Monolignol oxidative coupling is the primary process in lignin biosynthesis, which requires free phenol of a monolignol to generate active radicals. A 4-O-methyltransferase was created via protein engineering that masks the 4-hydroxyls of monolignols. Expressing this enzyme in Arabidopsis led to a substantial reduction of lignin content and concomitant production of novel phenolic esters.
Abstract
Although the practice of protein engineering is industrially fruitful in creating biocatalysts and therapeutic proteins, applications of analogous techniques in the field of plant metabolic engineering are still in their infancy. Lignins are aromatic natural polymers derived from the oxidative polymerization of primarily three different hydroxycinnamyl alcohols, the monolignols. Polymerization of lignin starts with the oxidation of monolignols, followed by endwise cross-coupling of (radicals of) a monolignol and the growing oligomer/polymer. The para-hydroxyl of each monolignol is crucial for radical generation and subsequent coupling. Here, we describe the structure-function analysis and catalytic improvement of an artificial monolignol 4-O-methyltransferase created by iterative saturation mutagenesis and its use in modulating lignin and phenylpropanoid biosynthesis. We show that expressing the created enzyme in planta, thus etherifying the para-hydroxyls of lignin monomeric precursors, denies the derived monolignols any participation in the subsequent coupling process, substantially reducing lignification and, ultimately, lignin content. Concomitantly, the transgenic plants accumulated de novo synthesized 4-O-methylated soluble phenolics and wall-bound esters. The lower lignin levels of transgenic plants resulted in higher saccharification yields. Our study, through a structure-based protein engineering approach, offers a novel strategy for modulating phenylpropanoid/lignin biosynthesis to improve cell wall digestibility and diversify the repertories of biologically active compounds.
INTRODUCTION
Lignin is a major biopolymer in secondary cell walls of terrestrial plants. However, its presence presents a formidable obstacle to the efficient use of cellulosic fibers in agricultural and industrial applications, particularly the conversion of cellulosic biomass to liquid biofuels (Weng et al., 2008). Although lignin biosynthesis has been studied for decades and considerable efforts have been dedicated to modulating lignin content or its composition to more economically use cell wall fibers, novel biotechnology/methodology is still required for efficiently manipulating plant lignification.
Lignin is generally considered to arise from three monolignols, p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, which differ in their degree of meta-methoxylation (methoxylation ortho to the phenol) (see Supplemental Figure 1 online). After their synthesis in the cytosol, they are transported across the cell membrane and deposited into the cell wall, where they are oxidized and polymerized to yield p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, respectively, when incorporated into the lignin polymer. The composition of lignin varies between different plant species and tissues. Lignin from gymnosperms and other lower vascular plants is rich in G-units and contains low amounts of H-units, and dicot lignin is mainly composed of G- and S-units (Weng and Chapple, 2010). The G-unit is monomethoxylated (via methylation of the 3-hydroxyl group in caffeyl intermediates), whereas the S-unit is dimethoxylated (via additional methylation of the 5-hydroxyl moieties in 5-hydroxyguaiacyl intermediates). The ratio of S:G varies from 0:100 to nearly 100:0, resulting in alterations in lignin structures that dictate the degree of lignin condensation, reactivity, and recalcitrance to delignification (Boerjan et al., 2003; Ralph et al., 2004b).
The monolignols are synthesized from a branch of the phenylpropanoid pathway, where ∼10 catalytic enzymes constitute a linear pathway converting the aromatic amino acid l-Phe to the three hydroxycinnamyl alcohols (see Supplemental Figure 1 online). In members of the Brassicaceae, including Arabidopsis thaliana, the biosynthetic intermediates, hydroxycinnamaldehydes, on this pathway can be diverted to yield methanolic soluble phenolic esters, primarily sinapoyl malate in leaves, and sinapoyl choline in seeds, which act as a UV screen and pest deterrent, respectively (Landry et al., 1995; Milkowski et al., 2004; Nair et al., 2004) (see Supplemental Figure 1 online).
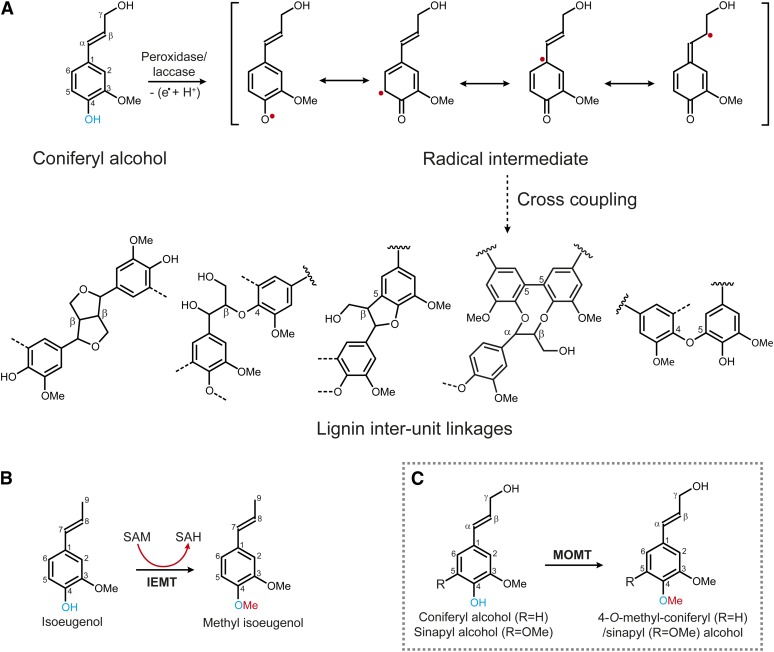
Earlier studies revealed that monolignols undergo dehydrodimerization or polymerization in an in vitro horseradish peroxidase–hydrogen peroxide (H2O2) system to form synthetic lignins (i.e., dehydrogenation polymers), with most of the structural features found in plant lignins (Freudenberg, 1959). Consequently, it is envisioned that lignin polymerization occurs via one-electron oxidation of phenols by oxidative enzymes (peroxidases/laccases) to yield resonance-stabilized phenolic radicals; subsequently, the coupling of phenoxy radicals with each other or, more importantly, with the growing polymer, forms lignin (Figure 1A) (Freudenberg, 1968; Boerjan et al., 2003; Ralph et al., 2004b; Davin et al., 2008). The cross-coupling of phenoxy radicals from the three monolignols (i.e., p-coumaryl, coniferyl, and sinapyl alcohols) within the plant cell wall generates different lignin interunit linkages, including the most frequent β-aryl ether (8–O–4) units and less frequent β–β (8–8) and β–5 (8–5) units; coupling between preformed oligomer/polymer units forms 5–5- and less frequent 5–O–4-linked units (Boerjan et al., 2003; Ralph et al., 2004b) (Figure 1A). In this oxidative coupling process, an unsubstituted phenol, the free para-hydroxyl of the monolignol, was critically implicated in generating phenoxy radical intermediates and in forming different types of lignin subunits (Harkin, 1967, 1973; Ralph et al., 2004b, 2009). Therefore, we rationalized that a chemical modification, for example, the methylation of the phenol (the para-hydroxyl), would prevent radical generation and deny the derived monolignols any participation in the subsequent coupling process, thus reducing the quantity of lignin produced.
Figure 1.
Schema of the Proposed Oxidative-Dehydrogenation and Phenoxy-Coupling, and the Reactions Catalyzed by Phenolic O-Methyltransferases.
(A) The active radical intermediates are generated from the monolignol (coniferyl alcohol) by oxidation of its phenol. The phenoxy radicals couple together or, more importantly, with the growing lignin oligomer/polymer yielding units with different interunit linkages in lignins.
(B) The 4-O-methylation of phenylpropenes catalyzed by IEMT.
(C) The engineered monolignol 4-OMT (MOMT) (in gray box); note the structural analogy between the monolignol coniferyl alcohol and the phenylpropene isoeugenol.
[See online article for color version of this figure.]
Monolignol biosynthesis requires the activities of two types of O-methyltransferases, the caffeoyl CoA 3-O-methyltransferase (EC. 2.1.1.104), which specifically catalyzes the methylation of cinnamoyl CoA conjugates (Franke et al., 2002; Boerjan et al., 2003), and a caffeate/5-hydroxyferulate 3/5-O-methyltransferase (COMT) (EC. 2.1.1.6), which preferentially methylates hydroxycinnamaldehydes or hydroxycinnamyl alcohols (Inoue et al., 2000; Parvathi et al., 2001) (see Supplemental Figure 1 online). However, both enzymes exclusively methylate the meta-hydroxyls, and none of them display activity toward 4-O-methylation (Parvathi et al., 2001; Bhuiya and Liu, 2010). However, an O-methyltransferase in fairy fans (Clarkia breweri), namely, isoeugenol O-methyltransferase (IEMT) (EC. 2.1.1.146), catalyzes the 4-O-methylation of volatile compounds isoeugenol and eugenol (Wang and Pichersky, 1998, 1999) (Figure 1B). These allyl/propenylphenols are structural analogs of monolignols, differing only in their propanoid tail (Figure 1B). IEMT exhibits strict substrate specificity, showing none or only negligible activity toward monolignols (Bhuiya and Liu, 2010). At the amino acid level, it shares more than 83% identity with lignin COMT from the same species. Phylogenetic analysis suggests it evolved from the parental enzyme COMT through functional diversification (Wang and Pichersky, 1998). Previously, Wang and Pichersky showed that swapping sequence fragments of IEMT and COMT, or converting some residues around their putative active sites, invariably and simultaneously alters their substrate preference and regio-selectivity of methylation (i.e., the mutations change the activity of IEMT from the 4-O-methylation of allyl/propenylphenols to the 3/5-O-methylation of caffeic acid and vice versa), although the catalytic efficiency of the resulting variants was greatly compromised (Wang and Pichersky, 1998, 1999). The evolutionary relationship of IEMT with the lignin biosynthetic enzyme and its functional amenability suggest the potential of using IEMT as template to create a novel enzyme conferring the ability to 4-O-methylate monolignols through protein engineering.
Protein engineering has proven a powerful strategy for enhancing the stability, activity, and/or selectivity of enzymes. During the last decade, the practice of rational design and laboratory-directed protein evolution has become firmly established as a versatile tool in biochemical research by enabling molecular evolution toward desirable phenotypes or for the detection of novel structure-function interactions. This strategy has led to a number of successes in creating novel biocatalysts for industrial applications. However, the practice of protein engineering for plant metabolites is rare. The most successful cases in altering the properties of plant secondary metabolic enzymes are on sesquiterpene synthases. By systematically exploring the relationship between sequence and function, Yoshikuni et al. (2006) identified and mutated a set of hot spots near the active site of sesquiterpene synthases that results in biocatalysts selectively catalyzing the formation of a spectrum of sesquiterpene products). Furthermore, O’Maille et al. (2008) systematically investigated the relationship between a combination of nine naturally occurring amino acid substitutions and the resulting sesquiterpene product spectra. However, the application of the resulting catalysts was not attempted in planta for diversifying plant metabolism.
Based on the IEMT homology modeling analysis and using the available COMT crystal structure (Zubieta et al., 2002), we previously identified a subset of evolutionarily useful hot spot residues in the active site of IEMT (Bhuiya and Liu, 2010). Site saturation mutagenesis yielded a few single to triple mutant variants that confer the ability to regiospecifically methylate the para-hydroxyl of monolignols; we termed them monolignol 4-O-methyltransferases (MOMTs) (Bhuiya and Liu, 2010). Although the triple mutant variants displayed considerable in vitro activity in etherifying monolignols, when overexpressed in planta they essentially did not result in any of the anticipated effects on phenylpropanoid and lignin biosynthesis. To further improve the catalytic properties of MOMT to obtain a physiologically effective enzyme, in this study, we explored the structural basis of the created MOMT variant for its regiospecific methylation by determining the x-ray crystal structure of a triple mutant variant; subsequently we refined its catalytic efficiency via additional iterations of saturation mutagenesis. We then expressed the created artificial enzyme in Arabidopsis, resulting in a substantial reduction of the lignin quantity in the cell wall and an increase in the yield of biomass saccharification concomitant with the production of novel phenylpropanoid metabolites.
RESULTS
Expressing the Early Set of MOMT Mutant Variants Does Not Affect Phenylpropanoid Metabolism in Planta
Our previous study created a subset of single (E165F) to triple (T133L-E165I-F175I) mutant variants of IEMT with significant catalytic efficiencies (kcat/Km of up to 1500 M−1 s−1) for methylating the para-hydroxyl of monolignols (Bhuiya and Liu, 2010) (see Supplemental Table 1 online). We expressed both the single and triple mutant variants in either tobacco (Nicotiana tabacum) or Arabidopsis, driven by either the constitutive double-35S promoter or the bean (Phaseolus vulgaris) phenylalanine ammonia lyase-2 (PAL2) promoter, which controls the expression of the first key enzyme in the phenylpropanoid pathway in vascular tissues (Cramer et al., 1989; Reddy et al., 2005). However, the resulting transgenic plants showed essentially no detectable change in the lignin content of cell wall and no change in the accumulation of other soluble phenolics. Among the possible reasons for the failure of the created enzymes to function in planta, one obvious obstacle probably lies in less than optimal catalytic efficiency.
