP (left). Diagram of the intron/exon organization of rps3 (right). Exons (E1 and E2) are shown as black bars and the intron as a black line. The rps3 fragment found to be associated in vivo with MPPR6 is shown below as a black bar (+8 to −330, with respect to the ATG).
(B) Positions of the rps3-specific probes (black lines) used in the dot (B) and slot blot (C) analysis.
(C) Dot blot analysis of the coimmunoprecipitated RNAs. For the dot blot analysis, 1 μL of the RNA extracted from the pellet and the supernatant was double applied to a nylon membrane and hybridized with a 32P-labeled probe specific for rps3 (rps3-1, see [B]). RNA from the Co-IP with preimmune serum was used as negative control. P, pellet; S, supernatant.
(D) Slot blot hybridization. One-third of the RNA purified from the pellet and one-twelfth of the supernatant RNA obtained by Co-IP using wild-type (negative control) and mppr6-strep overexpressing plants were applied to slot blots and hybridized with the indicated 32P-labeled probes (see also [B]). WT, the wild type.
An additional RNA Co-IP experiment was performed, in which we took advantage of the Strep-tag fused in frame to the MPPR6 protein. Crude mitochondrial protein extracts from the overexpressor mppr6-Strep and the wild type as negative control were subjected to a Co-IP using a strep-tactin column. Recovered RNAs were investigated by slot-blot analysis using two rps3-specific probes recognizing its 5′ UTR and ORF (Figure 9B), respectively, and two additional, randomly picked mitochondrial gene probes, rps7 and nad9. This experiment showed a marked enrichment of the 5′UTR of rps3 and, as expected, a slight enrichment of the middle region of the rps3 ORF (Figure 9D). By contrast, rps7 and nad9 RNAs were not enriched to a similar extent, confirming the specificity by which MPPR6 recognizes rps3 RNA.
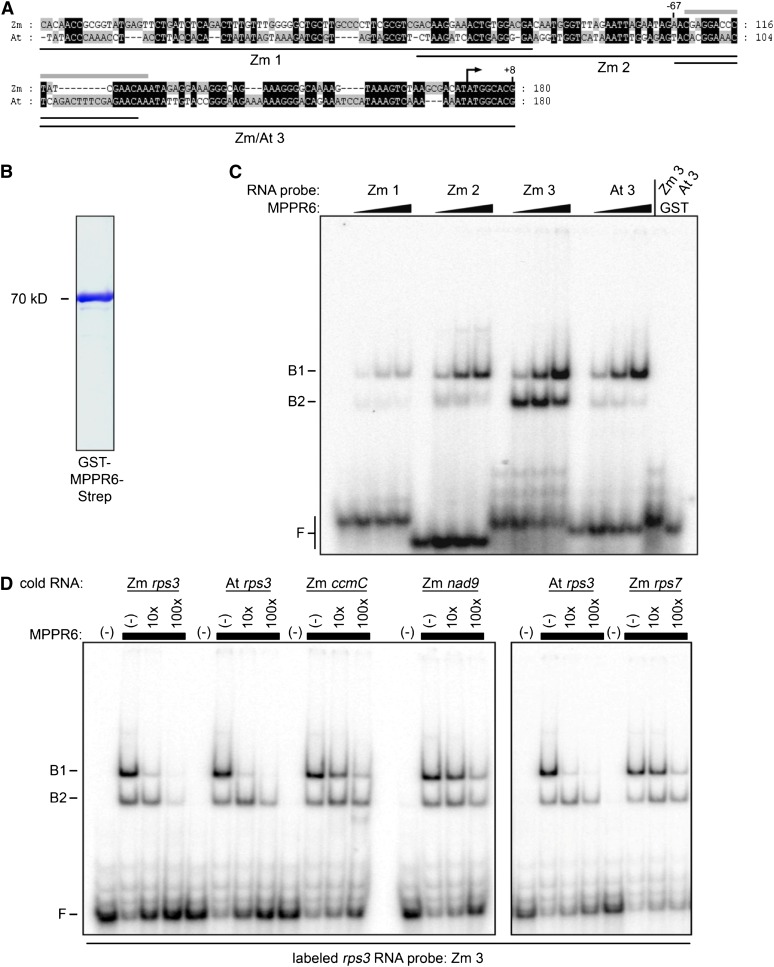
Direct binding of MPPR6 to the 5′ UTR of rps3 mRNA was further investigated by electrophoretic mobility shift assay (EMSA) using purified recombinant glutathione S-transferase (GST)-MPPR6-Strep fusion protein. In initial EMSA experiments, the binding site of MPPR6 within the coimmunoprecipitated 5′ region of rps3 (consisting of 338 nucleotides) was narrowed down to 180 nucleotides (data not shown). This part was divided further into three overlapping regions, which were then used as probes (Figure 10A). MPPR6 most efficiently bound to the probe nearest the start codon (Figure 10C; see Supplemental Figure 3 online), indicating that this region, mapping from +8 to −67 with respect to the ATG start codon, most likely comprises the binding site of MPPR6. Taking into account that this region shows the highest sequence similarity to the rps3 transcript in Arabidopsis, we additionally investigated binding of MPPR6 to the corresponding rps3 region in Arabidopsis. In agreement with the ability of the MPPR6 protein to restore the function of the Arabidopsis ortholog, the Arabidopsis probe showed a comparably strong binding affinity to the MPPR6 protein. In addition, unlabeled maize and Arabidopsis rps3 RNAs were able to compete efficiently with the bound maize rps3 probe when using a 10-fold molar excess over the bound probe. Unspecific RNA probes of the mitochondrial rps7, ccmC, and nad9 genes of similar size failed to compete efficiently (Figure 10D; see Supplemental Figure 4 online). These results clearly demonstrate that MPPR6 is specifically associated with the 5′ UTR of rps3 in vivo and is directly responsible for physical interaction as shown in vitro.
Figure 10.
In Vitro Binding of Recombinant MPPR6 Protein to rps3 RNA.
(A) Sequence alignment of the rps3 5′ region from Arabidopsis (At) and maize (Zm). Identical areas are highlighted in black. The sequence region of the overlapping probes (Zm 1, 2, and 3 and At 3) used in the EMSA experiment (C) are diagrammed as black lines. The estimated MPPR6 binding site is marked by a gray bar above the sequence. The translational start is indicated by an arrow.
(B) Coomassie blue staining of a SDS-PAGE gel of the purified protein. Recombinant GST-MPPR6-Strep protein was overexpressed in E. coli and purified by one-step affinity chromatography via strep tag.
(C) RNA binding of purified MPPR6 protein (B) was demonstrated by EMSA using the radiolabeled probes indicated in (A). Binding reactions contained RNA at 50 pM and MPPR6 at 0, 50, 250, and 500 nM. The concentration of purified GST protein (negative control) was 500 nM. B1 and B2, bound RNA; F, free RNA.
(D) RNA competition experiments. Diverse nonlabeled RNAs (cold RNA: Zm rps3, ccmC, nad9, rps7, and At rps3) were used in 10× and 100× molar excess over the radiolabeled rps3 probe (see [A], Zm 3) using 500 nM MPPR6 protein (black boxes). B1 and B2, bound RNA; F, free RNA.
[See online article for color version of this figure.]
MPPR6 Is Necessary for 5′ Maturation and Translational Initiation of rsp3 mRNA
PPR proteins are assumed to represent sequence-specific factors that guide various posttranscriptionally active effector enzymes to their targets rather than being direct effectors themselves (Barkan, 2011). To gain insight into MPPR6 function, the effect of the mutation on rps3 processing and gene expression was analyzed in the maize mutant.
Editing of maize rps3 has not been examined so far. Ten editing sites were found in rice rps3 (http://biologia.unical.it/py_script/log.html), which are conserved in maize as demonstrated by sequence analysis. RT-PCR and subsequent sequencing of the rps3 coding region revealed that all 10 editing sites within rps3 are transmethylated in the mutant excluding a potential function in RNA editing.
