The authors demonstrate that parasite gene-specific silencing signals originating from a transgenic host are transferred into the invading parasite, leading to reduced parasite yield, stature, and infectivity. This article also refreshes the debate on the origin of haustoria as the authors use morphological and molecular evidence to show that haustoria have both stem and root characteristics.
Abstract
Infection of crop species by parasitic plants is a major agricultural hindrance resulting in substantial crop losses worldwide. Parasitic plants establish vascular connections with the host plant via structures termed haustoria, which allow acquisition of water and nutrients, often to the detriment of the infected host. Despite the agricultural impact of parasitic plants, the molecular and developmental processes by which host/parasitic interactions are established are not well understood. Here, we examine the development and subsequent establishment of haustorial connections by the parasite dodder (Cuscuta pentagona) on tobacco (Nicotiana tabacum) plants. Formation of haustoria in dodder is accompanied by upregulation of dodder KNOTTED-like homeobox transcription factors, including SHOOT MERISTEMLESS-like (STM). We demonstrate interspecific silencing of a STM gene in dodder driven by a vascular-specific promoter in transgenic host plants and find that this silencing disrupts dodder growth. The reduced efficacy of dodder infection on STM RNA interference transgenics results from defects in haustorial connection, development, and establishment. Identification of transgene-specific small RNAs in the parasite, coupled with reduced parasite fecundity and increased growth of the infected host, demonstrates the efficacy of interspecific small RNA–mediated silencing of parasite genes. This technology has the potential to be an effective method of biological control of plant parasite infection.
INTRODUCTION
The negative effects of parasitic plants on agricultural crop production worldwide cannot be overstated. Parasitic plants use specialized organs called haustoria to attach to host tissues and extract host nutrients and water for parasite growth and reproduction. It has recently been estimated that parasitic plants impact over 300 million farmers throughout the world (Ejeta, 2007). Farm productivity on the African continent has been disproportionately impacted by this loss. Particularly deadly are the witchweeds of the family Orobanchaceae, genus Striga. Striga is a major pest infecting the staple crops maize (Zea mays), sorghum (Sorghum bicolor), and legumes (such as cowpea [Vigna unguiculata]), impacting food resources that are already under considerable strain. Current estimates place annual yield losses at greater than $7 billion in sub-Saharan Africa alone (Ejeta, 2007).
Parasitic plants are notoriously difficult to control because their life cycles are intricately coupled to that of their hosts. The growth of both host and parasite is dependent on the host, which leads to substantial losses in yield of parasitized crops. Indeed, the total biomass of the host and parasite combined has been shown by modeling to always be less than the mass of the unparasitized host (Hautier et al., 2010). Additionally, increased use of nitrogen fertilizers may exacerbate dodder (Cuscuta pentagona) infections. Comparisons of fertilized and unfertilized plots has revealed that supplementation with nitrogen fertilizers can double the cover of Cuscuta salina even without any evident increase in host mass (Pennings and Simpson, 2008). High intensity farming practices used in the developed world may only serve to worsen parasitic losses if introduced into fields with parasitic loads, which underscores the need to explore other methods of parasitic plant control. Traditional breeding approaches have shown some promise in the control of some plant parasites (Ejeta, 2007) but have to be pursued on a case-by-case basis due to the multiple independent evolutionary origins of parasitism (Ejeta et al., 2000; Grenier et al., 2001; Gurney et al., 2006; Ejeta, 2007; Westwood et al., 2010). Key to developing novel strategies for parasitic plant control is an understanding of the molecular mechanism of host–parasite associations. It is therefore of great interest to advance our biological understanding of plant parasitism in several host–parasite systems, look for factors that are shared between them, and apply this knowledge to the development of resistant crops.
This work focuses on plant parasites in the genus Cuscuta (dodder). The genus Cuscuta consists of ∼150 species and is among the most successful and devastating plant parasites, causing serious damage by suppressing the growth of its host and, in some cases, leading to host death. Its members exhibit a broad host range, parasitizing both domesticated and nondomesticated plants (Gaertner, 1950; Kuijt, 1969; Malik and Singh, 1979; Dawson et al., 1994). Dodder seeds germinate and form a thread-like stem. Seedlings have a short-lived rudimentary root system, lack enough reserves for sustained growth, and contain little chlorophyll and thus are completely dependent on finding a host within a few days for survival. At points of contact with the host, the coiled dodder stem produces haustoria that penetrate host tissues and form vascular connections (Kuijt, 1983; Vaughn, 2003). Haustoria produce long, unicellular searching hyphae that make many plasmodesmata at the contact point with host parenchyma (Vaughn, 2003; Birschwilks et al., 2006). Eventually, phloem–phloem and xylem–xylem connections formed between dodder and the host are used to transfer water and assimilates to the parasite (Jeschke et al., 1994; Vaughn, 2003; Birschwilks et al., 2006).
To date there is no single dodder control strategy available that is both effective and sustainable; thus, spread of the parasite continues unabated (Rispail et al., 2007). Strategies for dodder management include crop rotation, flooding or flaming some perennial crops, tilling, timing of crop planting, and use of selective herbicides (Mishra, 2009; Nadler-Hassar et al., 2009; Sandler, 2010). Many of these may be required in combination to provide effective protection. Additionally, the sensitivity to various herbicides can differ significantly between seedling dodder and dodder that is already attached to a host (Nadler-Hassar et al., 2009). The use of transgenic crops with herbicide resistance has been shown to afford some protection, but dodder plants attached to these transgenic plants can recover rapidly and resume growth, benefiting from the resistance mechanisms encoded by the host (Nadler-Hassar et al., 2009). Control of this parasite is complex and highly crop dependent; therefore, increasing our repertoire of control strategies to complement existing methods should be fruitful.
Macromolecules are naturally transported into the parasite from the host plant, and earlier reports have implicated haustorial connections in protein and RNA translocation between host and parasite (Mower et al., 2004; Roney et al., 2007; David-Schwartz et al., 2008). RNA movement between host and parasite has even been implicated in horizontal gene transfer between the host and parasite genomes. For example, a gene of unknown function appears to have been transferred into Striga hermonthica from one of its grass hosts by retrointegration of a host mRNA transcript (Yoshida et al., 2010), illustrating the intimate nature of the connection that allows RNA trafficking even between host–parasite interactions spanning the monocot-dicot divergence.
Several reviews on host–parasite interaction have proposed that cross-species RNA interference holds promise for parasitic plant management (Yoder et al., 2009; Runo et al., 2011). Downregulation of marker genes in the parasite through RNA interference (RNAi) in the host has been documented (Tomilov et al., 2008). One study reported downregulation of a parasite metabolic gene through ubiquitously expressed (cauliflower mosaic virus 35S promoter) host-based RNAi but did not detect the presence of small interfering RNAs (siRNAs) in the parasite (Aly et al., 2009). In this study, we prospected for candidate dodder genes involved in haustorial development that could be targeted for downregulation through the host.
An increase in cytokinin precedes induction of haustoria, and cytokinin is required for their development (Haidar et al., 1998; Furuhashi et al., 2011). Members of the KNOTTED1-like HOMEOBOX1 (KNOX1) gene family are known to promote cytokinin biosynthesis in the shoot apical meristem (SAM) (Jasinski et al., 2005), and KNOX1 genes are well established as regulators of indeterminate identity in the SAM throughout angiosperms (Chuck et al., 1996, 1998; Long et al., 1996; Kerstetter et al., 1997; Hake et al., 2004). Furthermore, KNOX1 expression has been modified repeatedly over evolutionary time to confer prolonged indeterminate identity during organogenic processes within lateral organs (Sinha et al., 1993; Bharathan et al., 2002; Pham and Sinha, 2003; Tsiantis, 2005; Hay and Tsiantis, 2006). Haustoria are initiated on stems of dodder, grow through the host, and establish vascular connection with the host, by differentiation of searching hyphal cells into vascular tissue. Finally, it has been reported that isolated dodder haustoria embedded in the host have the capability to regenerate new shoots, suggesting that these haustoria may retain indeterminate features and meristematic capacity (Truscott, 1958). This prompted us to explore the role of genes regulating indeterminacy in haustorium formation. The KNOX1 genes provided one potential set of candidate genes that might play a role in the development of haustoria in dodder. Since the parasite dodder is known to establish vascular connections with the host, we used a highly specific vascular promoter to generate the siRNAs to selected dodder KNOX1 genes and high-throughput sequencing to detect siRNAs in the host and the parasite. Furthermore, we analyzed the effect of this downregulation of parasite KNOX1 genes in the host on parasite establishment and growth.