Crystal Structure of MOMT3 and Improvement of Its Catalytic Efficiency
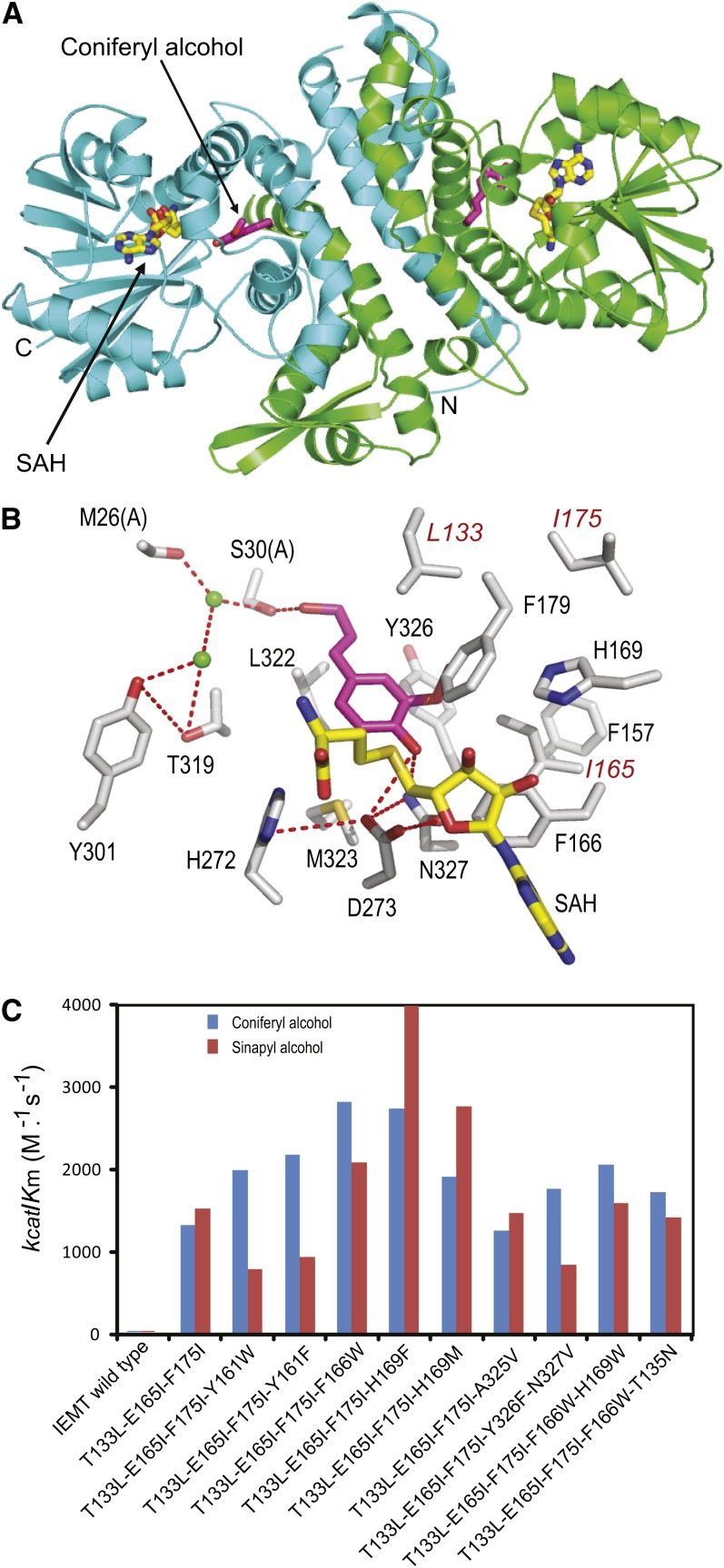
To improve the MOMT mutant enzymatic properties and to explore the structural basis for the altered substrate specificity of those variants (Bhuiya and Liu, 2010), we determined the crystal structure of a triple mutant variant, MOMT3 (T133L-E165I-F175I), complexed with the substrate coniferyl alcohol, and S-adenosyl homocysteine (SAH), the product of methyl donor S-adenosyl Met (SAM) (Figure 2A). The initial structure model was solved by molecular replacement using the well-defined caffeic acid 3-O-methyltransferase structure (Zubieta et al., 2002) and refined to a resolution of 2.4 Å (see Supplemental Table 2 online). Similarly to the previously characterized plant phenolic O-methyltransferases (Zubieta et al., 2002; Louie et al., 2010), MOMT3 forms a homodimer, and each monomer consists of a large C-terminal catalytic domain involved in SAM/SAH and phenolic substrate binding and a small N-terminal domain that primarily mediates dimerization (Figure 2A). The N-terminal domain also contributes to the formation of the active site of the dyad molecule by providing residues (Met-26 and Ser-30) lining the back wall of the substrate binding cavity and helping enclose the recognition surface (Figures 2A and 2B). As depicted, coniferyl alcohol bound in the active site is tethered by three hydrogen bonds formed via the γ-hydroxyl of its propanoid tail with Ser-30 of the dyad molecule of MOMT3 dimer and its 4-hydroxyl (phenol) with the side chains of Asn-327 and Asp-273 (Figure 2B). Moreover, its meta-methoxy moiety is buried in a hydrophobic pocket constituted by Leu-133 and Ile-165 (as established via mutations of Thr-133 and Glu-165 of IEMT), as well as by Leu-139, Phe-179, and Tyr-326 (Figure 2B). These H-bond linkages, the hydrophobic interactions, and probably the steric effect generated by the mutation (e.g., Thr-133 to Leu) enable monolignol bound in the active site with its para-hydroxyl proximal to both the methyl-donor and the catalytic base His-272 for transmethylation.
Figure 2.
Structure and Catalytic Efficiency of the Engineered MOMTs.
(A) The tertiary structure of the fashioned MOMT3 mutant variant. MOMT3 forms a dimer with an interface contributed by the small N-terminal domains from dyad molecules. Two chains are presented in green and cyan. The phenolic substrate (coniferyl alcohol) bound in the active site is depicted in magenta, and the methyl donor product SAH is color coded as C in yellow, N in blue, and O in red.
(B) A close-up view of the active site of MOMT3 in complex with the substrate coniferyl alcohol and the product of the methyl donor SAH. The three amino acid substitutions are labeled in red. The image is acquired from chain B of the dimer.
(C) The catalytic efficiency (kcat/Km) of the created MOMT variants in methylating coniferyl alcohol and sinapyl alcohol. For the detailed kinetic parameters, see Supplemental Table 1 online.
Based on the crystal structure information, 20 amino acid residues directly in contact with, or proximal to, the bound coniferyl alcohol were unequivocally identified (see Supplemental Figure 2 online). To further enhance the catalytic efficiency of MOMT for 4-O-methylation of monolignols, we performed an iterative saturation mutagenesis of those identified residues (Phe-130, Ala-134, Val-138, Leu-139, Phe-157, Tyr-161, Asn-164, Phe-166, Tyr-168, His-169, His-173, Phe-179, Met-183, Asn-186, Thr-319, Leu-322, Met-323, Ala-325, Tyr-326, and Asn-327) using the created triple mutant variant MOMT3 (T133L-E165I-F175I) as the template. This allowed us to screen out several mutant variants that bear an additional substitution(s) primarily at Tyr-161, Phe-166, or His-169, besides the three original mutations (Thr-133L, Glu-165I, and Phe-175I) of the MOMT3 parental enzyme (Figure 2C; see Supplemental Table 1 online). These tetra- and penta-mutants exhibited higher enzymatic activity than the parental MOMT3. Kinetics determination revealed that the catalytic efficiency (kcat/Km) of the mutants T133L-E165I-F175I-H169F (MOMT4) to both coniferyl alcohol and sinapyl alcohol reached up to 4000 (M−1 s−1), an ∼200-fold increase over those of the IEMT wild-type enzyme (Figure 2C; see Supplemental Table 1 online). Such catalytic properties are comparable to those of the native phenolic O-methyltransferases (Inoue et al., 2000; Parvathi et al., 2001). The MOMT4 enzyme predominantly reacted with coniferyl and sinapyl alcohols and their corresponding aldehydes but showed negligible activity toward p-coumaryl alcohol, its aldehyde, and caffeyl alcohol, which lack the meta-O-methyl moiety (Table 1). This highlights the importance of the observed interaction of the meta-methoxyl of a monolignol with the hydrophobic residues constituted by the mutations in the active site of MOMT (as seen in Figure 2B) for constraining the proper orientation of the substrate to ensure efficient 4-O-methylation. When a broader range of potential substrate phenolics was tested, the created enzyme showed only negligible activity on ferulic and sinapic acids, and none for feruloyl- and sinapoyl-CoAs and sinapoyl esters (sinapoyl glucose and malate) (Table 1), pointing out its relatively narrow substrate specificity.
Table 1. Substrate Specificity of MOMT4 on Different Phenolics.
| Substrate | Specific Activity (nmol mg−1 min−1) | Km (µM) | Vmax (nmol mg−1 min−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Coniferyl alcohol | 66.8 ± 0.7 (100)a | 192.6 ± 24.5b | 793.0 ± 3.4b | 2738.7b |
| Sinapyl alcohol | 79.6 ± 0.9 (119) | 68.1 ± 11.1b | 407.5 ± 16.5b | 3999.3b |
| Coniferaldehyde | 28.7 ± 0.8 (43) | 6.4 ± 0.5 | 22.0 ± 0.4 | 2291.8 |
| Sinapaldehyde | 14.6 ± 0.5 (22) | 3.0 ± 0.5 | 9.3 ± 0.2 | 2099.3 |
| P-coumaryl alcohol | 5.5 ± 0.1 (8.2) | 190.3 ± 12.0 | 13.7 ± 0.4 | 48.0 |
| P-coumaraldehyde | 1.9 ± 0.0 (2.8) | 171.0 ± 20.6 | 3.8 ± 0.2 | 14.6 |
| Caffeyl alcohol | 1.7 ± 0.0 (2.5) | |||
| Ferulic acid | 3.6 ± 0.0 (5) | 533.2 ± 32.3 | 14.2 ± 0.4 | 17.8 |
| Sinapic acid | 1.8 ± 0.0 (3) | 503.6 ± 12.2 | 20.3 ± 0.2 | 26.9 |
| Feruloyl CoA | NDc | |||
| Sinapoyl CoA | ND | |||
| Sinapoyl glucose | ND | |||
| Sinapoyl malate | ND |
The relative activity was measured from the reaction at 30°C for 30 min. Kinetic parameters were obtained from the reactions at 30°C for 10 min. The data represent the mean ± se of two replicates.
Data in parentheses represent relative activity.
Data obtained from radioactive assays.
ND, not detectable; no data shown indicates the kinetics were not tested.
4-O-Methylation of Monolignols Eliminates Their Oxidative Coupling in Vitro
To reevaluate the effect of substitution of the para-hydroxyl of monolignols on their coupling and cross-coupling propensities, we produced 4-O-methylated coniferyl and sinapyl alcohols using the created MOMT4 and subjected them to the peroxidase-catalyzed dehydrogenative polymerization. As expected, and in sharp contrast with conventional monolignols (see Supplemental Figures 3A and 3B online), no dimeric or oligomeric products from the 4-O-methyl-monolignols were detected (see Supplemental Figures 3C and 3D online), confirming that methylation of the phenol (para-hydroxyl) indeed deprives the oxidative dehydrogenation and the subsequent coupling of the modified compounds in vitro.
To examine whether the 4-O-methyl-monolignol interferes with the oxidative dehydrogenation and coupling of conventional lignin monomers, we first incubated an equal amount of coniferyl alcohol and 4-O-methyl-coniferyl alcohol in the presence of horseradish peroxidase and H2O2 and found that essentially the same amount and types of oxidative coupling products were produced as those derived from coniferyl alcohol alone (see Supplemental Figure 4A online). This suggests that 4-O-methylation of the monolignol prevents the participation of only the modified compound into the coupling or polymerization process and does not impair the activity of oxidative polymerization of conventional monolignols. For more comprehensive analysis, we mixed a fixed concentration of coniferyl alcohol with a series of concentrations of 4-O-methyl-coniferyl alcohol in the presence of peroxidase and H2O2 and quantified the major oxidative dimeric products. Compared with those derived from the polymerization reaction of coniferyl alcohol alone, the quantity of the detected coupling products in all the reactions coincubating both conventional and 4-O-methylated monolignols did not show substantial changes (see Supplemental Figure 4B online), confirming that 4-O-methylated monolignols do not inhibit or compete with the peroxidase-catalyzed oxidative dehydrogenation of conventional monolignols.