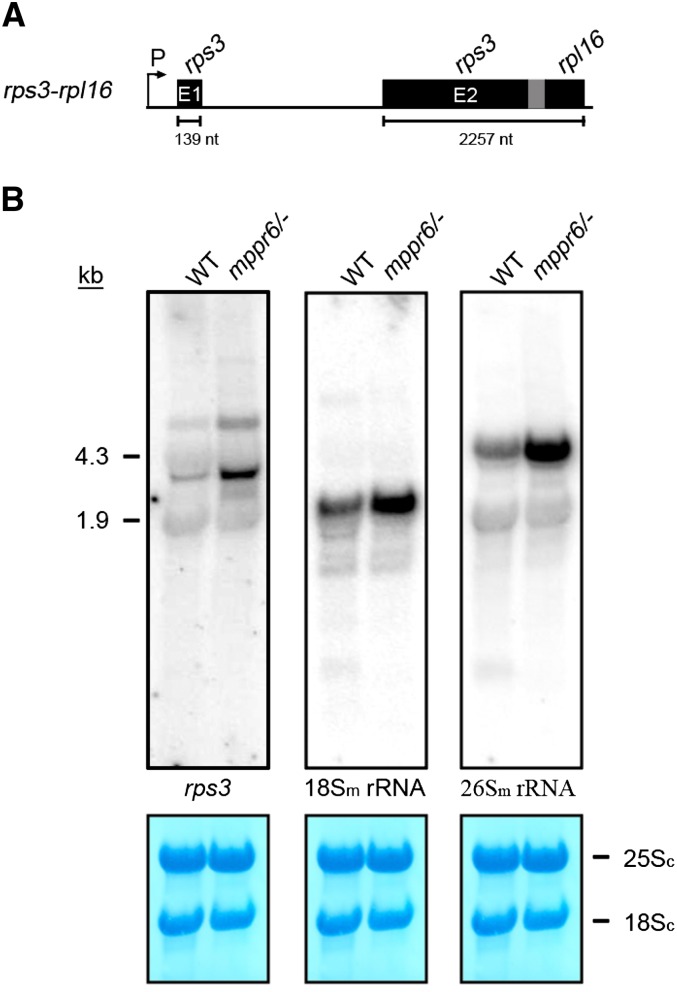
Total RNA extracted from wild-type and mutant embryos were hybridized with an rps3-specific probe. We found that the hybridization signal of ∼2.4 kb corresponding to the dicistronic rps3-rpl16 transcript (Figure 11A) accumulated in the mppr6 mutant in size comparable to that of the wild type, indicating that transcription and splicing of rps3 are not affected by the mppr6 mutation (Figure 11B). By contrast, the abundance of the rps3-rpl16 transcript was considerably increased in the mppr6 mutant compared with the wild type. This observation was confirmed by RT-PCR (see Supplemental Figure 5 online). A similar increased transcript abundance was also observed for the 18S and 26S mitochondrial rRNAs (Figure 11B), hinting at a general effect on transcript accumulation in the mutant.
Figure 11.
RNA Gel Blot Analyses of rps3-rpl16, 18Sm, and 26Sm rRNAs in Wild-Type and Mutant Embryos.
(A) Organization of the dicistronic rps3-rpl16 gene cluster (not to scale). The overlap of rps3 and rpl16 coding regions is shown in gray. The length of the exons is indicated below. E, exon; nt, nucleotides; P, promoter.
(B) Ten micrograms of total RNA obtained from wild-type (WT) and mppr6 embryos was analyzed by RNA gel blot hybridization using radioactive-labeled probes specific for the second rps3 exon and the mitochondrial 18Sm or 26Sm rRNAs. Methylene blue staining of cytoplasmic ribosomal 25Sc and 18Sc RNA is shown below. The probes used are indicated.
[See online article for color version of this figure.]
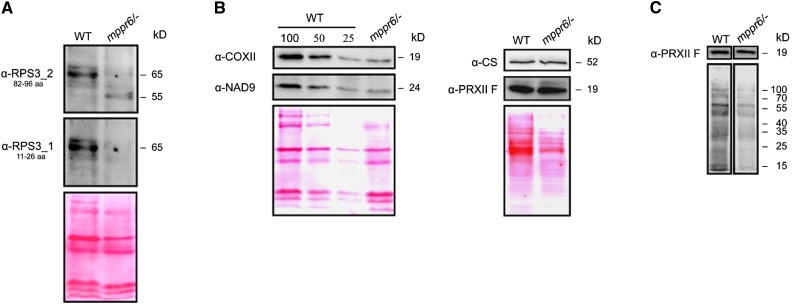
The changed transcript abundance is presumably due to translation defects in agreement with the situation in chloroplasts where inhibition of translation increases levels of many transcripts (Klaff and Gruissem, 1991; Meurer et al., 2002; Kato et al., 2006). Therefore, we checked a possible role of MPPR6 in translation of rps3. To achieve this goal, we generated antibodies against amino acid residues 82 to 96 of RPS3 and analyzed mitochondrial protein extracts from mutant and wild-type plants. The RPS3 protein appeared ∼10 kD smaller in the mutant compared with the wild-type form of 65 kD (Figure 12A). Furthermore, levels of this truncated protein were significantly reduced in the mutant. If translation initiation was affected, we would expect that the N-terminal part of the protein would be lacking in the truncated form and that instead use of an internal in frame start codon would be responsible for the smaller version. All attempts to analyze the RPS3 protein by mass spectrometry failed. Therefore, a second antipeptide antibody against the very N terminus of RPS3 (amino acid residues 11 to 26) was generated, and mitochondrial proteins from callus tissue were subjected to immunodetection. The serum recognized the RPS3 protein of 65 kD in the wild type as expected but failed to decorate the truncated mutant version, indicating that the N terminus rather than the C terminus is lacking (Figure 12A). This speaks in favor of the idea that the MPPR6 protein binds to the 3′ end of the rps3 5′ UTR to support translation initiation at its ATG start codon.
Figure 12.
Disruption of MPPR6 Causes Truncation of the RPS3 Protein and a General Translational Deficiency.
(A) Immunodetection of RPS3. Twenty micrograms of mitochondrial protein extracted from wild-type (WT) and mppr6/− callus tissue were separated by SDS-PAGE, blotted on a nitrocellulose membrane, and consecutively probed with affinity purified anti-RPS3 antibodies raised against amino acids 82 to 96 (top panel) or 11 to 26 (middle panel). Ponceau staining of the membrane-bound proteins is shown in the panel below.
(B) Left: Dilutions of membrane protein extracts from wild-type mitochondria were compared with the mppr6/− mutant using antibodies raised against the mitochondria-encoded proteins NAD9 and COXI. 100% corresponds to 20 µg of mitochondrial protein. Right: Immunodetection of the nucleus-encoded mitochondrial proteins PRXII F and CS in soluble extracts from mppr6 mutant and wild-type mitochondrial.
(C) In organello translation of wild-type and mppr6/− mitochondria after inhibition of cytoplasmic translation traces using cycloheximide. The same filter was probed with anti-PRXII F antibody.
[See online article for color version of this figure.]
It is likely that a nonfunctional RPS3 protein generally results in impaired mitochondrial translation. For that reason, we investigated the steady state levels of mitochondrial proteins. The amounts of the mitochondrial-encoded proteins NAD9 and COXII were reduced to ∼25% in the mutant, whereas peroxiredoxin-2F (PRXII F) and citrate synthase (CS), nuclear-encoded mitochondrial proteins, accumulated at levels comparably to the wild type (Figure 12B). In organello protein synthesis in the presence of [35S]Met revealed a similar reduction of newly synthesized mitochondrial proteins in the mppr6 mutant (Figure 12C). Collectively, our results indicate that absence of MPPR6 causes initiation of translation downstream and in frame with the natural start codon of RPS3, which in turn results in a truncated RPS3 protein and reduced mitochondrial translation efficiency.