RESULTS
Dodder Infection and Early Haustorial Development
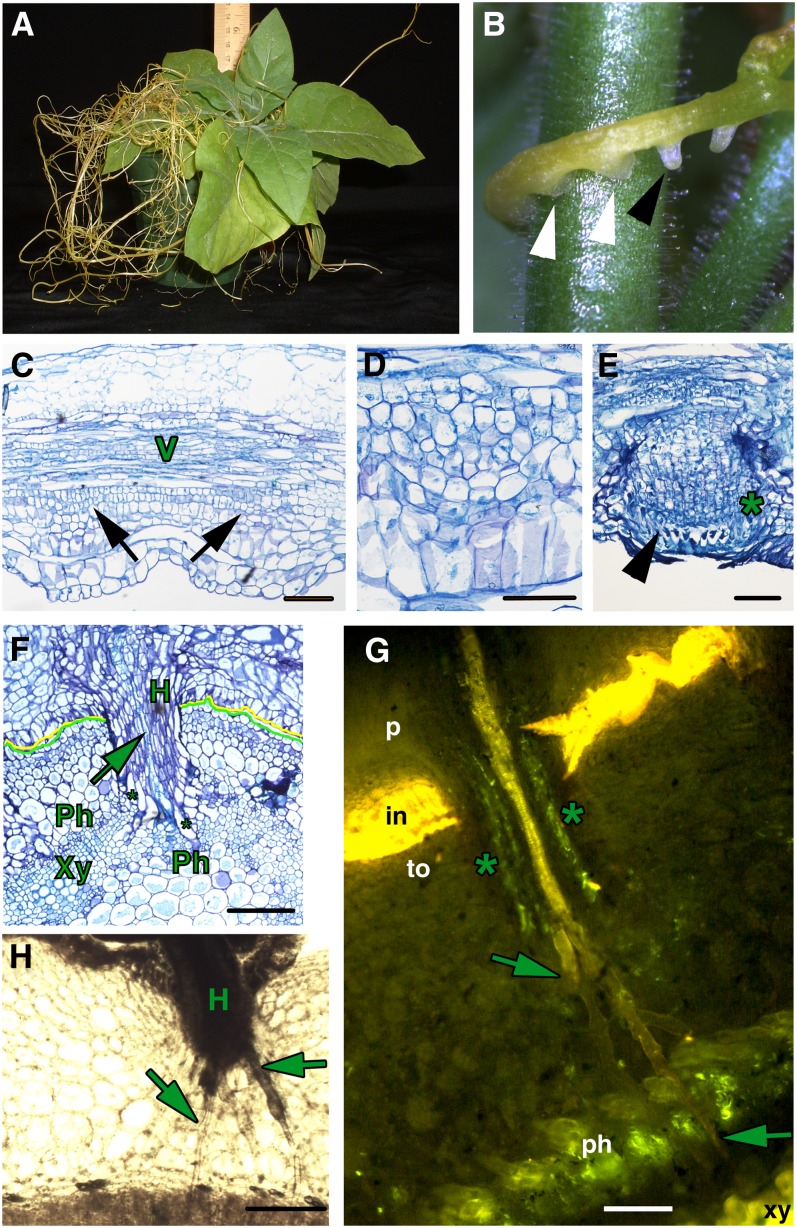
Dodder appears as a stringy, tangled mass encircling the host plant and lacks both a clear root system and expanded leaves (Figure 1A), making it dependent on finding a host species to acquire water and nutrients. This interaction is accomplished through the establishment of host-parasite connections termed haustoria (Figure 1B). As a first step in understanding the dodder infection process, we characterized the early stages of haustoria development. Stem tissues initiating haustoria show evidence of increased cell division activity adjacent to the vascular bundles (Figures 1C and 1D). Subsequent growth is supported by continued cell division activity in this region, and in the developing prehaustorium, three zones are visible. The vascular proximal cells are followed by a zone of dividing cells called file cells (marked with an asterisk in Figure 1E) (Lee, 2007). A distal-most zone of large cells with prominent nuclei called digitate cells (Lee, 2007) is evident at the tip of the prehaustoria (arrowhead in Figure 1E). These cells are likely to be the precursors of searching hyphae. The developing prehaustoria then rupture the epidermis and elongate into mature prehaustoria with a central column of elongated cells that will develop into vascular tissue upon attachment to the host vascular system (Figures 1F and 1G). Finally, the haustoria attach to the host plant and establish direct connections to the host vascular system (Figures 1F to 1H). The tips of haustoria develop searching hyphae, long cells that grow intercellularly toward the host vascular system (Figures 1G and 1H). The searching hyphae form unique unicellular strands that are either xylem (Xylic; Figure 1G, arrows) or phloem specific (Phloic; Figure 1G, asterisks) depending on their location within the haustorium and contact with the host vasculature. Upon contacting the host phloem and xylem, these cells acquire vascular identity (Figures 1F and 1G; see Supplemental Figure 1 online). Concomitant with this contact, we see vascular differentiation in the center of the haustorium (Figures 1F and 1G). This establishes a continuous vascular connection between the host and the parasite. The increased cell division activity in dodder shoot vascular-adjacent cell files at the time of haustorium initiation indicates that these regions of the stem possess sufficient indeterminate identity to initiate haustoria. The haustorium-adjacent region is also associated with the development of unusual lateral shoots, lending further credence to the idea that these regions have acquired some indeterminate potential. These shoots give rise to vegetative or floral branches later in development and form only subsequent to haustoria development, suggesting a possible link between these two processes (see Supplemental Figure 2 online). Reports that isolated dodder haustoria embedded in the host can regenerate into new shoots also suggest indeterminate features and meristematic capacity in these organs (Truscott, 1958).
Figure 1.
Parasitism by Dodder.
(A) Tobacco infected with dodder.
(B) Dodder strand showing embedded haustoria (white arrowheads) and prehaustoria (black arrowhead).
(C) Longitudinal section of a dodder strand with vascular tissue (V) and initiating haustoria (black arrows).
(D) Longitudinal section of a dodder strand in high magnification showing developing haustoria.
(E) Dodder haustoria emerging from the stem epidermis; the asterisk marks file cells, and the arrowhead marks digitate cells in the prehaustorium.
(F) Transverse section of a tobacco stem with an attached dodder strand. Mature haustoria (H) of dodder (marked with a yellow line) are embedded in the tobacco stem (demarcated with a green line) and Xylic searching hyphae (arrow) have contacted the host xylem (Xy), while Phloic hyphae (asterisks) contact the host phloem (Ph).
(G) Transverse section of a tobacco stem (to) with attached dodder parasite (p) stained with Aniline Blue. Mature haustorium shows a central column of Xylic hyphae (arrows) that have contacted the host xylem, and peripheral to that, Phloic hyphae (marked with asterisks) that contact the host phloem. in, host-parasite interface.
(H) Transverse section of a tobacco stem with attached dodder parasite showing a haustorium sending out numerous searching hyphae (arrows) toward the host vascular tissue.
Bars = 100 µm.
The Role of KNOX1 Genes in Haustorium Development
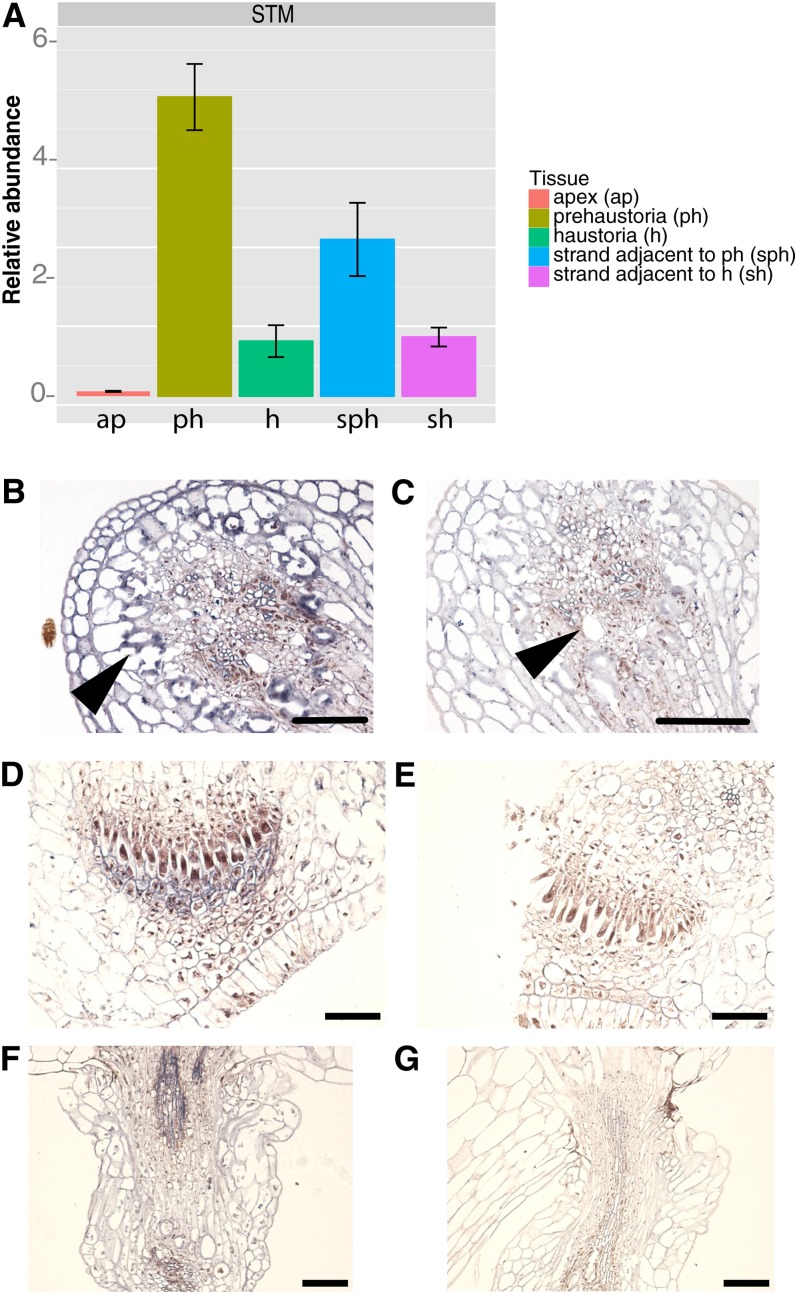
Because of their well-described role in establishment and maintenance of indeterminacy, we hypothesized that KNOX1 gene expression might play a role in the initiation of dodder haustoria. To examine this possibility, we cloned sequences representing a dodder SHOOT MERISTEMLESS-like gene (STM) and a KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1-like (KNAT1)-like gene using degenerate PCR and confirmed the identity of the cloned sequences by phylogenetic analysis (see Supplemental Figure 3 and Supplemental Data Set 1 online). Dodder infecting a tobacco (Nicotiana tabacum) host showed highest expression of these genes in strands wrapped around the host with developing prehaustoria and adjacent strands without prehaustoria when compared with the mature strands and fully formed haustoria attached to the host (Figure 2A; see Supplemental Figure 4 online). The least expression of the KNOXI genes investigated was seen in the dodder shoot apex (Figure 2A; see Supplemental Figure 4A online). Since these KNOX1 genes are usually downregulated in stem tissues compared with shoot meristems, such high expression in dodder strands compared with the apex was unexpected. To determine whether this high expression might be associated with haustorium development, we performed in situ hybridizations on dodder using both sense and antisense probes to the KNOX1 gene STM. Using the antisense probe, we detected strongest expression of STM in dodder strand in cells adjacent to the vascular tissues and in the vascular column (Figures 2B and 2C). In developing haustoria, expression was detected in the dividing group of cells and the digitate cells near the haustorial tip (Figures 2D and 2E). Expression was also detected in the developing haustorial vascular strand (Figures 2F and 2G; see Supplemental Figures 5A and 5B online). Expression in dodder SAMs was surprisingly low compared with haustoria though detectable compared with the control (see Supplemental Figures 5C and 5D online). Hybridizations of tissues to sense control probes showed no expression (Figure 2; see Supplemental Figure 5 online).