Expressing MOMT4 in Arabidopsis Lowers Lignin Content in the Cell Walls
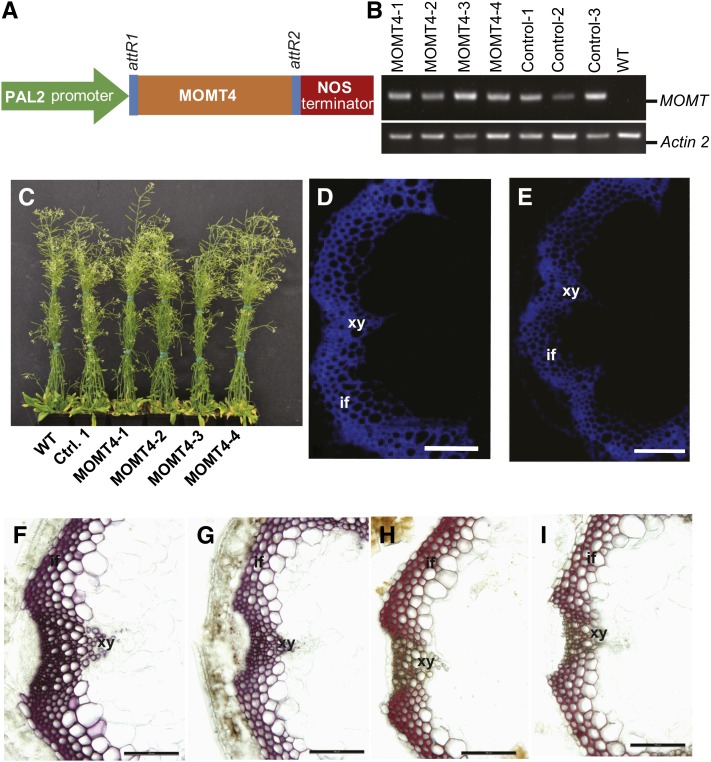
To explore the potential effects of 4-O-methylation of monolignols on lignin polymerization in planta, we overexpressed MOMT4 (T133L-E165I-F175I-H169F) in Arabidopsis. A loss-of-function IEMT variant (E165R) (Bhuiya and Liu, 2010) was included as the control. This E165R variant with substitution of a Glu residue by a bulkier Arg that appreciably projects into the enzyme active site completely abolished catalytic activity (Bhuiya and Liu, 2010); therefore, it serves an ideal transgenic empty vector control. A bean PAL2 promoter (Reddy et al., 2005) was used to drive the transgenes (Figures 3A to 3C). The T2 transgenic lines and their progenies were used for histochemical and chemical compositional analyses. In MOMT4 overexpression lines, cross sections of the basal internode of inflorescent stems (10 weeks old), under epifluorescence microscopy, exhibited weaker autofluorescence and fewer numbers of fluorescent cells within their interfascicular fibers and xylem bundles, compared with those of the control plants (Figures 3D and 3E). Furthermore, the conventional Wiesner (phloroglucinol-HCl, wherein a violet-red color is indicative of lignins) and Mӓule (which stains syringyl-containing lignins red) histochemical stains (Nakano and Meshitsuka, 1992) exhibited a less intense coloration in the vasculatures of MOMT4 overexpression lines than in the controls (Figures 3F to 3I). These data point to a reduction in lignin content in the cell walls of the MOMT transgenic lines. To assess the quantitative alteration in lignin deposition as a consequence of the expression of MOMT4, we measured the total lignin content of 10-week-old T2 generation transgenic plants using the acetyl bromide method (Iiyama and Wallis, 1990; Chang et al., 2008). Lignin content in all MOMT4 transgenic plants was reduced, and the maximum reduction reached∼24% in transgenic line MOMT4-3, compared with the control plants (Table 2).
Figure 3.
Expression of MOMT4 in Arabidopsis and Histochemical Analysis of Transgenic Plants.
(A) MOMT4 overexpression cassette driven by a bean Phe ammonia lyase promoter.
(B) RT-PCR analysis on the transgenes of MOMT4 and loss-function variant IEMT (E165R); both transgenes were expressed in the selected transgenic lines.
(C) Morphological phenotype of wild-type, control, and MOMT4 expression plants. WT, the wild type.
(D) UV autofluorescence of a stem section from the first node of a 10-week-old Arabidopsis control plant. if, interfascicular fiber; xy, xylem.
(E) UV autofluorescence of a stem section from the first node of a 10-week-old MOMT4-3 transgenic line.
(F) Phloroglucinol-HCl staining of a stem section from the second basal node of the control line, which indicates the total lignin in violet-red.
(G) Phloroglucinol-HCl staining of a stem section from the second basal node the MOMT4-3 transgenic line.
(H) Mӓule staining of a stem section from the second basal node of a control plant. The staining indicates syringyl lignin subunit in red.
(I) Mӓule staining of a stem section from the second basal node of a MOMT4-3 transgenic plant.
Bars = 50 µm
Table 2. Cell Wall Composition and Digestion Efficiency Analysis of MOMT4 Transgenic Plants.
| Lines | Total Acetyl Bromide Lignin (mg/g CWR) | Lignin Composition |
Cellulose (mg/g CWR) | Digestion Efficiency (Weight Loss %) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Monomers from β-O-4–Linked H, G, and S Conventional Lignin Units (µmol/g CWR)a |
Monomers from β-O-4–Linked H, G, and S Conventional Lignin Units (µmol/g lignin)b |
S/G | ||||||||
| H | G | S | H | G | S | |||||
| Control 1 | 152.37 ± 7.9 | NDc | 145.31 ± 8.51 | 84.41 ± 5.53 | ND | 1008.35 ± 54.95 | 583.27 ± 27.37 | 0.58 | 591.18 ± 54.7 | 39.8 ± 0.82 |
| Control 2 | 154.71 ± 10.85 | ND | 140.90 ± 2.62 | 76.07 ± 2.62 | ND | 931.22 ± 69.55 | 505.20 ± 64.28 | 0.54 | 580.97 ± 3.23 | 39.97 ± 3.90 |
| Control 3 | 143.64 ± 5.43 | ND | 140.05 ± 18.89 | 72.56 ± 11.99 | ND | 898.22 ± 85.73 | 464.94 ± 58.62 | 0.52 | 553.90 ± 28.22 | 39.07 ± 2.25 |
| MOMT4-1 | 128.81 ± 3.25** | ND | 123.54 ± 5.92* | 59.15 ± 3.89** | ND | 939.62 ± 46.22 | 449.66 ± 28.98 | 0.48 | 582.57 ± 26.78 | 44.05 ± 0.36* |
| MOMT4-2 | 130.63 ± 2.32* | ND | 118.57 ± 4.80** | 64.29 ± 2.64** | ND | 879.73 ± 48.29 | 476.93 ± 25.88 | 0.54 | 532.12 ± 8.99 | 44.9 ± 1.76* |
| MOMT4-3 | 113.47 ± 1.90 ** | ND | 92.36 ± 7.82** | 51.46 ± 4.95** | ND | 803.04 ± 76.55 | 447.35 ± 46.78 | 0.56 | 570.10 ± 8.28 | 47.62 ± 1.42** |
| MOMT4-4 | 126.62 ± 2.2 ** | ND | 113.30 ± 4.29** | 60.37 ± 2.42** | ND | 898.08 ± 51.02 | 478.48 ± 29.56 | 0.53 | 582.89 ± 14.11 | 42.99 ± 1.28 |
The data represent the means and se from triplicate or more biological analyses (n ≥ 3), except the data for lignin monomer and cellulolysis of control plants are from duplicate analyses. Asterisks indicate significant difference compared to the average value of control lines from three independent transgenic events: *Student’s t test, P < 0.05; **Student’s t test, P < 0.01.
Yield in main lignin-derived thioacidolysis monomers (expressed in micromoles per gram of extract-free cell wall residues [CWR]) recovered from conventional H, G, or S β-O-4–linked lignin units.
The monomers of b were expressed in micromoles per gram acetyl bromide lignin.
ND, not detectable.
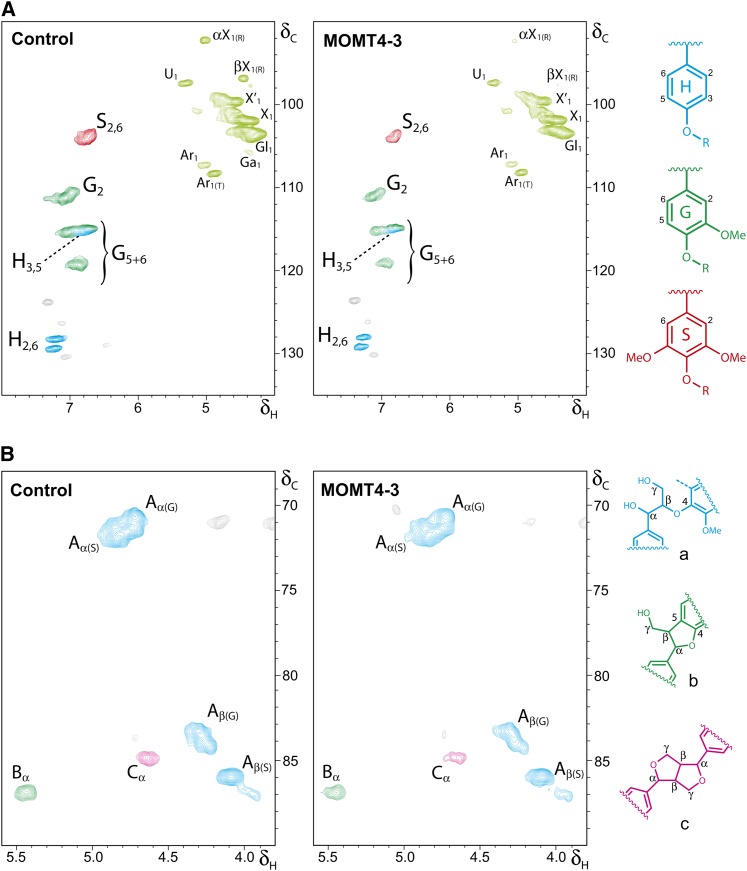
To evaluate the alteration of lignin composition and, to some extent, structure in the MOMT expression lines, we analyzed the products of analytical thioacidolysis by gas chromatography. Thioacidolysis primarily cleaves β–O–4-ether linkages interconnecting the lignin subunits and therefore enables the direct determination of the monomeric composition of noncondensed lignin (Lapierre, 1993). Calculating the yield of both the guaiacyl and syringyl subunits on the basis of the cell wall dry mass revealed a significant reduction in lignin-derived monomers from the MOMT transgenic lines compared with the controls (Table 2), which is consistent with the decrease in total lignin in their cell walls. However, the ratio of syringyl to guaiacyl units displayed only slight alteration in the overexpression plants (Table 2), suggesting that the structure of lignin in the MOMT overexpression lines is not modified drastically, although the quantity of total lignin was reduced. For a more detailed investigation into the effects on lignin structure, whole-cell wall gel samples in DMSO-d6/pyridine-d5 and cellulolytic enzyme lignins (CELs) were further prepared from both the control and overexpression lines and analyzed by NMR. The aromatic regions of two-dimensional (2D) 13C–1H correlation (heteronuclear single quantum coherence [HSQC]) NMR spectra of the equal amount of cell wall samples from overexpression and control lines showed that the signals of both G- and S-units were much lower in the cell wall of overexpression lines (Figure 4A; see Supplemental Figure 5 online), confirming the reduction of total lignin, but the signals of the p-hydroxyphenyl (H) unit remained the same or were only slightly reduced in overexpression lines, suggesting that MOMT modifies predominantly the precursors for guaiacyl and syrigyl lignin in vivo, which is consistent with its in vitro activity (Table 1). Furthermore, the HSQC spectra of both aliphatic side chain and aromatic regions of prepared lignin (CEL) samples exhibited no dramatic differences between the overexpression and control lines, except for the slight increase of the relative amount of H-subunit due to the decrease of G- and/or S-units in the lignin (Figure 4B; see Supplemental Figure 6 online). As expected, in 2D-HSQC NMR spectra, no 4-O-methylated guaiacyl or syringyl structures as the end units were identified (Figure 4B; see Supplemental Figure 6 online), confirming that aromatics without a free phenol (the para-hydroxyl) for initiating radical generation have no bypass way of entering into polymerization. Together, these data imply that the 4-O-methylation of monolignols by MOMT reduced the incorporation of the modified lignin precursors (primarily for G- and S-units) into the polymers but that the polymers produced were structurally normal regarding their linkages.
Figure 4.
NMR Spectra Analysis of Lignin Structure in MOMT Overexpression Plants.