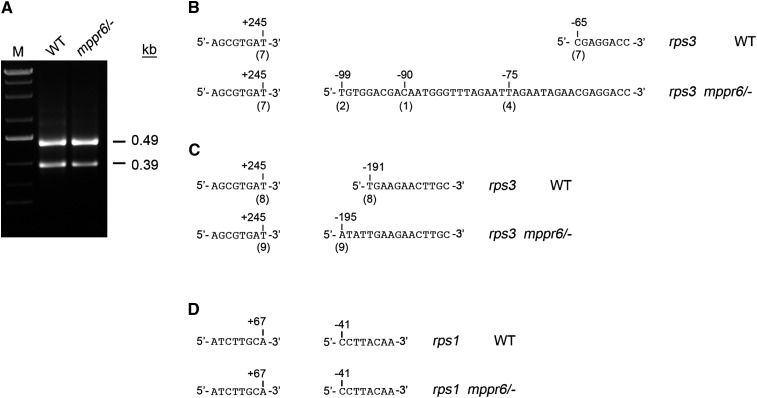
The question arises whether 5′ processing of rps3 is also affected in the mppr6 mutant. Since RNA gel blot analysis does not provide sufficient resolution to assess small size differences, we mapped the transcript ends of rps3 by circular RT-PCR (cRT-PCR). This assay yielded two distinct amplification products of roughly 490 and 390 bp in wild type and mppr6/− (Figure 13A; see Supplemental Figure 6 online). Sequencing of independently cloned cRT-PCR products finally revealed size variations between the mutant and wild type. In the 390-bp products, the rps3 5′ end of the wild type always mapped to base position −65 nucleotides (seven of seven clones) upstream of the start codon, which precisely coincides with the binding site of MPPR6. By contrast, in the mutant, the rps3 5′ end was found at quite different positions: −75 (four of seven clones), −90 (one of seven clones), and −99 (two of seven clones) (Figure 13B). In the large cRT-PCR products, all wild-type ends mapped again to the same position (−191 nucleotides, eight of eight clones), while the mutant products varied slightly in length (−194 nucleotides, seven of nine; −195 nucleotides, two of nine) (Figure 13C). As a control, the rps3-rpl16 3′ end position was exclusively found at +245 nucleotides downstream of the rpl16 stop codon in both the wild type and the mppr6 mutant (Figures 13B and 13C). In addition, rps1 termini in the mutant and the wild type were expectably unchanged (Figure 13D), indicating that the variations of the 5′ rps3 ends in the mutant are a specific consequence of MPPR6 absence.
Figure 13.
cRT-PCR Mapping of 3′ and 5′ rps3 Termini in the Wild Type and mppr6/− Mutant.
(A) rps3 transcript analysis using cRT-PCR. Products of circular RT-PCR were separated on 2% agarose gels, extracted, and cloned into pGEM T vector. Sequence results of independently cloned products are shown in (B) and (C). WT, the wild type.
(B) and (C) rps3 ends found in the 0.39- and 0.49-kb cRT-PCR products, respectively. The 5′ transcript ends are indicated relative to the start codon, whereas the position of the 3′ ends is shown in respect to the stop codon. The number of clones sequenced with each of the indicated ends is shown below in brackets.
(D) Control cRT-PCR of rps1transcripts. Obtained products were sequenced directly.
The transcript ends of rps3 were additionally determined by primer extension assay. A processing deficiency in the mppr6/− mutant was observed as a high molecular band likely to be the primary 5′ transcript end of rps3, which accumulated significantly more in the mutant compared with the corresponding primer extension product in the wild type (see Supplemental Figure 7 online), and the mature 5′ end determined by cRT-PCR (69 nucleotides upstream of the ATG start codon) failed to accumulate in the mppr6/− mutant. In addition, several other bands were absent in the mppr6/− mutant or accumulated less than in the wild type.
DISCUSSION
MPPR6 Is Essential for Seed Development
PPR mutants in land plants often result in quite diverse but not always lethal phenotypes (Lurin et al., 2004; Kocábek et al., 2006; Beick et al., 2008). Strong effects on seed development have been reported when mitochondrial localized PPR proteins are mutated (de Longevialle et al., 2007; Gutiérrez-Marcos et al., 2007). MPPR6 was shown to be important for maize seed development, since kernels are stunted and impaired in their ability to germinate in mppr6 mutants. Bright-field imaging revealed abnormalities of the WIG formation in the mppr6 BETL at an early stage of seed development. The BETL, the lowermost cell layer of the endosperm, is located in the immediate vicinity of the pedicel and mediates the connection between maternal and filial tissue. The most basal cell layer of the BETL is characterized by WIGs, which are labyrinth-like cell wall proliferations. By increasing the plasma membrane region, the WIGs are thought to facilitate the transfer of solutes from the maternal pedicel to the endosperm, required for seed development (Thompson et al., 2001). Therefore, normal differentiation and function of WIGs is a prerequisite for normal grain development and filling. It has been demonstrated that structural aberrations of the WIGs in the BETL at an early stage of seed development resulted later in severe seed phenotypes (Costa et al., 2003; Gutiérrez-Marcos et al., 2007; Kang et al., 2009). Furthermore, Kang et al. (2009) have shown an accumulation of mitochondria close to the WIGs, postulating a functional relationship between WIGs and mitochondria. Taking all this into account, we postulate that the observed reduction of translation efficiency in mitochondria of mppr6 plants and therefore decreased energy supply could result in deficient WIG formation, which in return could lead to a lack of nutrient transport and cause the described poor grain filling with concomitant germination impairments. As mitochondria are essential for development and growth, the pronounced dwarfism of rescued mppr6 plants could also be explained by aforementioned mitochondrial malfunction.
MPPR6 Is a Mitochondrial rps3 RNA Binding Protein
PPR proteins massively emerged during endosymbiosis in plant organelles and mainly exert their functions in fast evolving and defined posttranscriptional processes (Barkan, 2011) and are therefore expected to diverge rapidly. Sequence specificity is thought to be generally conferred by recognizing defined RNA segments in coding and noncoding regions. With exception of endonucleolytic activity of DYW motif–containing PPR proteins in vitro, an enzymatic function has not yet been described for members of this protein family (Okuda et al., 2006), indicating that they likely act in concert with RNA-modifying factors.
Co-IP experiments in combination with RT-PCR were performed to identify the mitochondrial RNA target of MPPR6 in vivo. Along with the mitochondrial rps3 5′ UTR, only plastid and cytoplasmic rRNAs as well as a sequence of unknown origin were obtained. Nonmitochondrial RNAs very likely represent contaminations due to their high abundance and/or similar target sequences. Several lines of evidence, such as dot and slot blot analysis, in vitro RNA binding studies, a failure to process the 5′ rps3 RNA efficiently, accumulation of 5′ truncated RPS3 protein, mitochondrial translation deficiencies, and seed lethality, demonstrate the specific function of MPPR6 in rps3 binding and expression. Misexpression of rps3 fully explains the mppr6 phenotype, again suggesting that rps3 is the only target of MPPR6, although we cannot rule out that additional mitochondrial RNAs of presumably low abundance are recognized by MPPR6.
The Structure and Function of MPPR6 Protein Is Conserved among Land Plants
In Arabidopsis, only one close homolog of MPPR6, termed APPR6, is present. According to a sequence-based phylogenetic analysis of rice and Arabidopsis gene families (O’Toole et al., 2008) for more than 80% of the PPR proteins orthologs exist in both species. Using the relational database POGs (http://plantrbp.uoregon.edu/), a putative orthologous protein in rice (OPPR6) was found. Owing to the high sequence similarity of nearly 97% between the rice and maize protein, we postulated that APPR6 represents the orthologous protein in Arabidopsis. In favor of this idea, a highly conserved PPR motif structure and a mitochondrial transit peptide was predicted for all three putative orthologous proteins and confirmed for MPPR6 (Figure 8).