Figure 2.
Expression of STM during Haustorial Development.
(A) qRT-PCR showing expression of STM in dodder apex and stem strands with haustoria (a, apex; h, haustorium; p, prehaustorium; sh, strand haustorium; sph, strand prehaustorium). Error bars represent sd from the mean.
(B) Dodder stem showing STM expression in vascular and adjacent tissue.
(C) Sense control.
(D) and (E) Prehaustorium antisense and sense. Prior to emergence from stem where (D) is showing expression in digitate cells and file cells compared with sense control (E).
(F) and (G) Developing haustoria. STM expression is seen in developing vascular strand (F) compared with sense control (G). Bars = 100 µm.
Cross-Species RNAi of the Dodder STM Gene
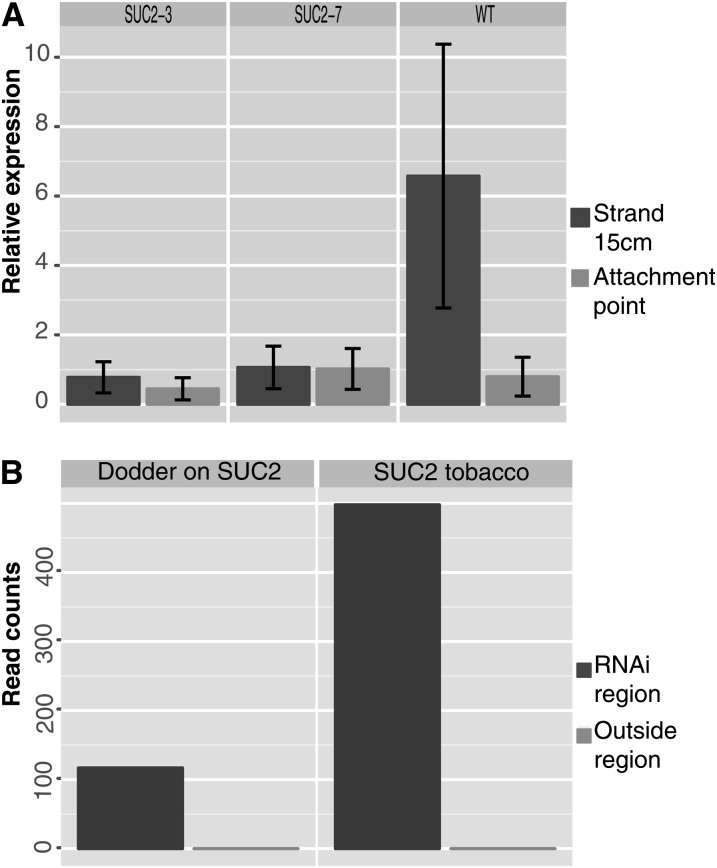
To elucidate the role of KNOX1 genes in haustorium formation, we used RNAi. RNAi signals are non-cell-autonomous and can act at both local (cell–cell) and systemic levels (spread through the vascular system). The RNA silencing signal in plants also moves across graft junctions (Palauqui et al., 1997; Voinnet and Baulcombe, 1997). RNAs have been shown to freely translocate between parasitic plants and their hosts (Roney et al., 2007; David-Schwartz et al., 2008). For example, a β-glucuronidase silencing signal generated by the host roots was translocated across the haustorium and was shown to be functional in the facultative root parasite Triphysaria (Tomilov et al., 2008). To examine the function of KNOX1 genes in dodder parasitism, we generated tobacco transgenics expressing an RNAi construct targeting the 3′ untranslated regions (UTRs) of two dodder KNOX1 genes that do not share significant sequence similarity with the host encoded orthologs (STM and KNAT1-3; see Supplemental Figures 6 and 7 online). We hypothesized that small RNAs generated from the host-derived transgene might cause downregulation of endogenous parasite transcripts, as has been suggested by experiments using the plant holoparasite Orobanche (Aly et al., 2009). However, in that study, the nature of the host parasite interactions was not described and no transgene specific siRNAs were found in the parasite. The transgenic tobacco plants expressed RNAi directed against the dodder STM and KNAT1-3 genes using the vascular-specific SUCROSE-PROTON SYMPORTER2 (SUC2) promoter (Truernit and Sauer, 1995). Dodder growing on plants transformed with this construct showed no downregulation of the KNAT1-3 homolog. This could be due to possible secondary structure that makes the target message inaccessible to the siRNAs, inefficient selection of the targeted region, or other as yet unknown reasons (McGinnis et al., 2007). The dodder strands at the attachment points with mature haustoria had low levels of STM expression and did not show differences between dodder grown on the wild type compared with transgenic hosts. However, strong downregulation of STM expression was observed in young developing dodder strands when compared with the parasite grown on untransformed plants (Figure 3A). These results demonstrate that effective downregulation of plant parasite endogenous transcripts can be achieved by SUC2-driven RNAi of target genes in the host.
Figure 3.
STM Downregulation in Dodder.
(A) qRT-PCR showing reduced dodder STM expression in both attached and independent RNAi transgenic lines SUC2-3 and SUC2-7.
(B) Distribution of the reads across the STM gene fragment in the region targeted for RNAi and reads outside the targeted region. Raw read counts are shown here to visualize the distribution.
Transgene-Specific siRNAs Traffic from the Host to the Parasite
To confirm the presence of siRNAs matching the STM (and KNAT1-3) gene, we compared small RNA sequences from dodder grown on both wild-type and transgenic tobacco (illustrated in Supplemental Figure 8 online). No small RNAs matching STM or KNAT1-3 were detected in dodder grown on wild-type tobacco, but such small RNAs were found in parasites infecting transgenic plants (Table 1; see Supplemental Table 1 online). Interestingly, all small RNAs in transgenic tobacco plants and in dodder grown on transgenic plants were confined to the sequence contained in the SUC2:RNAi construct, and no secondary siRNA production was observed (Figure 3B; see Supplemental Figure 9 online). This suggests that the KNOX1 siRNAs found in dodder might be mostly composed of the host primary transcripts and lack the transitive siRNAs that require RDR6 and SGS3 genes for their synthesis (Vaistij et al., 2002; Brodersen and Voinnet, 2006). However, the dodder seedling transcriptome being generated by us shows several RDR6 and SGS3-like genes (A. Ranjan, R. Kumar, Y. Ichihashi, and N. Sinha, unpublished data); thus, additional experiments are required to identify the reason for lack of transitive siRNAs in dodder. A majority of the reads that map to the target regions are 21 nucleotides long, though 22- and 24-nucleotide-long sequences are also observed (Blevins et al., 2006; Fusaro et al., 2006). A similar size distribution is seen in the reads mapped to the target sequences in the host as well as the parasite (see Supplemental Figure 10 online), indicating that these sequences are indeed molecules derived from the small RNAi pathway. Our results demonstrate non-cell-autonomous accumulation of mature small RNAs in the parasite. Since it is not clear if only the small RNAs move or if the precursor RNAs can also move through graft junctions (Brosnan et al., 2007; Molnar et al., 2010; Melnyk et al., 2011), the small RNA molecules detected in the dodder-infecting transgenic hosts can either be generated in the host or synthesized in the parasite from the host-derived precursor RNA molecules. The movement of RNA from the transgenic host plant to the parasite likely occurs through the haustorial vascular connection, though cell-to-cell movement that is nonvascular is also a possibility.
Table 1. Host-Derived Small RNAs in Dodder.
| Tissue | Total Reads | Total STM Reads | STM per 100k Reads |
|---|---|---|---|
| Wild-type tobacco | 1,131,135 | 0 | 0.000 |
| SUC2-3 tobacco | 1,525,178 | 485 | 31.800 |
| Dodder on the wild type | 3,655,552 | 0 | 0.000 |
| Dodder on SUC2-3 | 4,610,124 | 106 | 2.299 |
Columns display the total sequences recovered for each tissue, the total small RNAs with perfect matches to the STM region, and the number of mapped reads normalized to the sequencing depth.