(A) Partial short-range 2D 13C–1H (HSQC) NMR spectra (aromatic and carbohydrate anomeric region) of the equal amount of solubilized total cell walls of MOMT transgenic and control plants, after forming a gel in DMSO-d6:pyridine-d5 (4:1). The main structural units are colored to coincide with their structures on the right. αX1, α-d-xylopyranoside (R) [α-d-glucopyranoside (R)]; βX1, β-d-xylopyranoside (R) [β-d-glucopyranoside (R)]; U1, 4-O-methyl α-d-glucuronic acid, X´1, 2-O-acetyl-β-d-xylopyranoside; X1, β-d-xylopyranoside; Gl1, (1→4)- β-d-glucopyranoside; Ar1 α-l-arabinofuranoside; Ar1(T), α-l-arabinofuranoside (T). Reducing end; T, terminal (nonreducing end).
(B) Partial short-range 2D 13C–1H (HSQC) NMR spectra (aliphatic oxygenated region) of the equal amount of CELs with the main structural units colored to coincide with their structures on the right.
See Supplemental Figures 5 and 6 online for the complete set of spectra of the transgenic and control samples.
4-O-Methylation of a Monolignol Does Not Impair Its Transport across the Cell Membrane
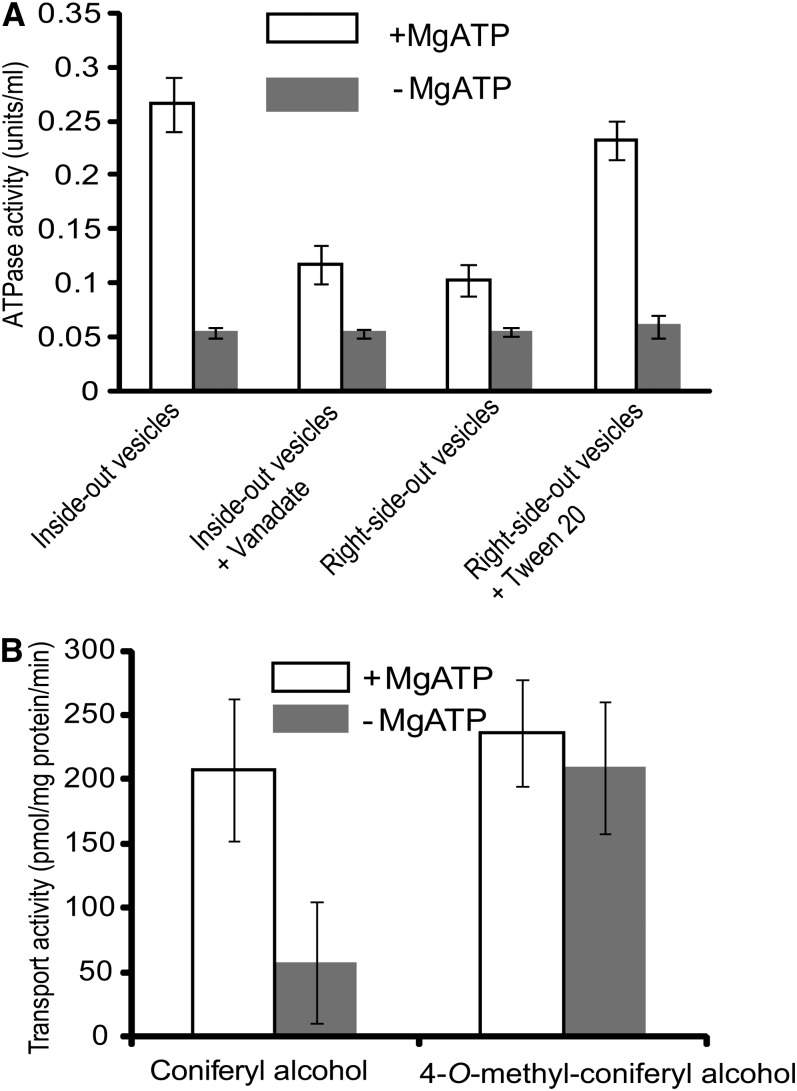
To examine whether the decline of lignin polymerization in vivo is due to the potential deprival of 4-O-methylated compounds from their deposition into the cell wall, we prepared the inverted membrane vesicles from wild-type Arabidopsis rosette leaves and primary bolting tissues (Miao and Liu, 2010) using an aqueous polymer two-phase partitioning system (Larsson et al., 1994). The quality of the prepared membrane fraction was monitored by measuring the vanadate-inhibited activity of the plasma membrane marker enzyme H+-ATPase. As depicted in Figure 5A, the detected ATP-dependent hydrolytic activity of the prepared inside-out vesicles was largely repressed with the effective plasma membrane ATPase inhibitor, sodium vanadate (Gallagher and Leonard, 1982), suggesting the majority of the inside-out membrane fraction represents plasma membrane. We then incubated the prepared inside-out vesicles with 4-O-methyl-coniferyl alcohol. In the presence or absence of ATP, the uptake of the 4-O-methyl monolignol by the inverted vesicles is comparable to, or even higher than, that of conventional monolignols, indicating that 4-O-methylation did not abrogate the compound’s transport across cell membranes (Figure 5B). Moreover, while the transport activity of the membrane vesicles to coniferyl alcohol was greatly impaired in the absence of ATP, transport of the 4-O-methyl monolignol remained largely unchanged, implying that 4-O-methylation enhances passive diffusion, presumably due to an increase in the compound’s lipophilicity with methylation.
Figure 5.
Membrane Preparation and Transport Assay.
(A) H+-ATPase activities of different membrane preparations in the presence or absence of MgATP and/or inhibitor.
(B) Transport activity of the prepared inside-out membrane vesicles to coniferyl alcohol and the 4-O-methylated coniferyl alcohol in the presence and absence of ATP.
Data are the means and sd of three replicates.
Expressing MOMT4 in Arabidopsis Yields Novel Wall-Bound Phenolics
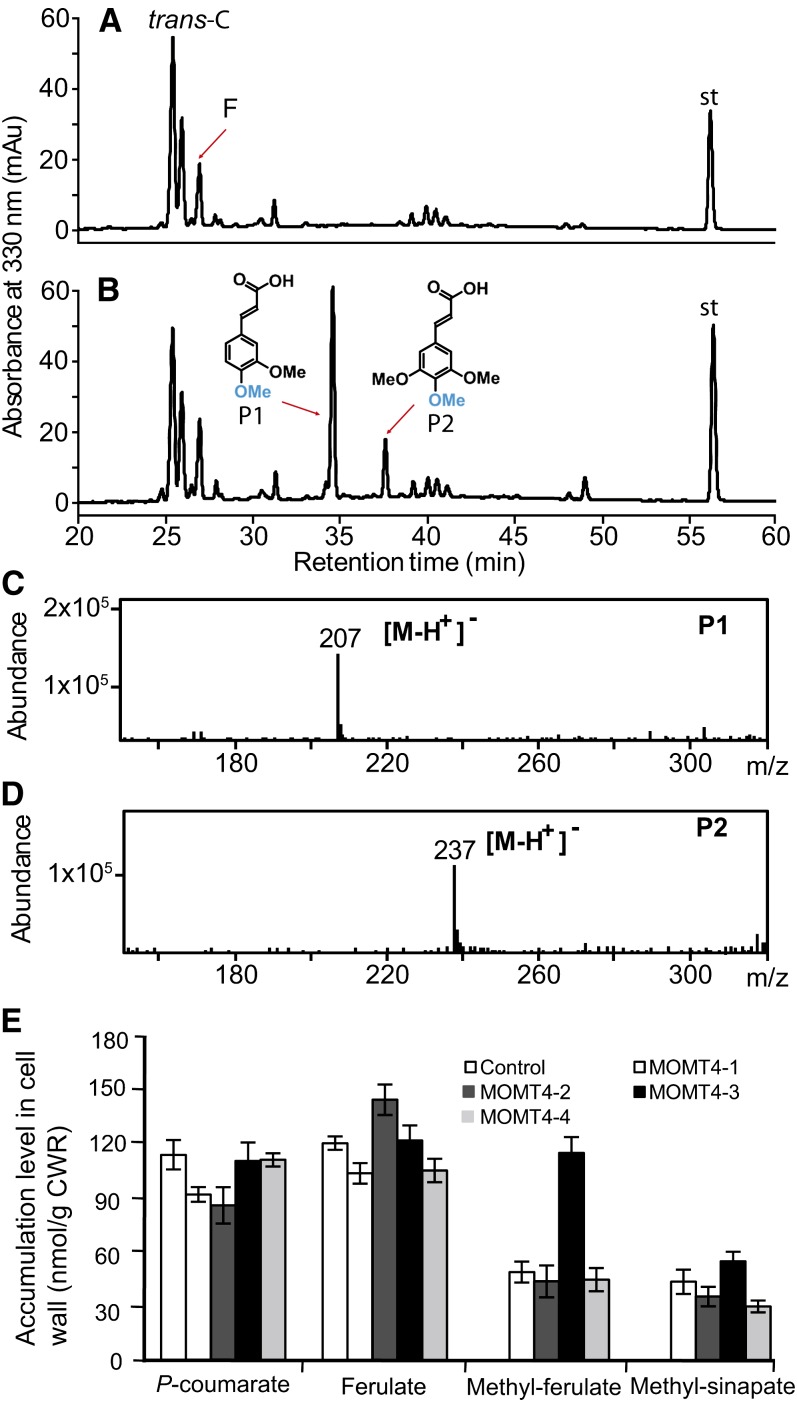
The cell walls of monocot grasses and some dicot species, including Arabidopsis, contain significant quantities of ester-bound hydroxycinnamates, primarily p-coumarate and ferulate (Ishii, 1997; Burr and Fry, 2009). As resolved by liquid chromatography–mass spectrometry (LC-MS; Figures 6A to 6D), expression of MOMT4 changed the profiles of those alkaline-released wall-bound phenolics. Besides two conventional phenolics, p-coumarate and ferulate, which were readily detected in the cell walls of both the control and MOMT4 overexpression lines, the bona fide phenolics were released from the walls of MOMT4 expression lines after mild alkaline treatment of the stem cell wall residuals (Figure 6B). LC-MS/MS analysis indicated that one compound (P1 in Figure 6B) yielded the molecular ion [M-H]− at a mass-to-charge ratio (m/z) of 207 (Figure 6C); its UV spectrum clearly showed its belonging to the cinnamic acid class of compounds that is identical to that of 4-O-methyl ferulate, the enzymatic product of MOMT4 with ferulic acid, and that of parental compound ferulic acid (see Supplemental Figure 7 online), suggesting P1 most likely represents 4-O-methyl-ferulate. Although the molecular mass of 207 also hints at 4-O-methyl-5-hydroxyconiferaldehyde, and sinapaldehyde, the UV spectrum of this novel wall-bound compound substantially differs from those of cinnamaldehyde type of compounds (as depicted in Supplemental Figure 7 online by the comparison with those of sinapaldehyde and 5-hydroxyconiferaldehyde, the parental compound of 4-O-methyl-5-hydroxyconiferaldehyde). Moreover, it is known that the alkaline-releasable wall-bound phenolics are incorporated into the cell wall via ester linkages, which chemically requires cinnamic acids but not aldehydes. Together, these data suggest the presence of a novel wall-bound phenolic ester, the 4-O-methyl-ferulate, in the cell walls of MOMT expression lines. Similarly, we identified the second phenolic P2 (Figure 6B) that has an m/z of 237 as 4-O-methyl-sinapate (Figure 6D). The accumulation of 4-O-methylated wall-bound phenolics correlated with the expression of the MOMT4 transgene; the line MOMT4-3 with the strongest gene expression (Figure 3B) and lignin reduction (Table 2) amassed the highest level of 4-O-methyl compounds, comparable to that of the endogenous p-coumarate or ferulate (Figure 6E).
Figure 6.
Accumulation of Wall-Bound Phenolics in MOMT Overexpression Plants.
(A) to (D) HPLC profiles of wall-bound phenolics from the cell wall residues of the control (A) and MOMT4 transgenic plants at the absorbance of A330 (B). P1 is tentatively identified as 4-O-methyl-ferulic acid, and P2 is tentatively 4-O-methyl-sinapic acid. The mass spectra of P1 and P2 are shown in (C) and (D). The UV spectra are shown in Supplemental Figure 7 online. F, ferulic acid; st, internal standard; trans-C, trans-p-coumaric acid. mAu, milli-absorbance unit.
(E) The quantification of novel 4-O-methyl wall-bound phenolic esters released from the cell wall of MOMT transgenic stems. The data are the means and se of three or four replicates. Me-F, 4-O-methyl-ferulate; Me-S, 4-O-methyl-sinapate. CWR, cell wall residue.
[See online article for color version of this figure.]