Similar to emp4 mutants affected in a mitochondrial PPR protein and showing pleiotropic effects on RNA levels (Gutiérrez-Marcos et al., 2007), mppr6 and appr6 mutations cause a conditionally lethal seed phenotype in both maize and Arabidopsis. A functional conservation between the maize and Arabidopsis protein was investigated by different methods. First, we showed that heterologous expression of MPPR6 in the corresponding Arabidopsis appr6 mutant restored the wild-type phenotype, demonstrating a conserved function of the orthologous proteins in monocots and dicots (Figure 6C). Second, the ability of the MPPR6 to bind to the conserved target efficiently in both maize and Arabidopsis 5′ UTR of rps3 was confirmed by gel shift assays, indicating that the structural features of MPPR6 and APPR6 are also conserved among land plants despite the recent burst of PPR proteins and fast evolving UTR sequences in plant organelles (Greiner et al., 2008). Third, predominant expression of both MPPR6 and APPR6 during early seed development demonstrates a conserved importance during this stage (Figure 7; see Supplemental Figure 8 online). Conservation of PPR function between relatively closely and distantly related species has already been shown for MRL1 and CRR4 (Okuda et al., 2008; Johnson et al., 2010). Here, we provide evidence for a conserved PPR function across a wide phylogenetic distance.
Binding of MPPR6 to the rps3 5′ UTR Assists 5′ Maturation
The 5′ transcript termini of rps3 were mapped by cRT-PCR and primer extension assay. Both methods showed a processing deficiency in the mppr6/− mutant. However, these methods seemed to differ in their sensitivity of mapping 5′ RNA isoforms. Whereas cRT-PCR appeared to amplify shorter rps3 transcripts preferentially, primer extension proved to detect much larger ends. Furthermore, cRT-PCR detects only processed termini containing 5′ monophosphates, whereas in the primer extension, all kinds of transcripts could serve as a template regardless of whether they have been processed or not; thus, the primary transcript ends and possible degradation products would also be detected explaining the complexity of the primer extension products obtained. Furthermore, both methods could be affected by complex secondary structure in the RNA, which would stop the reverse transcription, resulting in absence of expected products. As primer extension assay includes an extended primer annealing step, during which secondary structure formation could occur, this experiment seems to be more sensitive to RNA folding than cRT-PCR.
Since the mature 5′ UTR in the mutant was extended by only several nucleotides, we assume that the target site is recognized and cleaved by an endonuclease rather than protected against exonucleolytic cleavage. Alternatively, unspecific endo- or exonucleolytic cleavage might take place randomly at secondary structure-free regions resulting in 5′ ends of different lengths in the mutant (Stoppel and Meurer, 2012).Consistent with this, the fully processed 5′ end of the wild type precisely coincides with the MPPR6 binding site. We used m-fold (http://mfold.rna.albany.edu) to predict the secondary structures of the differently processed wild-type and mutant rps3 5′ region. Interestingly, a hairpin structure occurred in the correctly processed rps3 5′ UTR (see Supplemental Figure 9 online), which is recognized by MPPR6. The secondary structures of the three different 5′ UTR versions found in the mppr6 mutant proved to be more complex. So far, only for the plastid PPR protein, PPR10, the exact RNA binding site of 17 nucleotides within overlapping 5′ and 3′ regions of chloroplast mRNAs has been defined (Prikryl et al., 2011). Questions related to the precise physical interaction between MPPR6 and its target will be subjected in future studies.
MPPR6 Facilitates Translation Initiation of rps3
Factors involved in initiation and control of plant mitochondrial translation remain elusive. In contrast with the eubacterial ancestor, mitochondria have lost typical Shine Dalgarno sequences and ribosomal binding sites cannot be unambiguously classified (Hazle and Bonen, 2007). Loss of these binding sites has been presumably compensated for by the evolution of many more UTR-specific PPR and other proteins allowing individualized regulation of mitochondrial gene expression. It has recently been shown that an unfolded state of RNA near the ATG start codon in Shine-Dalgarno sequence-free mRNAs in bacteria and organelles is the major requirement for efficient start codon recognition and translation initiation (Scharff et al., 2011). Secondary structures in the rps3 5′ UTR are therefore likely to prevent translation initiation by masking the start codon. Thus, it is tempting to speculate that MPPR6 binding loosens up the secondary structure in the 5′ region of the rps3 mRNA, thereby facilitating ribosome recruitment and ensuring free access to the correct start codon. As a consequence of the absence of the MPPR6 protein in the mutant, the correct ATG is inaccessible; thus, an alternative start codon in a hairpin-free environment further downstream might be used, resulting in the observed truncated MPPR6 protein, which is lacking the N terminus. Since in mitochondria, ATG, GGG, AAU, GUG, or ACG is used as a start codon (Giegé and Brennicke, 2001), the N terminus of the mutant protein could not be determined with certainty.
Binding of the plastid PPR10 protein to the 3′ UTR of atpI transcript possibly hampers an exonuclease from continuing digestion at the binding site (Prikryl et al., 2011). Binding to the overlapping 5′ UTR of atpH mRNA is thought to unwind RNA to allow ribosomal binding. Similar to PPR10, a dual function in translational activation and 5′ maturation of a mitochondrial mRNA has been established for MPPR6. Interestingly, these dual functions evolved many times independently and in both plant organelles, hinting at a general principle of action for PPR proteins. Structure analysis also indicates that correct processing minimizes RNA folding around the start codon and therefore supports the action of MPPR6 in facilitating initiation of translation.
The observed mitochondrial translation deficiencies, reduced amounts of mitochondrial encoded proteins, and seed lethality in the mppr6 mutant are likely based on the production of only truncated RPS3 proteins, which do not appear in the wild type because of efficient translation initiation at the inherent start codon. RPS3 is essential in Escherichia coli and part of the30S ribosomal subunit (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp). It is added to the 16S rRNA at the last step of 30S subunit assembly (Culver and Noller, 1999). The plastid RPS3 protein was also shown to be essential in tobacco (Nicotiana tabacum; Fleischmann et al., 2011), indicating that the mitochondrial RPS3 version is essential as well. This is supported by the fact that deletion mutations within the rps3 gene in maize lead to a drastic reduction of mitochondrial protein synthesis along with massive growth impairments of heteroplasmic plants, whereas in homoplasmic individuals, the mutation proved to be kernel lethal (Hunt and Newton, 1991; Newton et al., 1996). Thus, absence of functional RPS3 is anticipated to result in mitochondrial translation deficiency. In the mppr6 mutant, mitochondrial translation is considerably reduced but not missing completely, as demonstrated by immunoblots and in organello protein synthesis. Residual activity of the truncated RPS3 protein and/or the production of traces of full-length RPS3 could account for the fact that basal mitochondrial translation principally occurs in mppr6.
The discovery of the essential and conserved functions of MPPR6-related proteins among land plants demonstrates the specificity by which mitochondrial rps3 gene expression is affected despite the rapid divergence of organellar UTRs. This implies an important role of MPPR6 during evolution. The recruitment of PPR and other proteins may offer the possibility of regulating expression in a gene-specific manner as has been demonstrated recently for the chloroplast PrfB3 protein (Stoppel et al., 2011). A dual role of a PPR protein in RNA processing and translation has been evolved independently and in different organelles, indicating a general principle of action during endosymbiosis. We anticipate that future research on PPR proteins will shed new light on the functional coevolution with their RNA targets.
METHODS
Plant Material and Growth Conditions
The mutant collection consisting of 27,500 maize (Zea mays) lines in which an endogenous Mu element had been allowed to transpose at high frequency was created by Biogemma using a Mu transposon line (provided by B. Taylor, Commonwealth Scientific and Industrial Research Organization, Canberra, Australia). The Mu line was crossed with maize hybrids developed by Limagrain to increase plant vigor and adaptation to European environmental conditions. Screening for defective kernels was performed visually on 22,000 F2 seeds; 6000 seed mutants were identified, 1200 of which showed an endosperm phenotype. Three hundred mutants showing a good 3:1 seed phenotype and stability of the phenotype over three generations were selected for further characterization and gene cloning. The maize lines A188 and H99 were additionally used in this study. Plants were grown in the field or in a greenhouse at 25°C, with 16 h light and a relative humidity of 55 to 95%. For DNA, RNA, and protein isolation, plant tissues were frozen in liquid nitrogen immediately after harvesting and stored at −70°C until needed.