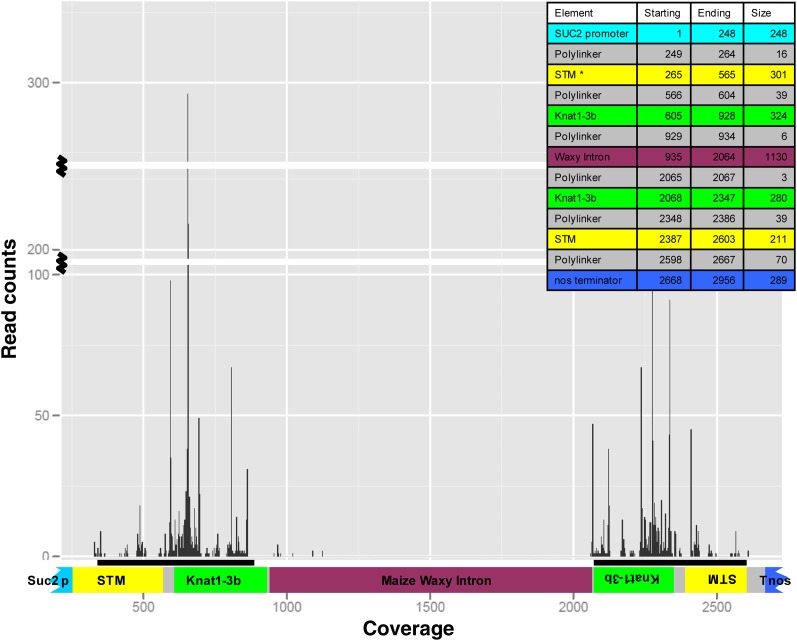
siRNA molecules derived from the SUC2:RNAi construct were well represented in siRNA libraries made from transgenic host plants and dodder grown on those plants but not from wild-type hosts or dodder grown on them. To better understand the relationship of the siRNAs to the transgene in the host, the siRNA reads from the dodder library were mapped onto the sequence of the RNAi construct in the host (Figure 4). As expected, the large majority of siRNAs map to the hairpin arms of the construct and include sequences spanning the KNOX1 and polylinker sequence (see Supplemental Figure 11 online). To confirm that no off-target siRNAs were generated against endogenous transcripts in the transgenic tobacco lines, we aligned the siRNA reads from transgenic and wild-type tobacco samples to 14 known tobacco genes, including STM and KNAT1 homologs (see Supplemental Table 2 online). We found some naturally occurring siRNAs mapping to tobacco homeobox23 in both the wild-type and transgenic lines but not in dodder parasitizing these hosts. The nature and origin of these siRNAs is unknown. No other siRNA counts to off-target genes tested were observed in tobacco or dodder samples. Additionally, to confirm that processes related to siRNA generation were not dramatically altered in dodder, we aligned reads to several additional dodder genes, including Class I and II KNOX genes, and found a low level background of siRNAs in the dodder orthologs of Liguleless, Phosphofructokinase, and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes common to dodder grown on wild-type and transgenic hosts. The predominant signal of mapping siRNAs came from the sequences included in our construct (see Supplemental Table 3 online).
Figure 4.
Coverage of Small RNA Reads across the Hairpin Construct in Dodder.
The read counts (peaks) have been overlayed on top of a graphic of the hairpin construct that has the SUC2 promoter and STM and Knat1-3b in forward and reverse orientation surrounding a maize waxy intron and the Nos terminator. The inset shows the starting, ending, and size of the various regions in the construct. Note that some reads are detected in the polylinker region as well as the intron.
It has been reported that there is a 1000-fold reduction in small RNA amounts when silenced shoot tissue is compared with the grafted root tissue (Molnar et al., 2010). Our result shows only ∼10-fold reduction in siRNA amounts in parasite compared with host tissue (Table 1; see Supplemental Table 1 online). While it is possible that host signal was diluted during RNA preparation as the promoter used is phloem specific but entire stem tissue was used for the small RNA analysis (though this was also the case for the dodder tissue), it is equally likely that phloem-specific expression allows for a higher proportion of siRNAs generated to gain access to the phloem stream compared with 35S, as well as better siRNA movement into the parasite, because of the direct haustorial vascular connection to this tissue.
STM Downregulation Alters Parasite Development
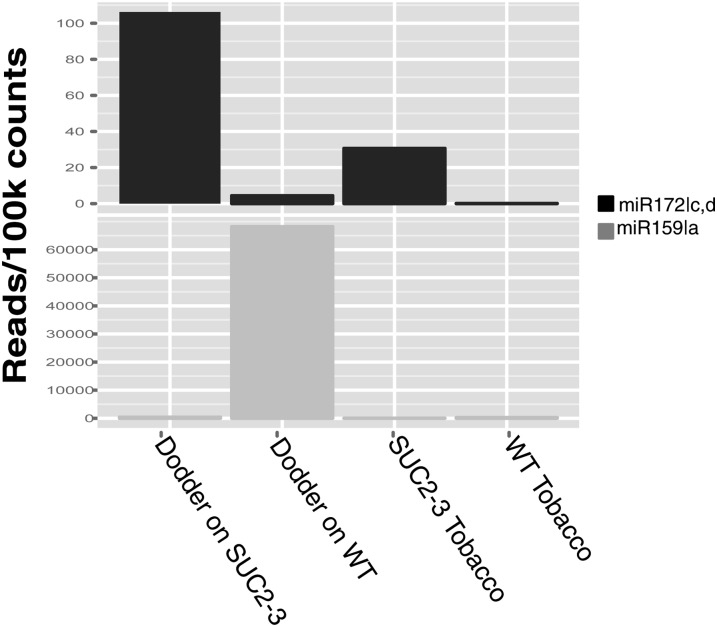
We next examined the phenotypic effect of the observed siRNA-mediated KNOX1 downregulation in dodder. Dodder grown on KNOX1 RNAi transgenics showed poor establishment and decreased growth when compared with plants grown on untransformed hosts (Figures 5A and 5B; see Supplemental Figure 12 online). This reduced parasite growth was accompanied by a significantly more rapid transition to flowering compared with dodder grown on nontransgenic hosts (Figure 5B; see Supplemental Figure 13 online). Rapid transition to flowering and loss of plant vigor are sometimes indicative of plants undergoing stress (Barnabás et al., 2008; Distelfeld et al., 2009). Despite the completion of its life cycle, dodder grown on the KNOX1 RNAi transgenics showed reduced seed set, as indicated by both seed weight and number (see Supplemental Figure 12 online). These results suggest an overall reduction in dodder vigor as a result of STM downregulation. Increase in miR172 abundance regulates expression of AP2-like genes and leads to accelerated flowering in Arabidopsis (Aukerman and Sakai, 2003). It is interesting to note there are >10-fold more miR172c,d reads in the dodder grown on transgenic tobacco compared with dodder grown on the wild type (Figure 6), possibly reflecting the flowering status of the parasite. Another small RNA molecule, miR159, which has been implicated in phase change and suppression of flowering (Achard et al., 2004), is ∼160 times more abundant on the dodder grown on the wild type (Figure 6). It will be interesting to see if the target genes of these miRNAs are downregulated at the transcriptional or translational level. Identification of these targets that relate to vigor, fitness, and floral induction in dodder will be possible once the complete dodder transcriptome has been sequenced and assembled.
Figure 5.
Dodder Grown on Transgenic Hosts Exhibits Reduced Vigor.
(A) Dodder grows vigorously on wild-type tobacco compared with SUC2 RNAi transgenic host. WT, wild type.
(B) Premature transition to flowering is seen in the dodder growing on transgenics.
[See online article for color version of this figure.]
Figure 6.
Normalized Read Counts of Small RNA Molecules.
Reads mapping to the microRNAs that regulate flowering, at-miR172c and d, and phase change, at-miR159a. WT, wild type.
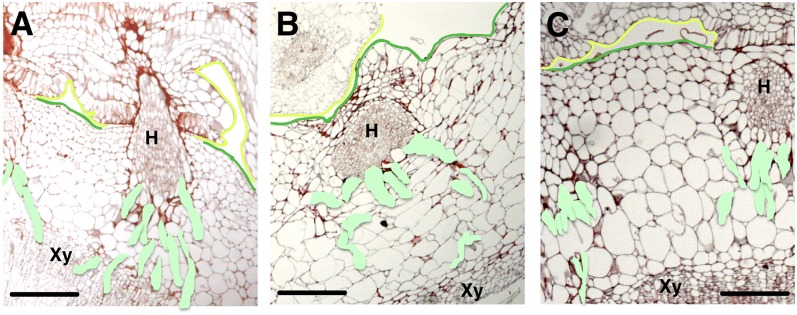
To determine whether this reduction in vigor might be associated with defects in haustorial attachment, we histologically examined mature haustoria formed on transgenic hosts (Figures 7A to 7C). Surprisingly, mature haustoria infecting transgenic hosts showed defects in vascular connection, with the searching hyphae wandering out of plane of section relative to the host vasculature rather than growing directly toward it, as seen on wild-type hosts (see Supplemental Figures 14 and 15 online). This indicates that STM expression is important for searching hyphae development. The in situ localization of STM in mature haustoria (see Supplemental Figure 5 online) is primarily seen in the central prevascular column of the haustorium. This part where STM expression is highest in haustoria is embedded in the host and not assayed quantitatively in dodder on transgenic hosts. The disruption of appropriate hyphal growth toward the vascular tissues in KNOX1 RNAi plants might suggest a role for the STM expression in haustoria in directing hyphal growth. Alternatively, STM expression might be important at earlier steps of haustorial development prior to host penetration, indirectly leading to disrupted hyphal function. Proper searching hyphal growth and development may require initiation from normal haustoria. In development, formative gene expression and morphogenetic events are usually thought to precede the occurrence of mature structures. Reduction of STM in young tissues, which show the highest expression of STM as well as the largest degree of silencing, likely leads to development of compromised prehaustoria and subsequent altered haustorial development.
Figure 7.
RNAi of STM Alters Haustorial Growth in Dodder.
Searching hyphae (colored light green) development on untransformed (A) and transgenic tobacco ([B] and [C]). The host is demarcated with a green line and the parasite with a yellow line. Host xylem is labeled Xy, and H is haustoria. Bars = 100 µm.
DISCUSSION
In this article, we show that developmental processes leading to the formation of haustoria in the plant parasite dodder involve the activation of foci of cell division in the parasite stem strand. We find that haustoria development is associated with a loss of the differentiated state of internal stem tissues and resumption of cell division activity. We further find that the expression of transcription factors, such as the Class I KNOX1 genes, which normally specify undifferentiated state in the SAM (Lincoln et al., 1994; Long et al., 1996) and cambium (Mele et al., 2003; Groover et al., 2006), are substantially upregulated during the period of haustorial development. This KNOX upregulation appears associated with prehaustorial and early haustorial development and is not seen in mature stage haustoria. Finally, we use this developmental knowledge to generate transgenic host lines that express parasite KNOX1 gene-specific small RNAs under the control of a phloem specific promoter. These plants show increased resistance to dodder growth, and the siRNAs delivered to the parasite through haustorial connections reduce STM expression and parasite fecundity.