Expressing MOMT4 Produces Novel Methanol-Soluble Phenolic Esters
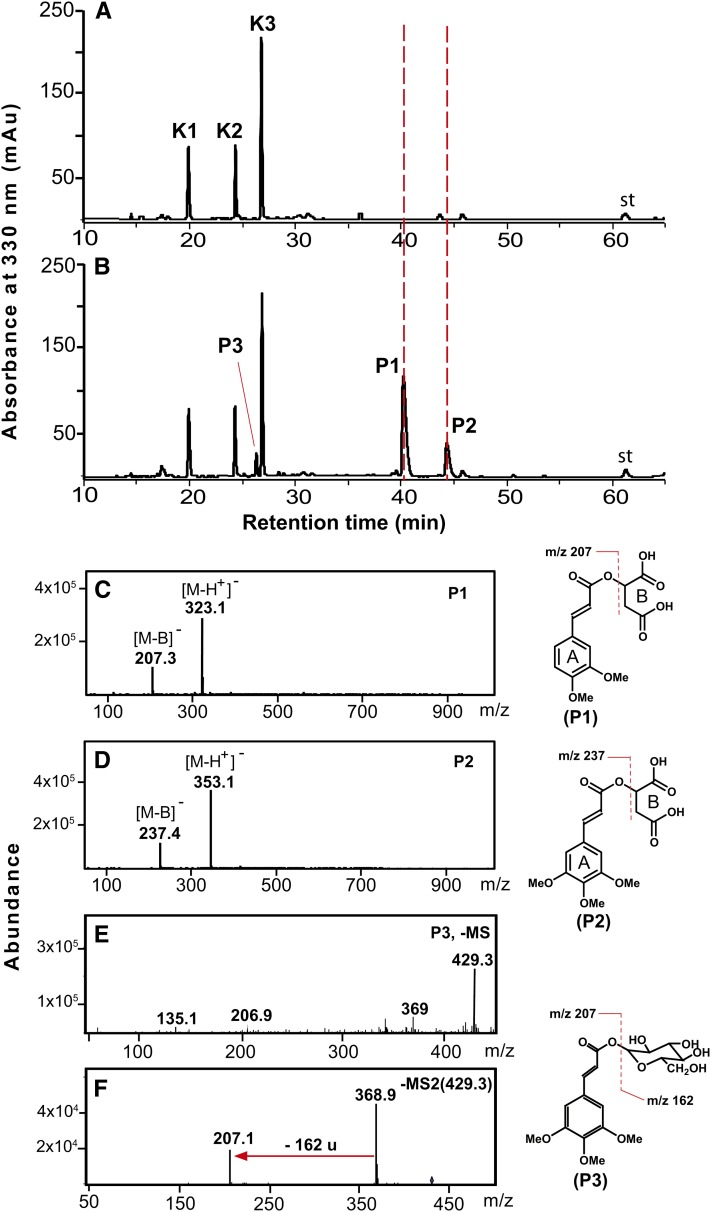
Profiling the methanolic extracts from the stems and leaves of MOMT4 transgenic lines also revealed the presence of bona fide phenolic conjugates (Figure 7). LC-MS/MS analysis indicated that three novel metabolites in extracts from the 10-week-old stems of those lines exhibited UV spectra similar to that of the enzymatic product 4-O-methyl-ferulate or sinapate, respectively, but different from those of cinnamaldehyde analogs (see Supplemental Figure 8 online). Two compounds, the P1 and P2, yielded the molecular ion [M-H]− at an m/z of 323 and 353, respectively, and a corresponding major fragment ion at m/z of 207 or 237, pointing to the presence of a 4-O-methylated ferulate or sinapate residue. The loss of 116 m/z suggests their conjugation with malate (Figures 7A to 7D). Similarly, a putative 4-O-methyl-feruloyl-1-O-glucose derivative accrued in the stems, leaves, and roots of the MOMT overexpression lines (Figures 7E and 8B; see Supplemental Figure 9 online). Although its precise structural feature needs to be further characterized, the MS/MS analysis revealed that this compound yielded a daughter ion [M-H]− of 207 (Figure 7F), and its UV spectrum is largely similar to that of 4-O-methyl-ferulate but differs from that of 4-O-methyl-5-hydroxyconiferaldehyde or sinapaldehyde (see Supplemental Figure 8 online), suggesting the presence of a core structure of 4-O-methyl-ferulate in this metabolite. The difference of m/z of 162 between fragment ions of 207 and 369 (Figure 7F) suggests the existence of a Glc moiety.
Figure 7.
LC-MS Analysis of Methanolic Soluble-Phenolic Compounds Accumulated in the Stems of MOMT4 Transgenic Plants.
HPLC profiles of phenolic extracts from stems of the control (A) and MOMT4 transgenic (B) plants. P1 is tentatively identified as 4-O-methyl-feruloyl malate, P2 is 4-O-methyl-sinapoyl malate, and P3 is a 4-O-methyl-feruloyl glucose derivative. The corresponding mass spectra of the compounds are shown in (C) to (F), and their UV spectra are shown in Supplemental Figure 8 online. No sinapoyl malate was detected in those mature stems. K1, kaempferol 3-O-[6′′-O-(rhamnosyl)glucoside] 7-O-rhamnoside; K2, kaempferol 3-O-glucoside 7-O-rhamnoside; K3, kaempferol 3-O-rhamnoside 7-O-rhamnoside; st, internal standard chrysin. mAu, milli-absorbance unit.
[See online article for color version of this figure.]
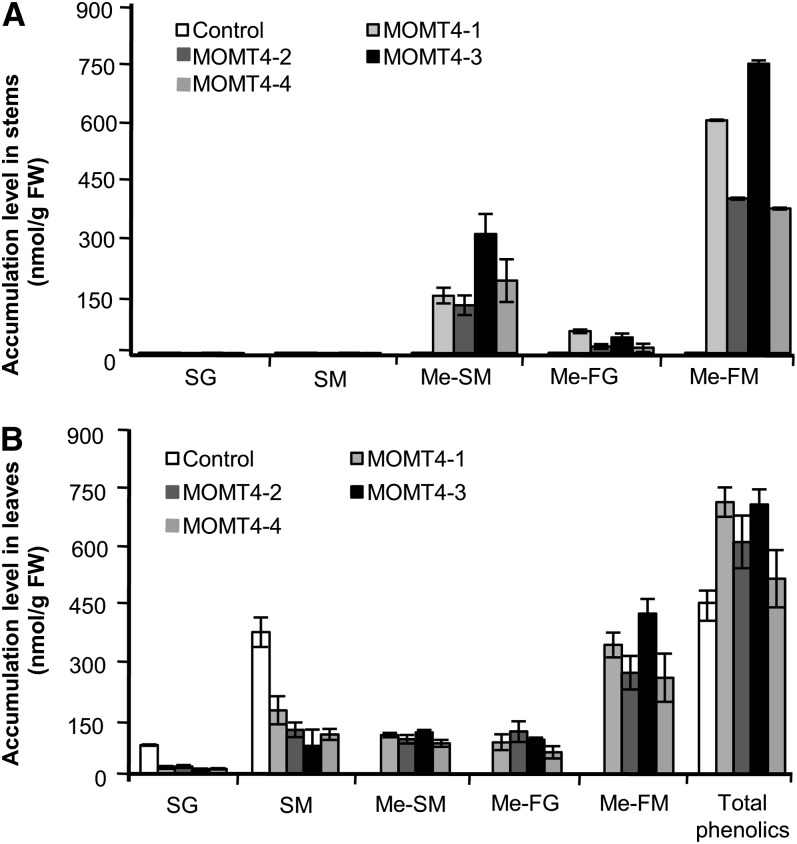
Figure 8.
Accumulation of Soluble Phenolic Esters in MOMT Overexpression Plants.
(A) The methanol-soluble 4-O-methyl compounds accumulated in the stems of MOMT transgenic plants. Note that no conventional sinapoyl esters accumulated in the mature stems of both the 10-week-old control and transgenic plants. The data are the means and se of four replicates. FW, fresh weight; Me-FG, 4-O-methyl-feruloyl glucose derivative; Me-FM, 4-O-methyl-feruloyl malate; Me-SM, 4-O-methyl-sinapoyl malate; SG, sinapoyl glucose; SM, sinapoyl malate.
(B) The methanol-soluble phenolic esters accumulated in the rosette leaves of 7-week-old control and MOMT transgenic plants.
The data represent mean and se of four or five replicates.
The level of accumulated 4-O-methyl-feruloyl malate was higher than that of 4-O-methyl-sinapoyl ester both in the mature stems and rosette leaves of these overexpression lines (Figures 8A and 8B; see Supplemental Table 3 online). As with the finding from previous studies (Nair et al., 2004), the conventional sinapoyl esters (sinapoyl malate and sinapoyl glucose) were predominant in the rosette leaves of the control Arabidopsis, whereas, along with production of 4-O-methylated compounds, their accumulations were compromised in the leaves of the MOMT overexpression lines; however, the total amount of quantified phenolic esters (summed for the native esters and nascent 4-O-methyalted esters) increased up to 60% in the overexpression lines compared with the control line (Figure 8B). In mature stems (10 weeks old), the endogenous sinapate esters were no long detected in either the control lines or MOMT overexpression lines; in the latter, the 4-O-methylated compounds were the predominant esters (Figure 8A; see Supplemental Table 3 online), implying that 4-O-methylation probably also slows down their decomposition or conversion in the mature tissues.
Expressing MOMT4 Does Not Significantly Affect the Plant’s Growth and Development
Despite changes in the synthesis of lignin and wall-bound phenolics, the growth and developmental characteristics of MOMT transgenic plants showed no drastic phenotypic abnormalities, compared with either wild-type or transgenic control plants (Figure 3C). Only at the late stages of growth (∼8 weeks), the leaf senescence of MOMT overexpression lines appeared slightly earlier (Figure 3C). However, the plant height and branching at this stage are nearly identical between control and MOMT expression lines, except the stem thickness of basal nodes of some overexpression lines was slightly decreased compared with the average basal node diameter of the control lines. However, the dry weight of total aboveground materials showed no significant difference (Table 3). Correspondingly, at a cellular level, we saw no dramatic anatomical differences in the vasculature between most of the overexpression and control lines (Figures 3G to 3J), except that slightly thinner secondary walls were occasionally observed in a few overexpression lines. Quantifying cellulose content in the stem cell wall residues revealed no significant differences in cellulose deposition between the control and most of the MOMT overexpression plants, although the amount is variable in different transgenic lines (Table 2). To further examine whether expressing the artificial MOMT and producing novel phenolics would potentially impose feedback regulation on global gene expression, we conducted transcriptomic analysis on MOMT transgenic lines. Among the more than 22,000 genes detected, only 12 showed a moderate change in their expression levels and none of them indicated a clear function in the central metabolic or regulatory pathways (Table 4; see Supplemental Data Set 1 online).
Table 3. Quantitative Growth Phenotypes of MOMT4 Transgenic Plants.
| Genotype | Height (cm) | Branches | Stem Diameter (mm) | Dry Weight (Grams/Three Plants) |
|---|---|---|---|---|
| Control-1 | 53.75 ± 1.07 | 5.25 ± 0.25 | 1.71 ± 0.07 | 1.89 ± 0.11 |
| Control-2 | 51.92 ± 1.10 | 5.0 ± 0.33 | 1.43 ± 0.03 | 2.05 ± 0.13 |
| Control-3 | 56.67 ± 1.2 | 5.10 ± 0.15 | 1.65 ± 0.04 | 2.13 ± 0.07 |
| MOMT4-1 | 53.39 ± 0.72 | 4.91 ± 0.18 | 1.45 ± 0.04* | 2.05 ± 0.11 |
| MOMT4-2 | 53.36 ± 0.72 | 5.00 ± 0.16 | 1.56 ± 0.04 | 2.17 ± 0.12 |
| MOMT4-3 | 52.42 ± 0.77 | 4.83 ± 0.22 | 1.47 ± 0.04* | 1.86 ± 0.09 |
| MOMT4-4 | 53.58 ± 0.93 | 4.83 ± 0.27 | 1.50 ± 0.04* | 2.0 ± 0.10 |
The data represent the means and se from eight or more biological analyses (n ≥ 8). Asterisks indicate significant difference compared to the average value of control lines from three independent transgenic events: *Student’s t test, P < 0.05.
Table 4. List of Genes Whose Expression Levels Changed in MOMT Transgenic Lines.
| Probe Set | Gene Name | Annotation | Transcript Level Related to Control |
|---|---|---|---|
| 263046_at | At2g05380 | Unknown protein | 2.59 |
| 250277_at | At5g12940 | Putative protein DRT100 protein precursor | 0.50 |
| 248681_at | At5g48900 | Putative pectate lyase | 0.50 |
| 259331_at | At3g03840 | Putative auxin-induced protein | 0.49 |
| 265443_at | At2g20750 | β-Expansin | 0.48 |
| 250012_x_at | At5g18060 | Auxin-induced protein-like | 0.46 |
| 254066_at | At4g25480 | Transcriptional activator CBF1-like protein | 0.45 |
| 261226_at | At1g20190 | Putative expansin S2 precursor | 0.45 |
| 249614_at | At5g37300 | Wax ester synthase and diacylglycerol acyltransferase | 0.43 |
| 260727_at | At1g48100 | Putative polygalacturonase PG1 | 0.40 |
| 264931_at | At1g60590 | Putative polygalacturonase | 0.40 |
| 249598_at | At5g37970 | SAM-dependent methyltransferase superfamily protein | 0.15 |
Expressing MOMT4 Enhances the Yield of Cell Wall Saccharification
We then assayed saccharification to evaluate the acceptability for biodegradation of the cell walls of the MOMT transgenic lines. In keeping with their low lignin levels, exposing the cell wall residues of these transgenic plants to a commercial cellulase mixture that specifically hydrolyzes cell wall carbohydrates, without pretreatment, raised their release of sugars by up to 22% (Table 2).