Maize BMS cell suspension culture was used for transient biolistic transformation. The cells were cultivated in MS medium (Murashige and Skoog, 1962) supplemented with 5.98 mM l-Gln, 1.3 mM Pro, 0.67 mM Asn, 2 mg/L 2,4-D, and 30 g/L Suc at 26°C in the dark with shaking (70 rpm). BMS cells were subcultured every week in fresh medium.
To obtain 7-d-old etiolated seedlings for mitochondria isolation, surface-sterilized kernels were germinated on water-soaked paper towels under sterile conditions in the dark at 26°C.
For embryo rescue experiments, embryos were dissected from surface-sterilized 18 DAP mutant and wild-type kernels and transferred to half-strength MS medium. Rescued individuals were transferred to soil once they reached the three-leaf stage and were grown in the greenhouse in the conditions described above.
Arabidopsis thaliana lines SALK_045714 (appr6-1), SALK_091917 (appr6-2), and SALK_061950 (appr6-3) obtained from the Salk Institute and Columbia-0 wild type were germinated on soil or half-strength MS medium supplemented with 1.5% Suc at 23°C, with a relative humidity of 50% and a 15-h photoperiod. Heterozygous plants were identified by screening immature siliques for the presence of 25% defective seeds following self-pollination and PCR genotyping. For microscopy studies, immature seeds were cleared with Hoyer’s solution as described by Liu and Meinke (1998) and examined using a Zeiss Axioscope microscope equipped with differential interference contrast optic. Mature dried seeds were visualized under dark-field illumination. Images were captured and processed using the Zeiss AxioVision camera and AxioVision software (release 4.6.3).
Fixation and Embedding of Maize Kernels
The 12 DAP mutant and wild-type kernels were trimmed on both sides parallel to the embryo axis and incubated for 16 h at 4°C in 100 mM phosphate buffer, pH 7.2, containing 4% (w/v) paraformaldehyde and 0.1% (w/v) glutaraldehyde. After two washing steps (15 min each) in ice-cold 100 mM phosphate buffer, pH 7.2, kernels were dehydrated in a graded ethanol series (40 min/change) to 100%, which was then replaced by xylene. Kernels were finally embedded in Paraplast Plus (Carl Roth). The samples were sliced into 12-µm-thick sections, placed onto silicon-coated glass slides, and incubated at 42°C overnight. After paraffin removal and rehydration, sections were stained either with Azure B, pH 4.0, for 3 min or periodic acid Schiff’s reagent (Carl Roth) according to the manufacturer’s instructions.
Isolation of an Additional mppr6 Allele and Allelism Test
An additional Mu insertion event was isolated at Biogemma. Screening for Mu insertions in MPPR6 was performed on F1 plant material, and genotyping was confirmed in the F2 progeny. A PCR-based method was performed, using a transposon terminal inverted repeat–specific primer, OmuA, and MPPR6-gene specific primers designed in the first half of the gene (PPR_R03 and PPR_F03). One additional mutant allele was identified, J0487, with insertion at nucleotide + 394 in the ORF of the MPPR6 gene. The putative allelic line J0487, which showed a mutant mppr6-1–like seed phenotype, was renamed mppr6-2.
To determine genetic allelism between mppr6-1 and mppr6-2, 20 phenotypically wild-type kernels from a segregating ear of each mutant line were sown on soil. Seedlings were genotyped by genomic PCR using the terminal inverted repeat–specific primer Muoligo2 in combination with mppr6_1fw (for mppr6-1) or mppr6_2rev (for mppr6-2). Some of the genomic DNA samples were additionally investigated by DNA gel blots. Crosses were then performed between mppr6-1/+ and mppr6-2/+ plants. The phenotype of the resulting ears was analyzed.
Cloning of MPPR6 and cDNA Isolation
Total RNA from immature mutant kernels of two segregating sibling ears and wild-type kernels of a single sibling wild-type ear was extracted as described previously (Kluth et al., 2002). cDNA synthesis was performed according to the SMART RACE cDNA amplification kit instructions (Clontech). For both 3′ and 5′ RACE, the universal primer provided with the kit was combined with the Mu-specific primer Muoligo1. The nested round PCR was performed using the nested universal primer from the kit and Muoligo2. RACE products were electrophoretically separated on agarose gel and candidate bands present in the mutant samples but absent in the wild-type were eluted using the NucleoSpin Extract II kit (Machery-Nagel). The DNA was finally cloned into the pCR 2.1-TOPO vector (Invitrogen) and sequenced.
To obtain a full-length mppr6 cDNA clone, a kernel cDNA population was generated using the SMART RACE cDNA amplification kit (Clontech) and total RNA isolated as described above. The mppr6_3fw and mppr6_3rev primers were designed on the genomic clone (MAGIv4_67802) and used to amplify the complete ORF of MPPR6. To obtain the 5′ and 3′ UTR, the universal primer was combined with mppr6_4fw and mppr6_4rev primers, respectively. A nested PCR round was performed using the nested universal primer in combination with mppr6_5fw and mppr6_5fw primers for the 5′ and 3′ RACE, respectively.
Cosegregation Analysis
Genomic DNA was extracted from leaf samples of wild-type and heterozygous mppr6 sibling maize plants using the Plant Genomic DNA Mini Kit (Avegene). Genomic PCR was performed using ∼100 ng genomic DNA and primers Muoligo2 and mppr6_1fw. For DNA gel blot analysis, 10 µg of SacI-digested genomic DNA was electrophoretically separated on 0.8% agarose gel. DNA was transferred to a neutral nylon membrane (Hybond-NX; GE Healthcare) by capillary blotting with 20× SSC. Digoxigenin (DIG)-labeled MPPR6-specific probe was generated with the PCR DIG probe synthesis kit (Roche). Hybridization was performed according to the DIG Application Manual for Filter Hybridization (Roche). Target-probe hybrids were detected in a chemiluminescent reaction with CSDP (Roche) as a substrate and visualized on x-ray film.
Complementation
The MPPR6 ORF was amplified using Pfu polymerase (Fermentas) and primers mppr6_3fw and mppr6_7rev. The PCR product was digested with BamHI and XbaI and cloned into the BamHI and XbaI sites of pCHF5 (gift of C. Fankhauser, University of Geneva, Switzerland). The resulting vector was transformed into Agrobacterium tumefaciens GV3101pMP90 (Koncz and Schell, 1986) and used for floral dip transformation of homozygous Arabidopsis plants as described by Clough and Bent (1998). Resulting seeds were sown on soil, and transgenic plants were selected by spraying the seedlings with aqueous solution of 250 mg/L phosphinothricin and 0.1% (v/v) Tween 20.
Generation of Transgenic Maize Plants
For generation of transgenic maize plants overexpressing mppr6-Strep, the ORF of MPPR6 was amplified (primers: mppr6-3fw and mppr6-9rev), digested with BamHI, and inserted into the BamHI sites of pUbi:cas (Dirk Becker, University of Hamburg). The resulting construct, pUbi:mppr6-Strep, was used for generation of transgenic maize plants by stable biolistic transformation of embryogenic calli from a cross between A188 × H99 as described previously (Brettschneider et al., 1997). T0 progeny of putative transgenic, phosphinothricin-resistant plants were verified by DNA and RNA gel blot analysis according to the DIG Application Manual for Filter Hybridization.