Origin of Haustoria
The origin and development of the haustorium has previously been studied in detail in Cuscuta australis and Cuscuta japonica (Lee, 2007). The haustorium initial originates from dedifferentiation of cortical parenchyma between the vascular stele and the epidermis (Lee, 2007), on the side of the dodder strand in contact with the host, in response to mechanical, light, and hormonal cues (Haidar et al., 1998; Furuhashi et al., 2011). The cells of the epidermis in contact with the host become elongated and branched (Lee, 2007), forming an adhesive junction above a region of meristematic cells (Lee, 2007). The digitate cells undergo periclinal cell divisions, generating growth directed toward and into the host tissues. These cells ultimately become the searching hyphal cells that invade the host vasculature and undergo differentiation into Xylic or Phloic hyphae.
Previous literature interprets the dodder haustorium as a highly modified adventitious root (Truscott, 1958; Kuijt, 1976; Lee and Chai, 1989; Lee, 2007). Ultrastructural analysis of the origin of adventitious roots in cuttings of the distant relative petunia (Petunia hybrida) show their origin from cell divisions within the cambium layer itself (Ahkami et al., 2009) in a manner distinct from the dedifferentiation of cortical parenchyma in dodder. At no point in the dodder life cycle does the plant possess a normal root. The dodder root is a transient and determinate structure, and in Cuscuta gronovii, it has been shown that the embryonic root never develops a root apical meristem and the structure dies back completely within a week of germination (Truscott, 1966). STM has not been shown to be involved in root development in any yet studied species, and the ectopic expression of this gene is typically associated with ectopic development of shoot meristems or with the acquisition of indeterminacy. Furthermore, loss of function of STM in Arabidopsis does not lead to abnormalities in the root (Barton and Poethig, 1993). The expression of STM and the dependence on cytokinin for haustorium development is suggestive of a shoot like origin for this organ in dodder. However, the anatomy of the meristematic region that forms the haustorium is clearly distinct from typical SAM architecture, as is the histology of the haustorium itself. Taken together, these observations suggest a mixed developmental origin using elements of both the shoot and root developmental programs.
KNOX1 Genes Fuel Developmental Innovation in Plants
Examples abound of modifications to STM-like KNOX1 gene expression in the evolutionary origin of developmental novelty. The role these genes play in the maintenance and establishment of indeterminate cell fate makes them targets of opportunity for the evolution of traits that require renewal or preservation of pluripotency. It has been shown that ectopic expression of KNOX1 genes can lead to dedifferentiation of cells and establishment of meristematic tissues, resulting in SAM formation directly on leaves (Sinha et al., 1993; Chuck et al., 1996; Nishimura et al., 2000). Expression of KNOX1 genes outside of the canonical domains of expression has been implicated in the evolution of novel modes of growth in numerous species. In Streptocarpus rexii, embryos fail to develop a SAM and one cotyledon grows continuously for the lifespan of the plant. In S. rexii postgermination, STM expression precedes the formation of a region of meristematic activity along the petiole and midvein of the dominant cotyledon where the inflorescence shoots will develop (Harrison et al., 2005). The gymnosperm Welwitschia mirabilis has a determinate SAM and spends the majority of its lifespan, which can last several hundred years, with just two continuously growing leaves. Its embryonic SAM aborts after initiating two vegetative leaves, which grow outward from KNOX1 expressing intercalary meristems at the base of the leaves (Pham and Sinha, 2003). KNOX1 gene expression has been shown to precede the formation of de novo meristems in the roots of the Podostemaceae (Katayama et al., 2010), aquatic angiosperms in which vegetative growth is produced entirely by indeterminate root systems and root-derived determinate shoot systems. The association of KNOX1 expression with the restoration of indeterminacy in haustorial tissues and establishment of de novo meristems in dodder add to the growing list of examples where KNOX1 genes play a role in the evolution of novel developmental modes that contribute to adaptation to new ecological niches (Garcês et al., 2007; Abraham-Juárez et al., 2010; Katayama et al., 2010). It is possible that acquisition of ectopic indeterminate identity is a common precursor to development of such evolutionarily novel structures, explaining the common involvement of KNOX1 pathways. It remains to be seen if haustorial development in other parasitic plants requires tissue reprogramming through KNOX1 expression. Identification of such common developmental pathways may provide insight into control avenues using transgenic and other strategies for some of the most devastating crop parasites.
The SUC2 Promoter Is Effective in Delivering RNAi Molecules from Host to Parasite
Precision in transgene expression is one key factor that determines success in generation of the right product. Tissue-specific gene promoters can target transgene expression to where the effect is desired and prevent undesirable affects in non-target tissues. The siRNAs that mediate RNA silencing have been shown to traffic in the phloem and effect silencing away from the site of transgene expression. The Arabidopsis SUC2 promoter expresses specifically in phloem companion cells and maintains vasculature-specific expression when used to drive reporter genes in a variety of species, including tobacco (Truernit and Sauer, 1995; Roberts et al., 2001; Zhao et al., 2004). In Arabidopsis, the SUC2 promoter has been used to drive an RNAi construct silencing the Arabidopsis homolog of the tobacco SULFUR (SUL) mRNA (Dunoyer et al., 2010). SUL is a nuclear-encoded magnesium chelatase protein expressed in the chloroplast and is essential for chlorophyll biosynthesis; loss of SUL function in tobacco causes chlorosis (Fitzmaurice et al., 1999). SUC2-driven RNAi of Arabidopsis SUL causes chlorotic regions in the leaf extending 10 to 15 cells out from the vasculature (Dunoyer et al., 2010). Since dodder has been shown to establish a vascular connection with its host upon parasitism and the proximity of incipient haustoria to the vascular cylinder, we used the SUC2 promoter to drive expression of a transgene for interspecific silencing of dodder KNOX1 genes. Successful delivery of siRNAs into the parasite and downregulation of one of the two targeted genes was achieved. In addition to the specific expression domain of the SUC2 promoter, in Arabidopsis and tobacco, SUC2 expression is strongest in plant tissues that act as source for sugar transport (Wright et al., 2003). This promoter has been used to show vascular continuity between roots of transgenic tomato (Solanum lycopersicum) expressing Arabidopsis SUC2:GFP (for green fluorescent protein) and Phelipanche aegyptiaca and transfer of soluble GFP, though not an endoplasmic reticulum–localized GFP, into the parasite vasculature and accumulation in shoots (Aly et al., 2011). However, it is possible that strongly expressing root-specific promoters may be even more useful than SUC2 in trans-species RNAi where the parasite targets root tissue.
The Usefulness of RNAi as a Cross-Species Parasitic Plant Control Platform
Some plant parasites are foliar and stem specialists, and others are adapted for root parasitism; within these categories, both obligate and facultative parasitism are found. Overall, parasitic species are found in at least 22 angiosperm families (Nickrent et al., 1998) and are distributed in a wide range of habitats. Most parasitic plants establish a vascular connection with their hosts, and this opens up strategies that can be used for their control. An important advantage of RNAi as a tool for crop improvement is that RNAi signals are non-cell-autonomous and can act at both local (cell–cell) and systemic levels (spread through vascular system) (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Bauchera et al., 1998). The RNA silencing signal in plants also moves across graft junctions (Palauqui et al., 1997). The gene silencing signal also transmits across many cells in worms, and the conduit for this transmission might require a membrane-associated protein (Winston et al., 2002). Furthermore, RNAi offers the advantages of being applicable to silencing of multigene family members and homologs in polyploids (Lawrence and Pikaard, 2003). It has been demonstrated that RNAs freely translocate between parasitic plants and their hosts (Roney et al., 2007; David-Schwartz et al., 2008). Thus, expression of RNAs that debilitate parasite development upon uptake, or RNAi in the host of target gene sequences specific to the parasite, are both strategies that are being explored for parasite control (Tomilov et al., 2007). Published results showed effective β-glucuronidase silencing in the parasite via RNAi in the host roots (Tomilov et al., 2008). Similar results have recently been reported for control of the root parasite Orobanche in transgenic tomato that expresses RNAi for the parasite mannose 6-phosphate reductase in the host (Aly et al., 2009).
However, these are strategies that require the establishment of an initial vascular connection between the host and the parasite through which the silencing signal can move at significant levels. Thus, while these strategies may somewhat debilitate the parasite, and permit some movement of macromolecules through plasmodesmatal connections in the ground tissue, they cannot offer complete control because an established vascular host parasite junction is required for any large-scale transfer of macromolecules. This suggests that parasite establishment and early development should be more normal, and continued transfer of silencing signals should lead to progressive debilitation of the parasite. In our study, we found that at terminal harvest, there were significant differences in overall biomass and seed production in dodder grown on transgenic hosts compared with wild-type hosts. For complete control, strategies that inhibit connection between the host and the parasite would have to be explored and will require a better understanding of the biology of host–parasite interactions.
Some intriguing host–parasitic plant interactions described include leaf shape mimicry in several Australian host–parasite species pairs (Barlow and Wiens, 1977). One of the factors in these interactions could possibly be the transfer of morphogenetic signals, including RNA and protein molecules, from host to parasite. Many instances of transfer of macromolecules from host to parasite have been described. These have suggested interesting avenues for control of parasite growth, such as the one described in our study. Small RNAs can be delivered to plant parasites from transgenic hosts, and this delivery can induce parasite specific phenotypic alterations. Our chosen target gene, upon downregulation in the parasite using a very specific promoter in the host, caused reduced biomass and seed production in the parasite, disrupted normal haustorial growth, and made the parasitized transgenic hosts more vigorous compared with their nontransformed parasitized siblings. This study opens the door for identifying more targets that can be used to develop parasitic plant–resistant lines using transgenic technologies.