DISCUSSION
Newly Engineered MOMT Mutant Variant Confers Effective Ability in Reducing Lignin Biosynthesis
The rapid development of protein engineering strategies opens avenues for effectively manipulating plant metabolism and creating desired agricultural traits. One early study by iterative gene shuffling of bacterial glyphosate N-acetyltransferase conferred an increased tolerance of Arabidopsis, tobacco, and maize (Zea mays) to the herbicide glyphosate when the fifth iteration and beyond mutants were expressed in planta (Castle et al., 2004). However, so far the majority of protein engineering studies have been performed within microorganisms, and translating from microbial platforms to plants remains largely unexplored (Jez, 2011).
Monolignol oxidative coupling, the primary process in lignin biosynthesis, proceeds via a free radical mechanism catalyzed in situ by peroxidases or laccases. Many in vitro chemical/biophysical studies implied the importance of phenol in generating active radicals for phenoxy coupling (Lloyd and Wood, 1974; Dewar and David, 1980; Russell et al., 1996). Therefore, a chemical protection (masking) of the phenol (the para-hydroxyl) would prevent radical generation and deny the derived monolignols the possibility of entering into the subsequent coupling process. This was confirmed in our study with the absolute deprival of cross-coupling of the 4-O-methylated monolignols in the horseradish peroxidase–H2O2-mediated polymerization system (see Supplemental Figure 3 online). Moreover, our analysis further revealed that the 4-O-methylated monolignol does not compete with, nor inhibit, the oxidative coupling or polymerization of the conventional monolignols, suggesting that 4-O-methylation removes the capability of the modified monomer to generate radicals but does not impair oxidative enzyme activity.
Despite substantial 4-O-methylation activity of the early set of MOMT mutant variants (the single to triple mutants) to monolignols in vitro, they were insufficient to confer detectable effects, when they were expressed in plants, on either lignin biosynthesis or formation of other related phenolics. The further iteration, guided by the crystal structure complexed with coniferyl alcohol, resulted in a physiologically active catalyst. When expressing the mutant enzyme with four amino acid residue substitutions under the same transgenic strategy as for expressing the triple mutants, the lignin content of the cell walls was substantially lowered (Table 2), suggesting that the action of 4-O-methyltransferase in situ indeed diminished the availability of monolignols for the one-electron oxidation and polymerization. Compared with the triple mutant variants, the newly created tetra mutation variant seemingly had only a slight improvement (approximately twofold increase) in its catalytic efficiency for methylating monolignols (see Supplemental Table 1 online); however, this limited increment is sufficient to overcome the metabolic barriers, leading to reprogramming of the phenylpropanoid-monolignol biosynthetic pathways. These data suggest that metabolic engineering of phenylpropanoid biosynthesis requires a minimal threshold activity of the exogenous enzyme to effectively compete with the intrinsic pathway(s)/enzymes, in this case, competing against oxidative enzymes in binding and converting available monolignols.
Expressing MOMT Diversifies Phenolic Ester Repertories
Expressing MOMT4 in Arabidopsis does not simply disrupt the endogenous monolignol biosynthetic pathway, but instead extends the pathway to produce “dead end” products, the para-etherified compounds, for which radical polymerization is impossible. Perturbing lignin biosynthesis by the created monolignol O-methyltransferase is concomitantly associated with a rerouting of metabolic flux to both soluble and insoluble novel phenolic compounds. Those non-natural para-methylated production of MOMT4 were found to efficiently enter into the intrinsic phenolic ester biosynthetic pathway (Figures 6 and 7), highlighting the extreme plasticity of phenylpropanoid metabolism and the highly promiscuous activities of the related enzymes that lead to a redirection in the flux of the 4-O-methylated metabolites, away from lignin biosynthesis.
In Brassicaceae, sinapate esters are synthesized via the phenylpropanoid-lignin biosynthetic pathway and are presumably stored in the vacuoles (Milkowski et al., 2004). A hydroxycinnamaldehyde dehydrogenase (aldehyde dehydrogenase [ALDH]) in Arabidopsis, namely REF1, reportedly can convert coniferaldehyde or sinapaldehyde to their corresponding hydroxycinnamates that, in turn, are sequentially transformed into malate- or choline-esters by specific glycosyltransferases and acyltransferases (Nair et al., 2004) (see Supplemental Figure 1 online). MOMT4 preferentially catalyzed the 4-O-methylation of coniferyl and sinapyl alcohols and their aldehydes in vitro. It showed negligible or no activity for other sinapoyl ester biosynthetic intermediates (Table 1). These data suggest the buildup of 4-O-methylated soluble phenolic esters must be de novo converted from the MOMT-produced 4-O-methyl-cinnamyl alcohols (or aldehydes). The fact that the effective transformation of 4-O-methylated monolignols to phenolic esters in transgenic Arabidopsis suggests an intrinsic cinnamyl alcohol dehydrogenase (CAD) activity is recruited, which is able to accommodate 4-O-methyl-cinnamyl alcohols and catalyze them in a reverse reaction to the corresponding cinnamaldehydes, and then the latter reaction further proceeded via the promiscuous activities of REF1 and other sinapoyl ester biosynthetic enzymes. The reversible catalytic property and substrate versatility of CADs in the monolignol biosynthetic pathway were well recognized (Kim et al., 2004) and were adopted for determining their functionality in vitro (Halpin et al., 1992). The tentative pathway for transforming those compounds is proposed in Supplemental Figure 1 online.
It is important to note that the described strategy, using 4-O-methyltransferases to modulate lignin biosynthesis, would not be restricted to the Brassicaceae. First, downregulation of lignin biosynthesis by para-methylation of monolignols does not necessarily rely on whether or not the modified monomers are able to reroute into sinapoyl ester biosynthetic pathways. Our in vitro polymerization and in vivo genetic studies here demonstrate that the para-methylation of monolignols prevents their participation in the oxidative coupling/polymerization process, therefore reducing lignin formation. Serendipitous production of novel wall-bound and soluble phenolic esters in transgenic Arabidopsis represents solely an intrinsic detoxification mechanism with respect to the versatile plasticity of phenylpropanoid metabolism in this species. When the monolignol biosynthetic pathway is genetically disrupted, the accumulated native intermediates are often found either rerouted into different endogenous pathways or transformed into more stable or storage forms. For example, Arabidopsis mutant plants deficient in cinnamoyl-CoA reductase produced excessive levels of ferulate esters in their cell walls in stems and feruloyl- and sinapoyl-malate in the soluble fraction of stem, indicating a redirection of accumulated feruloyl-CoA (resulting from the disruption of cinnamoyl-CoA reductase) to wall-bound phenolic and to sinapoyl ester biosyntheses (Mir Derikvand et al., 2008); similarly, downregulation of caffeoyl CoA 3-O-methyltransferase in poplar (Populus spp) caused an accumulation of glucosides of caffeic, vanillic, and sinapic acids (Meyermans et al., 2000; Morreel et al., 2004). These phenomena suggest that plants posses an amazing versatility, recruiting their intrinsic enzymes or pathways to evolve new metabolic capacity or detoxify biogenic products. Second, the Arabidopsis REF1 (ALDH) homologs were actually also reported to exist in non-Brassicaceae species like poplar (Nair et al., 2004). Presumably, the ubiquitous and versatile CAD and ALDH activities in non-Brassicaceae species could function in a comparable way as they do in Arabidopsis to transform 4-O-methylated monomers. Third, different species have developed diverse strategies to sustain their metabolic homeostasis and to transform aromatic compounds. For instance, poplars and other Salicaceae have evolved complicated hydroxycinnamate pathways, by which they accumulate a vast array of phenolic esters, and their Glc or glycosidic derivatives, such as populoside, salicin, tremuloidin, trichocarposide, etc. (Chen et al., 2010; Boeckler et al., 2011), whereas other major group of gymnosperms and angiosperms are able to produce monolignol esters through acylation (e.g., coniferyl acetate) then reduce them to a subset of volatile phenylpropenes (Koeduka et al., 2006). Therefore, the 4-O-methylated compounds in different species, predicatively, can have sharply different metabolic fates, depending on the promiscuities of intrinsic enzymes or pathways being recruited. The engineered MOMT preferentially catalyzes monolignols (in comparison with cinnamaldehydes), producing 4-O-methyl-cinnamyl alcohols (Table 1). If there is accumulation of 4-O-methyl-cinnamaldehydes in planta derived from the unfavorable activity of MOMT, and if there is no available ALDH activity to transform them to cinnamates, they can be, expectedly, converted to the corresponding cinnamyl alcohols, which is reminiscent of the classical monolignol synthesis, by versatile CADs, and then metabolized further by, for example, acylation and/or glycosylation (Koeduka et al., 2006). Taken together, the collective information suggests that perturbing lignin biosynthesis by expressing 4-O-methyltransferase would not be limited only to Brassicaceae.
Interestingly, Arabidopsis wild-type plants accumulate none or a negligible amount of soluble feruloyl esters (Nair et al., 2004), whereas the expression of MOMT diversified the phenolic profiles and led to the production of a perceivable amount of novel feruloyl- and sinapoyl-type esters (with 4-O-methylation) that persistently accumulated in the mature tissues (Figures 7 and 8). Those compounds exhibit the same or similar UV spectra as the corresponding endogenous phenolics (see Supplemental Figures 7 and 8 online); therefore, they would be predicted to offer plants an additive defense capacity against environmental threats, particularly in screening out UV irradiation.
Simple phenolics are often incorporated into cell walls of grass species and some dicot plants and linked to different cell wall polymers via ester bonds. Those conventional wall-bound phenolics, particularly ferulate, can dimerize or polymerize with each other or with other cell wall phenolics via free radical mechanism, forming ester-to-ether linkages so as to cross-link adjacent polysaccharides, lignins, and/or structural proteins (Ralph et al., 2004a; Ralph, 2010). They also potentially act as nucleation sites for lignin polymerization (Ralph et al., 2004a). Hence, the accumulation of conventional wall-bound phenolic esters might be a negative structural factor affecting cell wall biodegradation. Expressing MOMT in planta produced ester-bound 4-O-methylated compounds (Figure 6); the 4-O-methylation of phenols effectively prevents their radical cross-coupling reactions (see Supplemental Figure 3 online). Although more detailed analyses are required to determine their precise occurrence within the cell walls (e.g., whether they are bound to the noncellulosic matrix polysaccharides, lignin, or polyesters), incorporating 4-O-methylated esters in the cell wall polymers might afford a valuable strategy for mitigating the cross-linkage of biopolymers, thereby improving the digestibility of the cell wall.
MOMT-Mediated Reduction of Lignin Has Indiscernible Effect on Plant Growth and Development
Many attempts to manipulate lignin biosynthesis via the conventional genetic approach of regulating the monolignol biosynthetic genes have encountered the problem of severely compromising the plants’ fitness (Weng et al., 2008). Although the underlying mechanisms of the deleterious effect might be complicated and could be directly linked to dysfunction of the vasculature resulting from the dramatic reduction of lignin content; however, the potential accumulation of toxic metabolic intermediates due to disrupting particular monolignol biosynthetic step(s), and/or the disturbance of the potential interplay of lignin biosynthesis with other biological processes, might also play a role (Li et al., 2008; Weng et al., 2008). Although more comprehensive stress conditions remain to be examined, expressing MOMT, which acts on almost the last step of monolignol biosynthesis, in planta caused little interference with plant growth and development under normal and continuous light conditions. It can therefore be speculated that in transgenic plants, the monolignols diverted from polymerization are effectively sequestered into storage form metabolites or incorporated into the cell wall, which prevents their potential toxicity or their ability to act as potential feedback regulation signals triggering global changes of gene expression as is observed in the downregulation of laccases (Berthet et al., 2011) or CAD (Sibout et al., 2005). Consistent with this speculation, the global gene expression of MOMT transgenic lines in the normal growth condition showed only subtle changes (Table 3).
Overall, our studies offer a practical strategy to manipulate plant lignification, without severely affecting the plant’s growth and development, by introducing an artificial enzyme to etherify the para-hydroxyl of phenols. The approach functions in such a way that the modified monolignols (with 4-O-methylation via the action of the engineered enzyme) deprives the products of participation in oxidative dehydrogenation and therefore diminishes the level of monolignols available for lignin polymerization. The accumulated dead end 4-O-methyl lignin monomers serendipitously enter into endogenous metabolism via the diverse ability of plant detoxification mechanisms and the intrinsic metabolic plasticity of plant phenylpropanoid biosynthesis. Currently, we are transferring MOMT into poplar to evaluate the effects of expression of this enzyme on woody species’ physiology.