RT-PCR
Tissues from the inbred maize line A188 were used to obtain total RNA as described previously (Kluth et al., 2002). Removal of genomic DNA contamination was performed by treatment with RNase-free DNase I (Fermentas) according to the recommended protocol. One microgram of DNA-free RNA was reverse transcribed with PrimeScript reverse transcriptase (Takara Bio) according to the manufacturer’s directions of the SMART RACE cDNA amplification kit (Clontech). Three microliters of the 10-fold diluted reverse transcription reaction were used in a low-cycle (25) PCR with Taq DNA polymerase (Fermentas) and the MPPR6-specific primers: mppr6_6fw and mppr6_6rev. One-tenth of the RT-PCR products was separated by agarose gel electrophoresis and blotted on neutral nylon membrane (Hybond-NX; GE Healthcare) by capillary transfer with 20× SSC. DIG probe labeling, hybridization, and detection were performed as described above.
For cRT-PCR, 10 µg of total RNA obtained from wild-type and mppr6 mutant callus tissue were self-ligated using T4 RNA Ligase 1 (NEB). One-tenth of the circular RNA was subjected to reverse transcription reaction using the AMV RT (Fermentas) and gene-specific primer (rps3-rpl16_5fw), followed by two rounds of PCR with nested primers (for first-round PCR, rps3-rpl16_5fw and rps3_5rev; for second-round PCR, rps3-rpl16_6fw and rps3_6rev). Abundant cRT-PCR products were cut out from the agarose gel, purified, cloned into pGEM T vector, and sequenced. For better resolution, cRT-PCR products were additionally separated on denaturing 5% polyacrylamide gels containing 8 M urea. Electrophoresis was performed in 1% TBE buffer. cRT-PCR products were visualized after staining gels with ethidium bromide.
RNA Gel Blot Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Electrophoresis and RNA gel blotting were performed as described previously (Meurer et al., 2002). Hybridization was performed with ExpressHyb buffer (Clontech) and radioactive-labeled antisense 80-mers (see Supplemental Table 1 online) according to the manufacturer’s instructions. Blots were analyzed with a phosphor imaging system (Typhoon; Amersham Biosciences).
Protein Analysis and Antibody Production
For isolation of total protein, leaf material was ground in liquid nitrogen and resuspended in isolation buffer (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EGTA, 5 mM EDTA, 10 mM DTT, 0.5% [v/v] Triton X-100, and Complete Protease Inhibitor Cocktail Tablets [Roche]), and after centrifugation at 18,000g for 20 min, the supernatant was collected. To obtain mitochondrial proteins, crude mitochondria were resuspended in lysis buffer (30 mM HEPES-KOH, pH 7.7, 10 mM Mg acetate, 60 mM K acetate, 2 mM DTT, and 0.5% Nonidet P-40, supplemented with Complete Protease Inhibitor Cocktail Tablets) and lysed by a 30-min incubation on ice interrupted by occasional short vortexing. Membrane proteins were separated from the soluble ones by centrifugation at 20,800g for 30 min. Proteins were separated by SDS-PAGE, transferred into nitrocellulose membrane (Protran; Whatman) using semidry Electrophoretic Transfer Cell (Bio-Rad), and stained with Ponceau solution (0.1% [w/v] Ponceau S and 5% [v/v] acetic acid). Antibodies were used at the following dilutions: anti-MPPR6 rabbit, anti-RPS3_1 rabbit, and anti RPS3_2 rabbit 1:500 each; anti-NAD9 rabbit (Lamattina et al., 1993) 1:50,000; anti-COXII hen (Agrisera) 1:1000; anti-cpHSP70-2 rabbit (Agrisera) 1:2000; anti-PRXII F rabbit (Finkemeier et al., 2005) 1:5000; anti-CS rabbit (kindly provided by Iris Finkemeier, Ludwig-Maximillians-University Munich) 1:10,000. Signals were visualized using chemiluminescence. For antibody production, the amino acid residues 60 to 489 of MPPR6 were fused with a C-terminal His6 tag using the pET30a+ vector (Novagen) and expressed in Escherichia coli strain BL 21 (DE3). The recombinant protein was purified under denaturing conditions on a nickel-nitrilotriacetic acid agarose column (Qiagen) and used for the generation of antibodies in rabbits at the Eurogentec’s antibody facility.
Two antipeptide antibodies with specificity for RPS3 (amino acids 82 to 96, LKRRDKSRPGKDKGR, and amino acids 11 to 26, RLDLNRSSDPSRFSDY) were generated in rabbits and affinity purified by Eurogentec.
Subcellular Localization
The signal sequence of MPPR6 (amplified with primers mppr6_3fw/rev) and the ORF of EMP4 (amplified with primers emp4_1fw/rev) were inserted into Ubi:dsRed (constructed by R.B.) and Ubi:GFP (Lorbiecke et al., 2005), respectively. BMS cells or onion epidermis were biolistically cotransformed as described previously (Lorbiecke et al., 2005) using the resulting constructs Ubi:mppr6-dsRed and Ubi:emp4-GFP, or Ubi:mppr6-dsRed and Ubi:cpHSP70-2-GFP (kindly provided by Jantjeline Kluth, University of Hamburg, Germany), respectively. Eighteen hours later, the transient coexpression was visualized by fluorescence microscopy using a Zeiss AX10 ImagerZ1 ApoTome. The emission for dsRed was visualized at 570 to 640 nm and for GFP at 500 to 550 nm. The AxioVision 4.8 software was used for subsequent image processing.
Co-IP and Identification of MPPR6 Target Mitochondrial RNA
An enriched mitochondrial fraction was obtained from etiolated MPPR6-overexpressing maize seedlings as described previously (Takenaka and Brennicke, 2007). The crude mitochondrial pellet was resuspended in ice-cold lysis buffer (30 mM HEPES-KOH, pH 7.7, 10 mM Mg acetate, 60 mM K acetate, 2 mM DTT, and 0.5% Nonidet P-40, supplemented with Complete Protease Inhibitor Cocktail Tablets). Lysis of mitochondria was performed by two freeze/thaw cycles, followed by 30-min incubation on ice and several short vortexing steps. The lysate was then clarified by centrifugation at 20,800g for 15 min at 4°C and placed on ice.
Protein A Dynabeads (100 μL; Invitrogen) was washed twice in 0.5 mL of Co-IP buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.5% Nonidet P-40, and Complete Protease Inhibitor Cocktail Tablets) and finally resuspended in 100 μL of the same buffer. Forty microliters of anti-MPPR6 or preimmune rabbit serum was added to the washed Protein A Dynabeads and incubated for 2 h at 4°C with rotation. After three washes with Co-IP buffer, the beads were resuspended in 800 μL of Co-IP buffer supplemented with RNase Inhibitor (Fermentas). One hundred microliters of mitochondrial extract (∼1 mg protein) was added to the mixture from the previous step and incubated at 4°C overnight with rocking. Beads were collected by magnetic separation; the resulting supernatant was used for RNA extraction and subsequent dot blot hybridization. The beads were washed six times in 1 mL of Co-IP buffer, resuspended in 200 μL of Co-IP buffer supplemented with SDS to 1% and 10 mM EDTA, and incubated for 30 min at 55°C. RNA from each pellet or supernatant was phenol-chloroform extracted, ethanol-precipitated, and resuspended in 10 μL of water. Three microliters of the RNA obtained from the pellet was used in a poly(A) tailing reaction using E. coli Poly(A) Polymerase (NEB) according to the manufacturer’s instructions. The polyadenylated RNA was reverse transcribed and subsequently amplified by a long-distance RT-PCR following the Super SMART PCR cDNA Synthesis Kit protocol (Clontech). RT-PCR products were cloned into pCR 2.1-TOPO vector (Invitrogen) and sequenced.
Immunoprecipitation of strep-tagged proteins was performed with 1 mg of mitochondrial protein using a Strep-Tactin column (IBA) according to the manufacturer’s instructions. Washing and RNA extraction were performed as described above. For slot blot hybridizations, one-third of the pellet RNA and one-twelfth of the supernatant RNA were spotted onto nylon membranes and hybridized with the 32P-labeled probes.
For dot blot hybridization, 1 μL of RNA purified from the pellet or the supernatant was applied to a nylon membrane and hybridized with 32P-labeled rps3 probe (Sauter, 1997) generated by a random priming method using the Prime It II DNA labeling kit (Stratagene).