METHODS
Plant Materials
Cloning of Dodder KNOX1 Genes and Plasmid Construction
Total RNA extraction was performed with the Qiagen RNeasy kit according to the manufacturer’s protocol. cDNA was synthesized from 250 ng total RNA using Superscript III (Invitrogen) after RQ DNase treatment (Promega) using random hexamers. Degenerate PCR was used to clone dodder (Cuscuta pentagona) KNOX genes using dodder cDNA as template and consensus primer pairs (see Supplemental Table 4 online, oligonucleotides 1 to 5). More sequence information was required from the unconserved 3′ UTR of dodder KNOX genes for RNAi construct design. The 3′ rapid amplification of cDNA ends (RACE)-ready cDNA was used in conjunction with the KNOX gene–specific sense primers (see Supplemental Table 4 online, oligonucleotides 6 and 7) in a 3′RACE-PCR reaction according to the Clontech SMART RACE kit protocol and recommended cycling conditions (Clontech Laboratories). Following agarose gel electrophoresis, a PCR product (∼0.3 kb) was gel purified, subcloned, and sequenced. Regions of overlapping and identical sequences among the original degenerate PCR product and the 3′RACE products were joined using Contig Express software of VectorNTI (Invitrogen) to produce a contiguous sequence representing the longer length cDNA. New gene-specific primers (see Supplemental Table 4 online, oligonucleotides 7 to 11) were designed at the extreme 5′ and 3′ ends of the now longer fragment and subsequently PCR amplified using a high-fidelity Pfx proofreading polymerase (Invitrogen) and sequenced. Cloned sequences from dodder KNOX genes were aligned with the orthologous genes in tobacco (Nicotiana tabacum) using the VectorNTI Align X program. Unique sequences from the 3′ UTR KNOX gene fragments were identified (STMi and KNATi), amplified using PCR, and used for RNAi constructs. Overlap PCR was used to join sequences from both STMi and KNATi. Two PCRs were performed to generate linear fragments for the unique KNOX genes sequences. In the first PCR, ∼300-bp fragments were generated for STMi and KNATi gene fragments (see Supplemental Table 4 online, oligonucleotides 12 to 15). In this primary gene-specific PCR amplification reaction, defined overlapping regions were added to one of the STMi primers (STMi 3′ UTR Reverse). The products of these two PCRs were mixed together and used as a template for the second PCR reaction. In the second PCR, the overlap extension PCR, the product of the first PCR annealed to the DNA fragments and the two fragments were joined together by a pair of external primers (i.e., the 5′ end primer for STMi Forward and the 3′ end primer for KNAT1i Reverse). This resulted in a 600-bp fragment used for RNAi. The 600-bp fragment corresponding to the 3′ UTR of dodder KNOX genes was cloned into pCAMBIA 1300 to develop a double-stranded RNA vector containing a maize (Zea mays) WAXY intron separating the sense strand and the antisense strand. The DNA sequence of the phloem-specific Arabidopsis thaliana SUC2 gene promoter (Imlau et al., 1999) was retrieved from GenBank (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) under accession number X79702 and amplified from Arabidopsis genomic DNA. The identity of the amplified fragment was confirmed by sequencing, and it was cloned upstream of the KNOXi inserts to create pSUC:KNOXi construct.
Phylogenetic Analysis
To confirm the identity of the KNOX genes used in the construct, a phylogenetic tree was constructed using the Class I KNOX genes from tomato (Solanum lycopersicum), Arabidopsis, and dodder (see Supplemental Figure 3 online). To obtain sufficient sequence for phylogenetic analysis, tBLASTn was used to identify KNOXI sequences from a a draft dodder transcriptome assembly (A. Ranjan, R. Kumar, Y. Ichihashi, and N. Sinha, unpublished data) using the tomato and Arabidopsis KNOXI sequences as well as the cloned dodder KNOXI fragments as query. The genes were entered into GenBank and designated Knat1-1, Knat1-3b, Knat1-3c, Knat1-3a, STM, Liguleless, and GAPDH (see accession numbers below). The tree was generated using neighbor joining (Jukes-Cantor) using 597 unambiguously aligned nucleotides or for the incomplete STM sequence 473 unambiguously aligned nucleotides run for 10,000 replications and displayed as a bootstrap consensus. The sequence for Knat1-2 consists of only 141 bp and is insufficient for entry into GenBank or for phylogenetic analysis. The sequences designated Knat1-3a, Knat1-3b, and Knat1-3c represent contigs with nearly identical sequence, which could be alleles of the same Knat1-3 paralog; thus, we refer to this group of sequences as Knat1-3. The Knat1 fragment used in the RNAi construct corresponds to Knat1-3b. Because it might prove impractical to detect individually Knat1-3a, b, and c and, due to the high sequence similarity, the silencing would not be restricted to Knat1-3b.
Tobacco Transformation
Wild-type N. tabacum cv SR1 was transformed by Agrobacterium tumefaciens strain EHA105 carrying the RNAi constructs at the University of California Davis plant transformation facility. Plants were selected with hygromycin until the T2 generation where lines with 100% survival on Murashige and Skoog medium containing 300 μg/mL hygromycin and positively confirmed via PCR were assumed to be homozygous and used for further experiments. Ten tobacco lines were selected for a general screen, after which two SUC2 lines, SUC2-3 and 7, were selected for further experiments. In all our experiments, tobacco plants were infected with dodder at 4 weeks after planting.
Dodder Germination, Cultivation, and Synchronized Infection
Dodder seeds were rubbed with sandpaper before being dipped in concentrated sulfuric acid for 1 h, cleaned with water, and then treated for 20 min with bleach. Seeds were rinsed four times for 2 min each with distilled water followed by plating on moist filter paper in Petri dishes. After 7 d, germinated dodder seedlings were then placed next to 8-week-old Medicago sativa seedlings in 30-cm pots and left to establish for 6 weeks. M. sativa displays higher resistance to dodder and therefore provides more twines for use in synchronized infection. Bamboo sticks were erected in the M. sativa pots, and ∼10 cm of the auxiliary dodder strands coiled around them. The parasite developed prehaustoria within 72 h and was unwound from the stick and coiled around stems of 4-week-old transgenic and nontransgenic tobacco plants before being left to establish for 4 weeks.
Histology and in Situ Hybridization
Four weeks after infection, dodder growth behavior on tobacco was examined. Photographs showing the various phenotypes were taken using a digital Nikon camera. The connection points of dodder on tobacco plants were excised using a scalpel and fixed in 70% formaldehyde–acetic acid–alcohol, dehydrated through an ethanol series, and embedded in paraffin wax (Paraplast; McCormick Scientific). Blocks were sectioned at 10 µm on a Microme HM340E microtome and mounted on slides (Probe-On Plus; Thermo Fischer). Slides were deparaffinized in Histoclear (National Diagnostics), hydrated through an ethanol series, stained with 0.05% Ruthenium red or 0.05% Toluidine Blue O, and mounted in Permount (Fisher Scientific) (Kim et al., 2001; Bharathan et al., 2002). Bright-field images were taken on a Nikon Digital Sight DS SME E600 using the spot RT camera (Diagnostic Instruments) and images processed in Adobe Photoshop CS3 (Adobe Systems). In situ hybridization was performed as described (Long et al., 1996) with several modifications (Garcês et al., 2007). Aniline blue staining was performed on fresh sections using a solution of aniline blue and observed under UV florescence (Hulskamp et al., 1995).
Quantitative Real-Time PCR
To quantify the level of STM and KNAT1 like genes in different dodder tissues, dodder was attached to either transgenic or wild-type plants, and several dodder tissues were harvested from at least two different plants of each line for independent biological replicate pools. Tissue harvested included haustorial attachment tissue and 15 cm after the haustorial attachment tissue. All tissues were frozen in liquid nitrogen and stored at −80°C for further analyses. Tissues (20 to 100 mg) were later ground using a bead beater (Mini Beadbeater 96; BioSpec Products) in extraction buffer, and total RNA was extracted using the Qiagen RNeasy plant mini kit (Qiagen) with on-column DNase treatment. For some of the samples, mRNA was isolated directly from homogenized tissue using the Dynabeads mRNA direct kit (Invitrogen) using the manufacturer’s recommendations with a DNase (NEB) treatment prior to the second optional purification. All of the mRNA or 1 μg of total RNA was used to prepare single-stranded cDNA using Superscript III reverse transcriptase (Invitrogen) as described by the manufacturer using random primer mix (Invitrogen). The synthesized cDNA was diluted 10 times and the reactions conducted using an iCycler iQ real-time thermal cycler (Bio-Rad Laboratories) and IQ SYBR Green super mix (Bio-Rad Laboratories). Each experiment had four technical replicates per sample. All primers were designed using Beacon Designer version 3.0 (Bio-Rad). Specific primers for dodder STM and the three KNAT1-like genes were used and normalized to Cp-18S (see Supplemental Table 4 online, oligonucleotides 16 to 21).
The analyses were performed using GENEX and data presented as means of technical replicates. The sequence for GAPDH was identified from a local BLAST against a draft dodder transcriptome assembly.
Small RNA Library Preparation and Data Analysis
Small RNA libraries were prepared from dodder filaments 2 to 15 cm from haustorial attachment point (from dodder grown on wild-type and transgenic tobacco), and the wild-type and transgenic tobacco leaves and shoot apices were used for small RNA library preparation. Approximately 100 mg of tissue was frozen in liquid nitrogen. The tissue was homogenized in a bead beater (Mini Beadbeater 96; BioSpec Products) in extraction buffer containing 1 mL of Trizol (Invitrogen) and 100 μL of Plant RNA isolation aid (Applied Biosystems/Ambion). Total RNA was then extracted with the Trizol reagent RNA extraction protocol based on the manufacturer’s recommendations (Invitrogen). One microgram of total RNA was used as input for small RNA library construction for sequencing using the digital gene expression for small RNA sample prep kit based on Illumina small RNA sample preparation v1.5 protocol. The 5′ RNA adapter was modified and synthesized as follows to facilitate barcode-based 4× multiplexing (see Supplemental Table 4 online, oligonucleotides 22 to 25). Samples were purified and sent to the University of California Davis genome center expression analysis core for sequencing using Illumina’s GAII sequencing system.