METHODS
Saturation Mutagenesis and Mutant Library Screening
Saturation mutagenesis was performed by following the QuikChange site-directed mutagenesis strategy (Stratagene) using NNK degenerate primers (Bhuiya and Liu, 2010). The QuikChange PCR products were transferred and expressed in Escherichia coli BL21-Gold (DE3) (Stratagene). After induction by 0.2 mM isopropyl-β-d-1-thiogalactopyranoside, the cells were harvested and lysed with Bugbuster solution (Novagen). The lysate was either directly used for the enzymatic assay or further purified to obtain recombinant proteins.
For functional screening, the reaction was conducted in 50 mM Tris-HCl, pH 7.5, containing 1 mM DTT, 300 µM coniferyl or sinapyl alcohol, 500 µM SAM augmented with (for isotope activity detection) or without (for LC-MS detection) 0.005 µCi of S-adenosyl-L-Met-(methyl-14C), and 20 μL of lysate or purified protein. Reactions were incubated for 30 min at 30°C. The steady state kinetics of MOMT variants were determined using different concentrations of selected substrates at a fixed concentration of 500 µM SAM. Reactions proceeded for 10 min at 30°C. The Vmax and Km were determined by nonlinear regression analysis of the velocity concentration data fit to the Michaelis-Menten equation. When examining substrate specificity, purified MOMT4 (T133L-E165I-F175I-H169F) was used in the assay.
Crystallization and Structure Determination of MOMT3
Crystals of MOMT3 (T133L-E165I-F175I) in complex with SAH and coniferyl alcohol were obtained through the hanging-drop vapor diffusion method. Diffraction data (see Supplemental Table 2 online) were collected from single crystals at the X29- and X25-beamlines of National Synchrotron Light Source. The initial structure model was determined by the molecular replacement using COMT structure (Protein Data Bank code 1KYW) (Zubieta et al., 2002) via the Phaser program in CCP4i suite. The built model was refined using CNS 1.1 and Refmac5 (Vagin et al., 2004). The final model was built based on composite omit map to avoid structural bias.
In Vitro Polymerization
The 4-O-methylated coniferyl or sinapyl alcohol was produced in the enlarged reaction using MOMT4 enzyme. The enzymatic products were purified by Strata-X polymeric sorbents column (Phenomenex). Attempted in vitro polymerization and product examination by LC-MS was performed based on the method described (Bhuiya and Liu, 2010). For coincubation of monolignol and 4-O-methylated monolignol, the coniferyl alcohol was fixed at the concentration of 5 μM, and 0.5 to 50 μM 4-O-methyl-coniferyl alcohol was mixed into the reaction mixture containing 136 ng/mL horseradish peroxidase and 1.14 mM H2O2. The reactions were allowed to proceed for 5 min and then stopped by addition of equal volume of 50% acetonitrile. The products were separated by HPLC using 0.2% acetic acid (A) with an increasing concentration gradient of acetonitrile containing 0.2% acetic acid (B) for 0 to 2 min, 5%; 2 to 8 min, 5 to 39%; 8 to 10 min, 39 to 45%; 10 to 14 min, 45 to 50%; and 14 to 17min, 50 to 100%; at a flow rate of 1 mL/min. The peak areas of the major oxidative dimeric products were summed to quantify the polymerization efficiency.
RNA Extraction and RT-PCR
Stems of MOMT4 transgenic and control plants grown in the same growth chamber were collected at stage 6.2 (Boyes et al., 2001). For each line, we pooled the entire stems of three individual plants to constitute one sample. Samples were immediately frozen in liquid nitrogen and stored at −80°C until use for total RNA extraction.
Total RNA was extracted with the Qiagen RNeasy plant mini kit according to the manufacturer’s instructions. RNA samples were quantified with a Nanodrop spectrophotometer (ND-1000; Labtech). Reverse transcription was performed with 3 μg of total RNA and M-MLV (Promega) according to the manufacturer’s instructions. The MOMT4 transgene was detected using primers 5′-GCTCACGGCTACCGACAAGG-3′ and 5′-GCAATGCTCATCGCTCCAGT-3′. The internal standard ACTIN2 was amplified with primers 5′-GGTAACATTGTGCTCAGTGGTGG-3′ and 5′-CTCGGCCTTGGAGATCCACATC-3′.
In Vitro Transport Assay
Preparation of plasma membrane inside-out vesicles, their quality examination, and the uptake assay on coniferyl alcohol and 4-O-methylated coniferyl alcohol were performed essentially as described by Miao and Liu (2010).
Briefly, an Arabidopsis thaliana microsomal fraction was prepared from ∼4-week-old plants of the wild type (Columbia-0) according to Palmgren et al. (1990). Plasma membranes were then purified by partitioning the microsomal fraction into an aqueous polymer two-phase system as described (Larsson et al., 1994). The final upper phase containing the right-side-out plasma membranes was pelleted and resuspended in 5 mM potassium phosphate buffer, pH 7.8, containing 0.33 M Suc, 5 mM KCI, 1 mM DTT, and 0.1 mM EDTA (buffer A). The plasma membrane vesicles were then mixed with the detergent Brij 58 (Sigma-Aldrich) to a final concentration of 0.05% (w/v) and KCl to 50 mM, together with four cycles of freezing-thawing, to produce the sealed, inside-out vesicles as described (Johansson et al., 1995). The inverted plasma membrane vesicles were pelleted and gently resuspended in 10 mM MOPS-KOH buffer, pH 7.5, containing 0.33 M Suc, 0.1 mM EDTA, 1 mM DTT, and 1× protease inhibitor cocktail (Sigma-Aldrich). The quality of the prepared plasma membranes was monitored by measuring the vanadate-inhibited Mg2+-ATPase activity by an ATPase assay kit, according to the manufacturer’s instructions (Innova Biosciences).
The uptake assay was performed at 25°C for 30 min in a 500 μL assay mixture containing 25 mM Tris-MES, pH 8.0, 0.4 M sorbitol, 50 mM KCl, 5 mM Mg-ATP, 0.1% (w/v) BSA, and 100 μM of phenolic substrates. Mg-ATP was omitted from the nonenergized controls. Assays were initiated by the addition of membrane vesicles (100 μg of protein) and brief agitation. After incubation, the mixtures were subjected to vacuum filtration through prewetted nitrocellulose membrane filters (0.22-µm pore diameter; Millipore). After thorough washing, the filters were then extracted by 50% (v/v) methanol in an orbital shaker. The methanolic extracts were analyzed by HPLC. Quantification was made based on the standard curve of coniferyl alcohol.
Overexpression of MOMT4 in Arabidopsis
The open reading frames of MOMT4 and a loss-of-function mutant (E165R, as the control) (Bhuiya and Liu, 2010) were integrated into a binary vector pMDC32-PAL-GW that was constructed by replacement of the 35S promoter of pMDC32, a gateway destination vector (Curtis and Grossniklaus, 2003), by the PAL2 promoter. The constructs were transferred into Arabidopsis wild type (Columbia-0) mediated by Agrobacterium tumefaciens via the floral dip method (Clough and Bent, 1998). Arabidopsis plants were grown in a growth chamber at 22°C with 60% relative humidity under a 14-h-light (110 μmol m−1 s−1)/10-h-dark regime. The transgenic and control T2 or T3 generation plants were grown side by side for lignin and phenolic analysis.
Histochemical Analysis
Histochemical analysis was performed using 30-µm-thick sections from the same position of second basal node from 8-week-old stems of control and MOMT4 transgenic plants following the standard protocols for phloroglucinol (Wiesner) and Mӓule staining (Jensen, 1962; Nakano and Meshitsuka, 1992).
Analysis of Wall-Bound Phenolics and Lignin
Arabidopsis stems were harvested from 10-week-old plants at stage of 9.70 (Boyes et al., 2001). Stems from at least five individual plants for each transgenic line were grouped and ground into powder under liquid nitrogen. The powder was extensively extracted with 70% ethanol at 65°C for 3 h and repeated three times and then the residuals were treated with acetone at room temperature overnight. The extractive free residuals were dried at 45°C and then ball-milled into a fine powder. The wall-bound phenolics were extracted with 2 n NaOH in the dark at 37°C for 16 h and analyzed as described (Gou et al., 2008). Total lignin was quantified by the acetyl bromide method (Chang et al., 2008). To analyze the monomeric composition of lignin, we followed the thioacidolysis method (Rolando et al., 1992). Quantification of the corresponding monomers was done with a gas chromatography–flame ionization detector (Agilent) after an appropriate calibration relative to the tetracosane internal standard. The statistical analyses on the obtained data sets were performed with a Student’s paired t test, using two-tailed distribution and two-sample unequal variances.
For whole-cell wall NMR analysis, the ball-milled extractive-free residuals (55 mg) were mixed with 4:1 DMSO-d6/pyridine-d5 (500 μL) to form gel samples (Kim and Ralph, 2010). For a more complete and in-depth structural characterization of lignin, cell wall residuals were digested by Cellulysin (cellulase; Calbiochem) to obtain CEL. Ten milligrams of CELs were dissolved in 500 μL of DMSO-d6. 2D-HSQC NMR spectra were acquired on a Bruker Biospin Avance 500 MHz spectrometer fitted with a cryogenically cooled 5-mm triple-resonance gradient probe optimized for 1H and 13C observation with inverse geometry as previously described (Kim and Ralph, 2010).
Cellulose Quantification and Cellulolysis
Cellulose content was measured essentially according to the Updegraff’s method (Updegraff, 1969) at 620 nm on ball-milled cell wall materials. The efficiency of saccharification of cell wall biomass was determined using a described method (Sibout et al., 2005).
Phenotypic Analysis of MOMT4 Transgenic Plants
Eight-week-old plants between stages of 6.50 and 6.90 (Boyes et al., 2001) were used for phenotypic analysis. The shoot length of the main stem was measured by ruler as the height of the individual plant. The base of the main stem was measured using a Vernier caliper. The aboveground parts of three plants were sealed in paper bags and dried to constant weight (∼5 d) in an oven at 60°C.
Analysis of Soluble Phenolics
Roots, stems, leaves, and seeds of transgenic plants were ground into powder in liquid nitrogen and were extracted with 80% methanol containing 25 µM chrysin as the internal standard at 4°C overnight. The extracts were profiled by LC-MS and characterized by MSn analysis. Quantification of the detected compounds was based on the standard curves of ferulic acid and sinapic acid that were made in the same HPLC runs.
Accession Numbers
The atomic coordinates and structure factors, MOMT3-coniferyl alcohol-SAH with code 3TKY, have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics (Rutgers University, New Brunswick, NJ) (http/www.rcsb.org/). Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: JX287369 (MOMT4) and AT3G18780 (ACTIN2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Scheme of Simplified Phenylpropanoid-Lignin-Phenolic Ester Biosynthetic Pathways.
Supplemental Figure 2. Amino Acid Residues Directly in Contact with, or Proximate to, the Bound Coniferyl Alcohol in the MOMT3 Active Site.
Supplemental Figure 3. In Vitro Polymerization of Conventional Monolignols and 4-O-Methylated Monolignols.
Supplemental Figure 4. Coincubation of Conventional Monolignol with 4-O-Methylated Monolignol in the in Vitro Polymerization System.
Supplemental Figure 5. HSQC NMR Spectra (δc/δh 0 to 135/0 to 8.0 ppm) of the Total Cell Wall Samples of MOMT4 Transgenic and Control Plants after Gelling in DMSO-d6:Pyridine-d5 (4:1).
Supplemental Figure 6. Partial Short-Range HSQC NMR Spectra (Aromatic Region) of Cellulolytic Enzyme Lignins Isolated from Cell Walls of MOMT4 Transgenic and Control Plants.
Supplemental Figure 7. The UV Spectra of Novel Wall-Bound Phenolic Esters and the Related Phenolic Standards.
Supplemental Figure 8. The UV Spectra of Novel Soluble Phenolic Esters and the Related Phenolic Standards.
Supplemental Figure 9. LC-MS Analysis on Novel Methanolic Soluble-Phenolic Compounds Accumulated in the Root of MOMT4 Transgenic Plants
Supplemental Table 1. The Kinetic Parameters of MOMT Variants for Monolignols.
Supplemental Table 2. Crystallographic Data Sheet for the Structure of MOMT3 Mutant Variant.
Supplemental Table 3. Quantification of Soluble Phenolics in 10-Week-Old Stems of MOMT4 Transgenic Plants.
Supplemental Data Set 1. Complete Data Set of Transcriptome Study of MOMT Transgenic Line by cDNA Microarray Analysis.