Expression and Purification of Recombinant MPPR6
To generate a GST-MPPR6-Strep fusion protein, the coding sequence of MPPR6 corresponding to the mature protein (excluding the predicted transit sequence) was amplified by RT-PCR using primers mppr6_8fw and mppr6_8rev and inserted into the high-level expression vector pET41b+ after digestion with SpeI and XhoI. The resulting plasmid was introduced into E. coli strain Bl21pLysS (Novagen). Cells were grown in Luria-Bertani medium supplemented with kanamycin at 37°C to an OD600 of 0.7. After induction of protein synthesis by addition of isopropyl-β-d-thiogalactopyranoside, the culture was incubated for 4 h at 37°C. Cells were then lysed by sonification in strep-washing buffer (IBA) supplemented with 10% (v/v) Sarkosyl and Complete Protease Inhibitor Cocktail Tablets. The postcentrifugation supernatant was filtered through a 0.45-µm filter, diluted 1:5 with Strep-washing buffer supplemented with 2% Triton X-100 and 0.1% CHAPS, and incubated for 1 h in Strep-Tactin-Macroprep (IBA). Washing steps were performed with Strep-washing buffer lacking detergents. The GST-MPPR6-Strep-Fusion protein was eluted with strep-elution buffer supplemented with 30% glycerol. Protein concentration was determined by Bradford assay using BSA as standard. Protein purity was controlled by SDS-PAGE and subsequent Coomassie Brilliant Blue staining.
EMSA and in Organello Translation
RNAs were generated by in vitro transcription using T7 RNA Polymerase (Fermentas) according to the manufacturer’s instructions. PCR products containing the T7 promoter were used as template. Reactions (25 µL) were performed in binding buffer composed of 40 mM Tris, pH 7.5, 100 mM NaCl, 0.1 mg/mL BSA, 4 mM DTT, and 0.5 mg/mL heparin at 25°C for 20 min. Protein and RNA concentrations are indicated in Figure 10. Samples were separated on a nondenaturing 4% polyacrylamide (29:1 acrylamide:bis-acrylamide) gel prepared in 0.5× TBE buffer. Gels were dried and analyzed with a phosphor imaging system (Typhoon).
In organello translation was performed as described previously (Grohmann, 1996).
Sequence Analysis
cDNA clones and PCR fragments were sequenced using the ABI Prism Big Dye reaction kit and ABI Prism sequencer (Applied Biosystems). Sequence data were analyzed using the programs included in DNASTAR 4.05 software package. PPR domains were identified using the ScanProsite tool of the ExPASy Proteomics Server (http://www.expasy.ch/tools/scanprosite/). The subcellular localization was determined using TargetP (Emanuelsson et al., 2000) and Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html) (Small et al., 2004). For the search of putative orthologous protein groups, thePlantRBP/POGs database (http://plantrbp.uoregon.edu/) was used. Alignment was calculated with ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) (Larkin et al., 2007) and manually edited using GeneDoc 2.6.002 (Nicholas and Nicholas, 1997).
Accession Numbers
Sequence data for mppr6 cDNA can be found in the GenBank database under accession number HQ534069. Sequences of the maize, rice (Oryza sativa), and Arabidopsis PPR proteins shown in Figure 4 can be found in the GenBank database under accession numbers GI:313851107, GI:115455503, and GI:334183972, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotype and Segregation of mppr6.
Supplemental Figure 2. Molecular Analysis of mppr6-strep Transgenic Plants.
Supplemental Figure 3. MPPR6 Binds to 5′ UTR of rps3 RNA at High Salt Conditions.
Supplemental Figure 4. Unlabeled Competitor RNA.
Supplemental Figure 5. Analyses of rps3 Transcripts.
Supplemental Figure 6. Rps3 cRT-PCR Products Separated on Polyacrylamide Gel.
Supplemental Figure 7. Mapping of rps3 5′ Termini by Primer Extension Assay.
Supplemental Figure 8. APPR6 Expression Profile Determined by the Arabidopsis eFP Browser.
Supplemental Figure 9. Secondary Structure of rps3 5′ UTRs Determined by M-Fold.
Supplemental Table 1. Names and Sequences of Used Primers.
Supplementary Material
Acknowledgments
We thank Gregorio Hueros and Peter Rogowsky for developing the cDNA-based mutant screening strategy and for useful discussions. We also thank the gene machine team (Biogemma) for the screening for a second mppr6 allele, Sönke Jäger and Wyatt Paul for reading the article and suggesting changes, Susan Pusunc and Victoria Martens for performing the RNA extractions, Iris Finkemeier for providing peroxiredoxin and CS antibodies, Géraldine Bonnard for providing the NAD9 antiserum, and Katja Müller for help with the maize transformation. This work was financially supported by the Deutsche Forschungsgemeinschaft (SFB-TR1) to J.M. and the Bundesministerium für Bildung und Forschung (EraNet Grant FKZ 0313995) to N.M. V.G. was partially supported by the Agence Nationale de la Recherche (Grant ERAPG-001-01).
AUTHOR CONTRIBUTIONS
N.M., R.B., J.M., U.W., and V.G. designed the research. N.M. performed the research. N.M., J.M., R.B., and V.G. analyzed data. N.M. and J.M. wrote the article.
Glossary
- ESR
embryo surrounding region
- BETL
basal endosperm transfer layer
- PPR
pentatricopeptide repeat
- UTR
untranslated region
- DAP
days after pollination
- MS
Murashige and Skoog
- WIG
wall ingrowth
- RACE
rapid amplification of cDNA ends
- ORF
open reading frame
- GFP
green fluorescent protein
- BMS
Black Mexican Sweet
- Co-IP
coimmunoprecipitation
- EMSA
electrophoretic mobility shift assay
- cRT-PCR
circular RT-PCR
- DIG
to be defined
References
- Barkan A. (2011). Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A. (2008). The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S., Knight W., Hanson M. (2010). Natural variation in Arabidopsis leads to the identification of REME1, a pentatricopeptide repeat-DYW protein controlling the editing of mitochondrial transcripts. Plant Physiol. 154: 1966–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider R., Becker D., Lörz H. (1997). Efficient transformation of scutellar tissue of immature maize embryos. Theor. Appl. Genet. 94: 737–748 [Google Scholar]
- Cai W., Okuda K., Peng L., Shikanai T. (2011). PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 67: 318–327 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin A.L., des Francs-Small C.C., Delannoy E., Kahlau S., Tanz S.K., de Longevialle A.F., Fujii S., Small I. (2011). OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J. 65: 532–542 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin A.L., Ramos-Vega M., Guevara-García A., Andrés C., de la Luz Gutiérrez-Nava M., Cantero A., Delannoy E., Jiménez L.F., Lurin C., Small I., León P. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Costa L.M., Gutierrez-Marcos J.F., Brutnell T.P., Greenland A.J., Dickinson H.G. (2003). The globby1-1 (glo1-1) mutation disrupts nuclear and cell division in the developing maize seed causing alterations in endosperm cell fate and tissue differentiation. Development 130: 5009–5017 [DOI] [PubMed] [Google Scholar]
- Culver G.M., Noller H.F. (1999). Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA 5: 832–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle A.F., Meyer E.H., Andrés C., Taylor N.L., Lurin C., Millar A.H., Small I.D. (2007). The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L.J., Dietz K.J. (2005). The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 280: 12168–12180 [DOI] [PubMed] [Google Scholar]
- Fleischmann T.T., Scharff L.B., Alkatib S., Hasdorf S., Schöttler M.A., Bock R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: A developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé P., Brennicke A. (2001). From gene to protein in higher plant mitochondria. C. R. Acad. Sci. III 324: 209–217 [DOI] [PubMed] [Google Scholar]
- Greiner S., Wang X., Herrmann R.G., Rauwolf U., Mayer K., Haberer G., Meurer J. (2008). The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol. Biol. Evol. 25: 2019–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann L. (1996). In organello protein synthesis. Methods Mol. Biol. 49: 391–397 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Dal Prà M., Giulini A., Costa L.M., Gavazzi G., Cordelier S., Sellam O., Tatout C., Paul W., Perez P., Dickinson H.G., Consonni G. (2007). empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 19: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., des Francs-Small C.C., Takenaka M., Tanz S.K., Okuda K., Shikanai T., Brennicke A., Small I. (2011). The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J. Biol. Chem. 286: 21361–21371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Miyake H., Sugita M. (2007). A Pentatricopeptide repeat protein is required for RNA processing of clpP Pre-mRNA in moss chloroplasts. J. Biol. Chem. 282: 10773–10782 [DOI] [PubMed] [Google Scholar]