The sequence file generated in the fastq format was then sorted based on the four barcodes into the respective samples using a Perl script. For mapping, only the reads that had the first 12 bases of the 3′ adapter sequence (5′- ATCTCGTATGCCGTCTTCTGCTTGT-3′) and the ones that were within the range of 18 to 26 bp (after trimming the 3′ adapter and the barcode) were included. The 3′ adapter detection and trimming was done using the FASTQ clipper from the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/commandline.html). The reads were then aligned to the reference file that included the RNAi construct, the partial clones of the STM and KNAT1-like genes, and the Arabidopsis microRNAs and trans-acting small interfering RNA downloaded from the Arabidopsis small RNA project database (Gustafson et al., 2005; Backman et al., 2008). Alignment of the reads was performed using bowtie (Langmead et al., 2009) using the following parameters: -a–best–strata -v 0 -l 15 -p 10 –sam. The raw counts were then extracted from the aligned files using a Perl script and analyzed in Microsoft Excel or in R (R Development Core Team, 2011). The graphs were plotted either using Microsoft Excel or the ggplot2 package in R (Wickham, 2009). The statistical tests of analysis of variance and t test were done using the R core package (R Development Core Team, 2011), and the BH method (Benjamini and Hochberg, 1995) was used for multiple testing correction of the t test P values. All of the bioinformatics and statistical analyses were done either on our local servers or on the iPLANT Atmosphere cloud server (Goff et al., 2011).
Effect of STM Silencing on Dodder Vigor
The measures of dodder vigor determined were total dodder mass, total dodder seed weight, total dodder seed numbers, and dodder seed germination percentage. All the dodder mass on three tobacco plants per treatment were stripped off and separately dried in paper bags at 37°C for 14 d before weighing on a balance; these readings were averaged and means separated with se. To determine the total seed weight, all the dry dodder was separately crushed and seed separated from general dodder mass tissue by sieving. Total seed per plants of every treatment averaged and means separated as earlier described. Dodder seed yield per plant was estimated in three replicates per treatment. The weight of 100 seeds from each replicate was used to estimate total seed number.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Knat1-1 (JQ799050), Knat1-3b (JQ799051), Knat1-3c (JQ799052), Knat1-3a (JQ799053), STM (JQ799054), Liguleless (JQ799055), and GAPDH (JQ799056).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Searching Hyphae in Dodder.
Supplemental Figure 2. Adventitious Shoots Adjacent to Haustoria.
Supplemental Figure 3. Phylogenetic Tree Showing Relationships of Cuscuta pentagona Class I KNOX Genes to Representative Sequences from Ipomea, Tomato, Tobacco, and Arabidopsis.
Supplemental Figure 4. Dodder Haustorial Development.
Supplemental Figure 5. STM Expression the Dodder Shoot Apex.
Supplemental Figure 6. Alignment of STM Genes.
Supplemental Figure 7. Alignment of KNAT1 Genes.
Supplemental Figure 8. Graphic Representation of sRNA Movement from the Tobacco Host into Dodder via Haustorial Connections.
Supplemental Figure 9. KNAT1-3 Read Distribution.
Supplemental Figure 10. Percent Read Distribution.
Supplemental Figure 11. View of the Alignment of Reads across the Specific Regions of the Hairpin Template.
Supplemental Figure 12. Reduced Dodder Vigor on KNOX RNAi Transgenics.
Supplemental Figure 13. Early Flowering in Dodder Grown on SUC2 Transgenic Plants.
Supplemental Figure 14. Dodder Haustorial Growth on Tobacco Stem.
Supplemental Table 1. Host-Derived Small RNAs in C. pentagona.
Supplemental Table 2. Mapping of Dodder and Tobacco Small RNA Reads.
Supplemental Table 3. Mapping of Dodder and Tobacco Small RNA Reads.
Supplemental Table 4. Oligonucleotide Sequences.
Supplemental Data Set 1. KNOXI nucleotide alignment tpc099994SD.pir.
Supplementary Material
Acknowledgments
We thank Deborah Delmer for encouragement, members of the Sinha Laboratory for helpful discussions, Lauren Headland for help with plant growth and seed collection, Dan Chitwood for critical comments on the article, and Aashish Ranjan and Yasunori Ichihashi for kindly providing dodder sequences for our work. J.K. was funded by a Katherine Esau postdoctoral fellowship, S.K. by a Japan Society for the Promotion of Science postdoctoral fellowship, and R.D.-S. by a U.S.–Israel Binational Agricultural Research and Development Fund postdoctoral fellowship. This work was supported by a Rockefeller Foundation award (N.S. and J.M.) and National Science Foundation award 0820854 (N.S.).
AUTHOR CONTRIBUTIONS:
A.A., R.K., D.K., S.K., B.T., S.R., H.M.G, J.K., R.D.-S., and N.S. designed the research. A.A., R.K., D.K., S.K., B.T., S.R., H.M.G, J.K., A.Y., and N.S. performed the research. R.K. and D.K. contributed to new analytic and computational tools. A.A., R.K., D.K., S.K., B.T., S.R., H.M.G, J.K., R.D.-S, J.M., and N.S. wrote the article.
Glossary
- RNAi
RNA interference
- siRNA
small interfering RNA
- SAM
shoot apical meristem
- UTR
untranslated regions
- RACE
rapid amplification of cDNA ends
References
- Abraham-Juárez M.J., Martínez-Hernández A., Leyva-González M.A., Herrera-Estrella L., Simpson J. (2010). Class I KNOX genes are associated with organogenesis during bulbil formation in Agave tequilana. J. Exp. Bot. 61: 4055–4067 [DOI] [PubMed] [Google Scholar]
- Achard P., Herr A., Baulcombe D.C., Harberd N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Ahkami A.H., Lischewski S., Haensch K.T., Porfirova S., Hofmann J., Rolletschek H., Melzer M., Franken P., Hause B., Druege U., Hajirezaei M.R. (2009). Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 181: 613–625 [DOI] [PubMed] [Google Scholar]
- Aly R., Cholakh H., Joel D.M., Leibman D., Steinitz B., Zelcer A., Naglis A., Yarden O., Gal-On A. (2009). Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol. J. 7: 487–498 [DOI] [PubMed] [Google Scholar]
- Aly R., Hamamouch N., Abu-Nassar J., Wolf S., Joel D.M., Eizenberg H., Kaisler E., Cramer C., Gal-On A., Westwood J.H. (2011). Movement of protein and macromolecules between host plants and the parasitic weed Phelipanche aegyptiaca Pers. Plant Cell Rep. 30: 2233–2241 [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman T.W.H., Sullivan C.M., Cumbie J.S., Miller Z.A., Chapman E.J., Fahlgren N., Givan S.A., Carrington J.C., Kasschau K.D. (2008). Update of ASRP: The Arabidopsis Small RNA Project database. Nucleic Acids Res. 36(Database issue): D982–D985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow B.A., Wiens D. (1977). Host-parasite resemblance in Australian mistletoes: The case for cryptic mimicry. Evolution 31: 69–84 [DOI] [PubMed] [Google Scholar]
- Barnabás B., Jäger K., Fehér A. (2008). The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 31: 11–38 [DOI] [PubMed] [Google Scholar]
- Barton M.K., Poethig R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana - An analysis of development in the wild-type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Bauchera M., Montiesb B., van Montagu A.M., Boerjana W. (1998). Biosynthesis and genetic engineering of lignin. Crit. Rev. Plant Sci. 17: 125–197 [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Me. 57: 289–300 [Google Scholar]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Birschwilks M., Haupt S., Hofius D., Neumann S. (2006). Transfer of phloem-mobile substances from the host plants to the holoparasite Cuscuta sp. J. Exp. Bot. 57: 911–921 [DOI] [PubMed] [Google Scholar]
- Blevins T., Rajeswaran R., Shivaprasad P.V., Beknazariants D., Si-Ammour A., Park H.S., Vazquez F., Robertson D., Meins F., Jr, Hohn T., Pooggin M.M. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34: 6233–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22: 268–280 [DOI] [PubMed] [Google Scholar]
- Brosnan C.A., Mitter N., Christie M., Smith N.A., Waterhouse P.M., Carroll B.J. (2007). Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 14741–14746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Lincoln C., Hake S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Meeley R.B., Hake S. (1998). The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12: 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Schwartz R., Runo S., Townsley B., Machuka J., Sinha N. (2008). Long-distance transport of mRNA via parenchyma cells and phloem across the host–parasite junction in Cuscuta. New Phytol. 179: 1133–1141 [DOI] [PubMed]
- Dawson J.H., Musselman L.J., Wolswinkel P., Dorr I. (1994). Biology and control of Cuscuta. Reviews of Weed Science 6: 265–317 [Google Scholar]
- Distelfeld A., Li C., Dubcovsky J. (2009). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J.C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916 [DOI] [PubMed] [Google Scholar]
- Ejeta G. (2007). Breeding for Striga resistance in sorghum: Exploitation of an intricate host–parasite biology. Crop Sci. 47: S216–S227 [Google Scholar]
- Ejeta G., Mohammed A., Rich P., Melake-Berhan A., Housley T.L., Hess D.E. (2000). Selection for specific mechanisms of resistance to Striga in sorghum. In Breeding for Striga Resistance in Cereals, B.I.G. Haussmann, ed (Weikersheim, Germany: Margraf), pp 29–37
- Fitzmaurice W.P., Nguyen L.V., Wernsman E.A., Thompson W.F., Conkling M.A. (1999). Transposon tagging of the sulfur gene of tobacco using engineered maize Ac/Ds elements. Genetics 153: 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi T., Furuhashi K., Weckwerth W. (2011). The parasitic mechanism of the holostemparasitic plant Cuscuta. J. Plant Interact. 6: 207–219 [Google Scholar]