Supplementary Material
Acknowledgments
We thank Eran Pichersky (University of Michigan) for donating the Clarkia breweri IEMT clone. We also thank John Shanklin and William Studier (Brookhaven National Laboratory) for their valuable discussions on this work. This work is supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (DOE) through Grant DEAC0298CH10886 (BO-147 and 157) to C.-J.L. Transport measurement (by Y.M.) was supported in part by National Science Foundation through Grant MCB-1051675. J.R.P. was funded by a Spanish Ministry of Education (postdoctoral fellowship); J.R., J.R.P., and H.K. (NMR analysis) were funded in part by the DOE’s Great Lakes Bioenergy Research Center (Grant BER DE-FC02-07ER64494). The use of x-ray beamlines at X25 and X29 of National Synchrotron Light Source is supported by the DOE, Office of Basic Energy Sciences, under Contract DEAC0298CH10886.
AUTHOR CONTRIBUTIONS
C.-J.L., K.Z., and M.-W.B. designed the research. K.Z, M.-W.B, and C.-J.L. performed biochemical, structural, and genetic experiments. Y.M. performed the transport assay, and J.R.P. and H.K. performed NMR analysis. C.-J.L., J.R, K.Z., and M.-W.B. analyzed data. C.-J.L., J.R., K.Z., and M.-W.B. wrote the article. All authors contributed to the editorial efforts.
Glossary
- H2O2
hydrogen peroxide
- COMT
caffeate/5-hydroxyferulate 3/5-O-methyltransferase
- IEMT
isoeugenol O-methyltransferase
- MOMT
monolignol 4-O-methyltransferase
- SAH
S-adenosyl homocysteine
- SAM
S-adenosyl Met
- CEL
cellulolytic enzyme lignin
- 2D
two-dimensional
- HSQC
to be defined
- LC-MS
liquid chromatography–mass spectrometry
- m/z
mass-to-charge ratio
- ALDH
aldehyde dehydrogenase
- CAD
cinnamyl alcohol dehydrogenase
References
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., Lapierre C., Jouanin L. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiya M.W., Liu C.J. (2010). Engineering monolignol 4-O-methyltransferases to modulate lignin biosynthesis. J. Biol. Chem. 285: 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckler G.A., Gershenzon J., Unsicker S.B. (2011). Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses. Phytochem. 72: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr S.J., Fry S.C. (2009). Extracellular cross-linking of maize arabinoxylans by oxidation of feruloyl esters to form oligoferuloyl esters and ether-like bonds. Plant J. 58: 554–567 [DOI] [PubMed] [Google Scholar]
- Castle L.A., Siehl D.L., Gorton R., Patten P.A., Chen Y.H., Bertain S., Cho H.J., Duck N., Wong J., Liu D., Lassner M.W. (2004). Discovery and directed evolution of a glyphosate tolerance gene. Science 304: 1151–1154 [DOI] [PubMed] [Google Scholar]
- Chang X.F., Chandra R., Berleth T., Beatson R.P. (2008). Rapid, microscale, acetyl bromide-based method for high-throughput determination of lignin content in Arabidopsis thaliana. J. Agric. Food Chem. 56: 6825–6834 [DOI] [PubMed] [Google Scholar]
- Chen F., Liu C.J., Tschaplinski T.J., Zhao N. (2010). Genomics of secondary metabolism in Populus: Interactions with biotic and abiotic environments. Crit. Rev. Plant Sci. 28: 375–392 [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cramer C.L., Edwards K., Dron M., Liang X., Dildine S.L., Bolwell G.P., Dixon R.A., Lamb C.J., Schuch W. (1989). Phenylalanine ammonia-lyase gene organization and structure. Plant Mol. Biol. 12: 367–383 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin L.B., Jourdes M., Patten A.M., Kim K.W., Vassão D.G., Lewis N.G. (2008). Dissection of lignin macromolecular configuration and assembly: Comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat. Prod. Rep. 25: 1015–1090 [DOI] [PubMed] [Google Scholar]
- Dewar M.J.S., David D.E. (1980). Ultraviolet photoelectron-spectrum of the phenoxy radical. J. Am. Chem. Soc. 102: 7387–7389 [Google Scholar]
- Franke R., Humphreys J.M., Hemm M.R., Denault J.W., Ruegger M.O., Cusumano J.C., Chapple C. (2002). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30: 33–45 [DOI] [PubMed] [Google Scholar]
- Freudenberg K. (1959). Biosynthesis and constitution of lignin. Nature 183: 1152–1155 [DOI] [PubMed] [Google Scholar]
- Freudenberg K. (1968). The constitution and biosynthesis of lignin. In Constitution and Biosynthesis of Lignin, K. Freudenberg and A.C. Neish, eds (Berlin: Springer-Verlag), pp. 45–122 [Google Scholar]
- Gallagher S.R., Leonard R.T. (1982). Effect of vanadate, molybdate, and azide on membrane-associated ATPase and soluble phosphatase activities of corn roots. Plant Physiol. 70: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J.Y., Park S., Yu X.H., Miller L.M., Liu C.J. (2008). Compositional characterization and imaging of “wall-bound” acylesters of Populus trichocarpa reveal differential accumulation of acyl molecules in normal and reactive woods. Planta 229: 15–24 [DOI] [PubMed] [Google Scholar]
- Halpin C., Knight M.E., Grima-Pettenati J., Goffner D., Boudet A., Schuch W. (1992). Purification and characterization of cinnamyl alcohol dehydrogenase from tobacco stems. Plant Physiol. 98: 12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin J.M. (1967). Lignin - A natural polymeric product of phenol oxidation. In Oxidative Coupling of Phenols, W.I. Taylor and A.R. Battersby, eds (New York: Marcel Dekker), pp. 243–321 [Google Scholar]
- Harkin J.M. (1973). Lignin. In Chemistry and Biochemistry of Herbage, G.W. Butler, ed (London: Academic Press), pp. 323–373 [Google Scholar]
- Iiyama K., Wallis A.F.A. (1990). Determination of lignin in herbaceous plant by an improved acetyl bromide procedure. J. Sci. Food Agric. 51: 145–161 [Google Scholar]
- Inoue K., Parvathi K., Dixon R.A. (2000). Substrate preferences of caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferases in developing stems of alfalfa (Medicago sativa L.). Arch. Biochem. Biophys. 375: 175–182 [DOI] [PubMed] [Google Scholar]
- Ishii T. (1997). Structure and functions of feruloylated polysaccharides. Plant Sci. 127: 111–127 [Google Scholar]
- Jensen W.A. (1962). Botanical Histochemistry: Principles and Practice. (Berkeley, CA: W.H. Freeman and Company) [Google Scholar]
- Jez J.M. (2011). Toward protein engineering for phytoremediation: Possibilities and challenges. Int. J. Phytoremediation 13 (Suppl 1): 77–89 [DOI] [PubMed] [Google Scholar]
- Johansson F., Olbe M., Sommarin M., Larsson C. (1995). Brij 58, a polyoxyethylene acyl ether, creates membrane vesicles of uniform sidedness. A new tool to obtain inside-out (cytoplasmic side-out) plasma membrane vesicles. Plant J. 7: 165–173 [DOI] [PubMed] [Google Scholar]
- Kim H., Ralph J. (2010). Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d(6)/pyridine-d(5). Org. Biomol. Chem. 8: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.J., Kim M.R., Bedgar D.L., Moinuddin S.G., Cardenas C.L., Davin L.B., Kang C., Lewis N.G. (2004). Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T., et al. (2006). Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 103: 10128–10133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry L.G., Chapple C.C.S., Last R.L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C. (1993). Application of new methods for the investigation of lignin structure. In Forage Cell Wall Structure and Digestibility, H.G. Jung, D.R. Buxton, R.D. Hatfield, and J. Ralph, eds (Madison, WI: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America), pp. 133–166 [Google Scholar]
- Larsson C., Sommarin M., Widell S. (1994). Isolation highly purified plant plasma membranes and seperation of inside-out and right-side-out vesicles. Methods Enzymol. 228: 451–469 [Google Scholar]
- Li X., Weng J.K., Chapple C. (2008). Improvement of biomass through lignin modification. Plant J. 54: 569–581 [DOI] [PubMed] [Google Scholar]
- Lloyd R.V., Wood D.E. (1974). Free-radicals in an adamantane matrix. 8. EPR and INDO study of benzyl, anilino, and phenoxy radicals and their fluorinated derivatives. J. Am. Chem. Soc. 96: 659–665 [Google Scholar]
- Louie G.V., Bowman M.E., Tu Y., Mouradov A., Spangenberg G., Noel J.P. (2010). Structure-function analyses of a caffeic acid O-methyltransferase from perennial ryegrass reveal the molecular basis for substrate preference. Plant Cell 22: 4114–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyermans H., et al. (2000). Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275: 36899–36909 [DOI] [PubMed] [Google Scholar]
- Miao Y.C., Liu C.J. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 107: 22728–22733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkowski C., Baumert A., Schmidt D., Nehlin L., Strack D. (2004). Molecular regulation of sinapate ester metabolism in Brassica napus: Expression of genes, properties of the encoded proteins and correlation of enzyme activities with metabolite accumulation. Plant J. 38: 80–92 [DOI] [PubMed] [Google Scholar]
- Mir Derikvand M., Sierra J.B., Ruel K., Pollet B., Do C.T., Thévenin J., Buffard D., Jouanin L., Lapierre C. (2008). Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956 [DOI] [PubMed] [Google Scholar]
- Morreel K., Ralph J., Kim H., Lu F., Goeminne G., Ralph S., Messens E., Boerjan W. (2004). Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol. 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R.B., Bastress K.L., Ruegger M.O., Denault J.W., Chapple C. (2004). The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano J., Meshitsuka G. (1992). The detection of lignin. In Methods in Lignin Chemistry, S.Y. Lin and C.W. Dence, eds (Berlin: Springer Verlag), pp. 23–32
- O’Maille P.E., Malone A., Dellas N., Andes Hess B., Jr, Smentek L., Sheehan I., Greenhagen B.T., Chappell J., Manning G., Noel J.P. (2008). Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren M.G., Askerlund P., Fredrikson K., Widell S., Sommarin M., Larsson C. (1990). Sealed inside-out and right-side-out plasma membrane vesicles: Optimal conditions for formation and separation. Plant Physiol. 92: 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi K., Chen F., Guo D., Blount J.W., Dixon R.A. (2001). Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Ralph J. (2010). Hydroxycinnamates in lignification. Phytochem. Rev. 9: 65–83 [Google Scholar]
- Ralph J., Bunzel M., Marita J.M., Hatfield R.D., Lu F., Kim H., Schatz P.F., Grabber J.H., Steinhart H. (2004a). Peroxidase dependent cross-linking reactions of p-hydroxycinnamates in plant cell walls. Phytochem. Rev. 3: 79–96 [Google Scholar]
- Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P.F., Marita J.M., Hatfield R.D., Christensen J.H., Boerjan W. (2004b). Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3: 29–60 [Google Scholar]
- Ralph J., Schatz P.F., Lu F., Kim H., Akiyama T., Nelsen S.F. (2009). Quinone methides in lignification. In Quinone Methides, S. Rokita, ed (Hoboken, NJ: Wiley-Blackwell), pp. 385–420 [Google Scholar]
- Reddy M.S.S., Chen F., Shadle G., Jackson L., Aljoe H., Dixon R.A. (2005). Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. USA 102: 16573–16578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando C., Monties B., Lapierre C. (1992). Thioacidolysis. In Methods in Lignin Chemistry, S.Y. Lin and C.W. Dence, eds (Berlin, Heidelberg, Germany: Springer-Verlag), pp. 334–340 [Google Scholar]
- Russell W.R., Forrester A.R., Chesson A., Burkitt M.J. (1996). Oxidative coupling during lignin polymerization is determined by unpaired electron delocalization within parent phenylpropanoid radicals. Arch. Biochem. Biophys. 332: 357–366 [DOI] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Mouille G., Pollet B., Lapierre C., Jouanin L., Séguin A. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Vagin A.A., Steiner R.S., Lebedev A.A., Potterton L., McNicholas S., Long F., Murshudov G.N. (2004). REFMAC5 dictionary: Organisation of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60: 2284–2295 [DOI] [PubMed] [Google Scholar]
- Wang J., Pichersky E. (1998). Characterization of S-adenosyl-L-methionine:(iso)eugenol O-methyltransferase involved in floral scent production in Clarkia breweri. Arch. Biochem. Biophys. 349: 153–160 [DOI] [PubMed] [Google Scholar]
- Wang J., Pichersky E. (1999). Identification of specific residues involved in substrate discrimination in two plant O-methyltransferases. Arch. Biochem. Biophys. 368: 172–180 [DOI] [PubMed] [Google Scholar]
- Weng J.K., Chapple C. (2010). The origin and evolution of lignin biosynthesis. New Phytol. 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Weng J.K., Li X., Bonawitz N.D., Chapple C. (2008). Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19: 166–172 [DOI] [PubMed] [Google Scholar]
- Yoshikuni Y., Ferrin T.E., Keasling J.D. (2006). Designed divergent evolution of enzyme function. Nature 440: 1078–1082 [DOI] [PubMed] [Google Scholar]
- Zubieta C., Kota P., Ferrer J.-L., Dixon R.A., Noel J.P. (2002). Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.