- Hazle T., Bonen L. (2007). Comparative analysis of sequences preceding protein-coding mitochondrial genes in flowering plants. Mol. Biol. Evol. 24: 1101–1112 [DOI] [PubMed] [Google Scholar]
- Hölzle A., Jonietz C., Törjek O., Altmann T., Binder S., Forner J. (2011). A RESTORER OF FERTILITY-like PPR gene is required for 5′-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 65: 737–744 [DOI] [PubMed] [Google Scholar]
- Hunt M.D., Newton K.J. (1991). The NCS3 mutation: Genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 10: 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X., Wostrikoff K., Finazzi G., Kuras R., Schwarz C., Bujaldon S., Nickelsen J., Stern D.B., Wollman F.A., Vallon O. (2010). MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonietz C., Forner J., Hildebrandt T., Binder S. (2011). RNA PROCESSING FACTOR3 is crucial for the accumulation of mature ccmC transcripts in mitochondria of Arabidopsis accession Columbia. Plant Physiol. 157: 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonietz C., Forner J., Hölzle A., Thuss S., Binder S. (2010). RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell 22: 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B.-H., Xiong Y., Williams D.S., Pozueta-Romero D., Chourey P.S. (2009). Miniature1-encoded cell wall invertase is essential for assembly and function of wall-in-growth in the maize endosperm transfer cell. Plant Physiol. 151: 1366–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Ishikura K., Kasai S., Shinmyo A. (2006). Efficient translation destabilizes transcripts in chloroplasts of Chlamydomonas reinhardtii. J. Biosci. Bioeng. 101: 471–477 [DOI] [PubMed] [Google Scholar]
- Kiesselbach T.A. (1949). The Structure and Reproduction of Corn. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Klaff P., Gruissem W. (1991). Changes in chloroplast mRNA stability during leaf development. Plant Cell 3: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluth A., Sprunck S., Becker D., Lörz H., Lütticke S. (2002). 5′ Deletion of a gbss1 promoter region from wheat leads to changes in tissue and developmental specificities. Plant Mol. Biol. 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Kawabata M., Hisano K., Kazama T., Matsuoka K., Sugita M., Nakamura T. (2012). Identification and characterization of the RNA binding surface of the pentatricopeptide repeat protein. Nucleic Acids Res. 40: 2712–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocábek T., Repková J., Dudová M., Hoyerová K., Vrba L. (2006). Isolation and characterization of a novel semi-lethal Arabidopsis thaliana mutant of gene for pentatricopeptide (PPR) repeat-containing protein. Genetica 128: 395–407 [DOI] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396 [Google Scholar]
- Koprivova A., des Francs-Small C.C., Calder G., Mugford S.T., Tanz S., Lee B.R., Zechmann B., Small I., Kopriva S. (2010). Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J. Biol. Chem. 285: 32192–32199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Lamattina L., Gonzalez D., Gualberto J., Grienenberger J.M. (1993). Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217: 831–838 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Liu C.M., Meinke D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16: 21–31 [DOI] [PubMed] [Google Scholar]
- Liu Y., He J., Chen Z., Ren X., Hong X., Gong Z. (2010). ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J. 63: 749–765 [DOI] [PubMed] [Google Scholar]
- Lorbiecke R., Steffens M., Tomm J.M., Scholten S., von Wiegen P., Kranz E., Wienand U., Sauter M. (2005). Phytosulphokine gene regulation during maize (Zea mays L.) reproduction. J. Exp. Bot. 56: 1805–1819 [DOI] [PubMed] [Google Scholar]
- Lowe J., Nelson O.E. (1946). Miniature seed-a study in the development of a defective caryopsis in maize. Genetics 31: 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Lezhneva L., Amann K., Gödel M., Bezhani S., Sherameti I., Oelmüller R. (2002). A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell 14: 3255–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Heazlewood J.L., Kristensen B.K., Braun H.-P., Møller I.M. (2005). The plant mitochondrial proteome. Trends Plant Sci. 10: 36–43 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Newton K.J., Mariano J.M., Gibson C.M., Kuzmin E., Gabay-Laughnan S. (1996). Involvement of S2 episomal sequences in the generation of NCS4 deletion mutation in maize mitochondria. Dev. Genet. 19: 277–286 [DOI] [PubMed] [Google Scholar]
- Nicholas K.B., Nicholas H.B.J. (1997). GeneDoc: A tool for editing and annotating multiple sequence alignments. EMBNEW.NEWS 4: 14 [Google Scholar]
- Okuda K., Habata Y., Kobayashi Y., Shikanai T. (2008). Amino acid sequence variations in Nicotiana CRR4 orthologs determine the species-specific efficiency of RNA editing in plastids. Nucleic Acids Res. 36: 6155–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K., Nakamura T., Sugita M., Shimizu T., Shikanai T. (2006). A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 281: 37661–37667 [DOI] [PubMed] [Google Scholar]
- O’Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., Sugita M., Small I. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Rojas M., Schuster G., Barkan A. (2011). Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusnik M., Schneider A. (2012). A trypanosomal pentatricopeptide repeat protein stabilizes the mitochondrial mRNAs of cytochrome oxidase subunit 1 and 2. Eukaryot. Cell 11: 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo J., Gomez E., Hueros G. (2007). Transfer cells. In Endosperm, Plant Cell Monographs, Vol. 8 (Berlin: Springer-Verlag), pp. 73–89
- Sauter M. (1997). Differential expression of a CAK (cdc2-activating kinase)-like protein kinase, cyclins and cdc2 genes from rice during the cell cycle and in response to gibberellin. Plant J. 11: 181–190 [DOI] [PubMed] [Google Scholar]
- Scharff L.B., Childs L., Walther D., Bock R. (2011). Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 7: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R., Barkan A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R.E., Williams-Voelker P.M., Kroeger T.S., Vichas A., Barkan A. (2006). A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I., Peeters N., Legeai F., Lurin C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Small I.D., Peeters N. (2000). The PPR motif - A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Stoppel R., Lezhneva L., Schwenkert S., Torabi S., Felder S., Meierhoff K., Westhoff P., Meurer J. (2011). Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell 23: 2680–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R., Meurer J. (2012). The cutting crew - Ribonucleases are key players in the control of plastid gene expression. J. Exp. Bot. 63: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Sung T.Y., Tseng C.C., Hsieh M.H. (2010). The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J. 63: 499–511 [DOI] [PubMed] [Google Scholar]
- Takenaka M., Brennicke A. (2007). RNA editing in plant mitochondria: Assays and biochemical approaches. Methods Enzymol. 424: 439–458 [DOI] [PubMed] [Google Scholar]
- Thompson R.D., Hueros G., Becker H., Maitz M. (2001). Development and functions of seed transfer cells. Plant Sci. 160: 775–783 [DOI] [PubMed] [Google Scholar]
- Uchida M., Ohtani S., Ichinose M., Sugita C., Sugita M. (2011). The PPR-DYW proteins are required for RNA editing of rps14, cox1 and nad5 transcripts in Physcomitrella patens mitochondria. FEBS Lett. 585: 2367–2371 [DOI] [PubMed] [Google Scholar]
- Uyttewaal M., Mireau H., Rurek M., Hammani K., Arnal N., Quadrado M., Giegé P. (2008). PPR336 is associated with polysomes in plant mitochondria. J. Mol. Biol. 375: 626–636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.