- Fusaro A.F., Matthew L., Smith N.A., Curtin S.J., Dedic-Hagan J., Ellacott G.A., Watson J.M., Wang M.B., Brosnan C., Carroll B.J., Waterhouse P.M. (2006). RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner E.E. (1950). Studies of Seed Germination, Seed Identification, and Host Relationships in Dodders, Cuscuta spp. (Ithaca, NY: Cornell University Agricultural Experiment Station)
- Garcês H.M.P., Champagne C.E.M., Townsley B.T., Park S., Malhó R., Pedroso M.C., Harada J.J., Sinha N.R. (2007). Evolution of asexual reproduction in leaves of the genus Kalanchoë. Proc. Natl. Acad. Sci. USA 104: 15578–15583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., et al. (2011). The iPlant Collaborative: Cyberinfrastructure for plant biology. Front. Plant Sci. 2: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier C., Rich P.J., Mohamed A., Ellicott A., Shaner C., Ejeta G. (2001). Independent inheritance of LGS and IR genes in sorghum. In 7th International Parasitic Weed Symposium, A. Fer, P. Thalouarn, D.M. Joel, L.J. Musselman, C. Parker, and J.A.C. Verkleij, eds (Nantes, France), pp. 220–223
- Groover A.T., Mansfield S.D., DiFazio S.P., Dupper G., Fontana J.R., Millar R., Wang Y. (2006). The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 61: 917–932 [DOI] [PubMed] [Google Scholar]
- Gurney A.L., Slate J., Press M.C., Scholes J.D. (2006). A novel form of resistance in rice to the angiosperm parasite Striga hermonthica. New Phytol. 169: 199–208 [DOI] [PubMed] [Google Scholar]
- Gustafson A.M., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D. (2005). ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 33(Database issue): D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar M.A., Orr G.L., Westra P. (1998). The response of dodder (Cuscuta spp.) seedlings to phytohormones under various light regimes. Ann. Appl. Biol. 132: 331–338 [Google Scholar]
- Hake S., Smith H.M., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Harrison J., Möller M., Langdale J., Cronk Q., Hudson A. (2005). The role of KNOX genes in the evolution of morphological novelty in Streptocarpus. Plant Cell 17: 430–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautier Y., Hector A., Vojtech E., Purves D., Turnbull L.A. (2010). Modelling the growth of parasitic plants. J. Ecol. 98: 857–866 [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hulskamp M., Schneitz K., Pruitt R.E. (1995). Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A., Truernit E., Sauer N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jeschke W.D., Räth N., Bäumel P., Czygan F.-C., Proksch P. (1994). Modelling the flow and partitioning of carbon and nitrogen in the holoparasite Cuscuta reflexa Roxb. and its host Lupinus albus L.: I. Methods for estimating net flows. J. Exp. Bot. 45: 791–800 [Google Scholar]
- Katayama N., Koi S., Kato M. (2010). Expression of SHOOT MERISTEMLESS, WUSCHEL, and ASYMMETRIC LEAVES1 homologs in the shoots of Podostemaceae: Implications for the evolution of novel shoot organogenesis. Plant Cell 22: 2131–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R.A., Laudencia-Chingcuanco D., Smith L.G., Hake S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Kim M., Canio W., Kessler S., Sinha N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293: 287–289 [DOI] [PubMed] [Google Scholar]
- Kuijt J. (1969). The Biology of Parasitic Flowering Plants. (Berkeley, CA: University of California Press) [Google Scholar]
- Kuijt J. (1983). Tissue Compatibility and the Haustoria of Parasitic Angiosperms. (Waco, TX: Baylor University) [Google Scholar]
- Kuijt J., Toth R. (1976). Ultrastructure of angiosperm haustoria—A review. Ann. Bot. (Lond.) 40: 1121–1130 [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R.J., Pikaard C.S. (2003). Transgene-induced RNA interference: A strategy for overcoming gene redundancy in polyploids to generate loss-of-function mutations. Plant J. 36: 114–121 [Google Scholar]
- Lee K.B. (2007). Structure and development of the upper haustorium in the parasitic flowering plant Cuscuta japonica (Convolvulaceae). Am. J. Bot. 94: 737–745 [DOI] [PubMed] [Google Scholar]
- Lee K.B., Chai D.L. (1989). The structure and development of the haustorium in Cuscuta australis. Can. J. Bot. 67: 2975–2982 [Google Scholar]
- Lincoln C., Long J., Yamaguchi J., Serikawa K., Hake S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Malik C.P., Singh M.B. (1979). Physiological and biochemical aspects of parasitism in Cuscuta: A review. Annu. Rev. Plant Sci. 1979: 67–112 [Google Scholar]
- McGinnis K., et al. (2007). Assessing the efficiency of RNA interference for maize functional genomics. Plant Physiol. 143: 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele G., Ori N., Sato Y., Hake S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk C.W., Molnar A., Baulcombe D.C. (2011). Intercellular and systemic movement of RNA silencing signals. EMBO J. 30: 3553–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra J. (2009). Biology and management of Cuscuta species. Indian J. Weed Sci. 41: 1–11 [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Mower J.P., Stefanović S., Young G.J., Palmer J.D. (2004). Plant genetics: Gene transfer from parasitic to host plants. Nature 432: 165–166 [DOI] [PubMed] [Google Scholar]
- Nadler-Hassar T., Shaner D.L., Nissen S., Westra P., Rubin B. (2009). Are herbicide-resistant crops the answer to controlling Cuscuta? Pest Manag. Sci. 65: 811–816 [DOI] [PubMed] [Google Scholar]
- Nickrent D.L., Duff R.J., Colwell A.E., Wolfe A.D., Young N., Steiner K., DePamphilis C. (1998). Molecular phylogenetic and evolutionary studies of parasitic plants. In Molecular Systematics of Plants II: DNA sequencing, D. Soltis, P. Soltis, and J.J. Doyle, eds (Heidelberg, Germany: Springer), pp. 211–241
- Nishimura A., Tamaoki M., Sakamoto T., Matsuoka M. (2000). Over-expression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant Cell Physiol. 41: 583–590 [DOI] [PubMed] [Google Scholar]
- Palauqui J.C., Elmayan T., Pollien J.M., Vaucheret H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16: 4738–4745 [DOI] [PMC free article] [PubMed]
- Pennings S., Simpson J. (2008). Like herbivores, parasitic plants are limited by host nitrogen content. Plant Ecol. 196: 245–250 [Google Scholar]
- Pham T., Sinha N. (2003). Role of KNOX genes in shoot development of Welwitshchia mirabilis. Int. J. Plant Sci. 164: 333–343 [Google Scholar]
- R Development Core Team (2011). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing) [Google Scholar]
- Rispail N., Dita M.A., González-Verdejo C., Pérez-de-Luque A., Castillejo M.A., Prats E., Román B., Jorrín J., Rubiales D. (2007). Plant resistance to parasitic plants: molecular approaches to an old foe. New Phytol. 173: 703–712 [DOI] [PubMed] [Google Scholar]
- Roberts I.M., Boevink P., Roberts A.G., Sauer N., Reichel C., Oparka K.J. (2001). Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218: 31–44 [DOI] [PubMed] [Google Scholar]
- Roney J.K., Khatibi P.A., Westwood J.H. (2007). Cross-species translocation of mRNA from host plants into the parasitic plant dodder. Plant Physiol. 143: 1037–1043 [Google Scholar]
- Runo S., Alakonya A., Machuka J., Sinha N. (2011). RNA interference as a resistance mechanism against crop parasites in Africa: A ‘Trojan horse’ approach. Pest Manag. Sci. 67: 129–136 [DOI] [PubMed] [Google Scholar]
- Sandler H.A. (2010). Managing Cuscuta gronovii (swamp dodder) in cranberry requires an integrated approach. Sustainability 2: 660–683 [Google Scholar]
- Sinha N.R., Williams R.E., Hake S. (1993). Overexpression of the maize homeobox gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7: 787–795 [DOI] [PubMed] [Google Scholar]
- Tomilov A.A., Tomilova N.B., Wroblewski T., Michelmore R., Yoder J.I. (2008). Trans-specific gene silencing between host and parasitic plants. Plant J. 56: 389–397 [DOI] [PubMed] [Google Scholar]
- Tomilov A.A., Tomilova N.B., Yoder J.I. (2007). Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta 225: 1059–1071 [DOI] [PubMed] [Google Scholar]
- Truernit E., Sauer N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Truscott F. (1966). Some aspects of morphogenesis in Cuscuta gronovii. Am. J. Bot. 53: 739–750 [Google Scholar]
- Truscott F.H. (1958). On the regeneration of new shoots from isolated dodder haustoria. Am. J. Bot. 45: 169–177 [Google Scholar]
- Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [Google Scholar]
- Vaistij F.E., Jones L., Baulcombe D.C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn K.C. (2003). Dodder hyphae invade the host: A structural and immunocytochemical characterization. Protoplasma 220: 189–200 [Google Scholar]
- Voinnet O., Baulcombe D.C. (1997). Systemic signalling in gene silencing. Nature 389: 553. [DOI] [PubMed] [Google Scholar]
- Westwood J.H., Yoder J.I., Timko M.P., dePamphilis C.W. (2010). The evolution of parasitism in plants. Trends Plant Sci. 15: 227–235 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009). Ggplot2 Elegant Graphics for Data Analysis. (New York: Springer-Verlag) [Google Scholar]
- Winston W.M., Molodowitch C., Hunter C.P. (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295: 2456–2459 [DOI] [PubMed] [Google Scholar]
- Wright K.M., Roberts A.G., Martens H.J., Sauer N., Oparka K.J. (2003). Structural and functional vein maturation in developing tobacco leaves in relation to AtSUC2 promoter activity. Plant Physiol. 131: 1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.I., Gunathilake P., Wu B., Tomilova N., Tomilov A.A. (2009). Engineering host resistance against parasitic weeds with RNA interference. Pest Manag. Sci. 65: 460–466 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Maruyama S., Nozaki H., Shirasu K. (2010). Horizontal gene transfer by the parasitic plant Striga hermonthica. Science 328: 1128. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Liu Q., Davis R.E. (2004). Transgene expression in strawberries driven by a heterologous phloem-specific promoter. Plant Cell Rep. 23: 224–230 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.