Abstract
Bacterial production of polyunsaturated fatty acids (PUFAs) is a potential biotechnological approach for production of valuable nutraceuticals. Reliable method for screening of number of strains within short period of time is great need. Here, we report a novel simplified method for screening and isolation of PUFA-producing bacteria by direct visualization using the H2O2-plate assay. The oxidative stability of PUFAs in growing bacteria towards added H2O2 is a distinguishing characteristic between the PUFAs producers (no zone of inhibition) and non-PUFAs producers (zone of inhibition) by direct visualization. The confirmation of assay results was performed by injecting fatty acid methyl esters (FAMEs) produced by selected marine bacteria to Gas Chromatography-Mass Spectrometry (GCMS). To date, this assay is the most effective, inexpensive, and specific method for bacteria producing PUFAs and shows drastically reduction in the number of samples thus saves the time, effort, and cost of screening and isolating strains of bacterial PUFAs producers.
1. Introduction
Microbial lipids are a diverse group of compounds with a number of vital nutraceutical and pharmaceutical applications and utilized commercially since the 1980s. These microbial lipids or polyunsaturated fatty acids (PUFAs) are obtained from various sources. Now a day, microorganism-produced (algae/fungi/bacteria) PUFAs are commercially competitive with plant and fish oils.
PUFAs are the fatty acids having more than one double bond. Eicosapentaenoic acid (EPA, 20 : 5, n-3) and docosahexaenoic acid (DHA, 22 : 6, n-3) are the important n-3 fatty acids, while arachidonic acid (AA, 20 : 4, n-6) is a vital n-6 fatty acid. EPA and DHA are important for prevention of arthrosclerosis, cancer, rheumatoid arthritis, psoriasis, and diseases of old age such as Alzheimer's and age-related macular degeneration [1, 2]. AA and DHA are of special importance in the brain and blood vessels and are considered essential for pre- and postnatal brain and retinal development [3]. Eicosanoids such as prostaglandins, prostacyclins, and leukotrienes derived from n-3 PUFA are also important in new-born and infant development, modulatory vascular resistance, and wound healing [4–6]. PUFAs are either directly available as components of the diet or produced from precursors like linoleic acid (LA, C18 : 2 n-6) and a-linolenic acid (ALA, C18 : 3 n-3) [7].
Accordingly, PUFAs are highly important substances in the pharmaceutical, medical, and nutritional fields. Recent investigations have focused on microorganisms as alternative natural source for production of oil containing PUFAs. These are potentially promising lipid source because of their high growth rates in simple media and simplicity of their manipulation. As traditional sources of n-3 fatty acids such as fish oil continue to diminish, identification of alternate sources will become crucial. Marine microorganisms represent one of the less explored sources of biologically active natural products. Novel compounds with various bioactivities such as antibiotic, antitumor, cytotoxic, and anti-inflammatory, have been isolated and elucidated from this source [8].
Considering importance of PUFAs, many researchers have tried to isolate and screen marine organisms for these bioactive compounds. The next step after isolation of microorganisms is screening. Ideally, selective procedure would allow the detection and isolation of microorganisms producing the desired metabolite. This primary screening should be rapid, inexpensive, predictive, specific, but effective over a broad range and should be applicable to large scale. Sometimes primary screening is time consuming and labour intensive when a large number of isolates have to be screened to identify a few potential ones. However, this is possibly the most critical step since it eliminates the large bulk of unwanted isolates, which are either nonproducers or producers of known compounds.
Screening and isolation method for long chain PUFA by marine protistan was reported but judged unsuitable by Bowles et al. for high throughput screening. The H2O2 plate assay method can be applied to a wide range of bacterial samples collected from different regions or from different animal sources. During screening of random hundreds of different marine bacterial samples, we have successfully discovered new strains of marine microorganisms with the special characteristic to produce PUFAs.
Generally, PUFAs are the molecules which are most susceptible to oxygen and reactive oxygen species (ROS) [10]. There are facts that PUFAs are stable when they are in vivo against oxidative stresses caused by ROS. This study was based on application of antioxidative effect of PUFA against ROS (Figure 1), for rapid screening of large number of marine isolates. However, no information regarding the screening of PUFAs producing marine bacteria has been reported. In the following strategy, we have presented qualitative method for rapid screening of PUFA producers.
Figure 1.
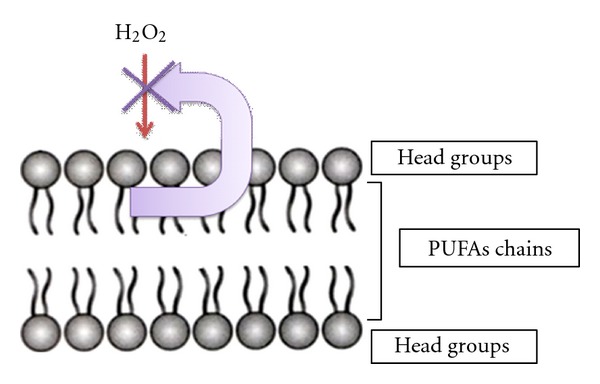
Proposed mode of action for the H2O2-plate assay method.
2. Experimental Procedures
2.1. Materials
All the chemicals and media components used in the present study were AR grade and purchased from Hi Media Ltd, Mumbai, India. Sodium azide was purchased from S.D. Fine Chemicals Limited, Mumbai, India.
2.2. Media and Culture Conditions for Marine Microorganisms
2.2.1. Sample Collection
Various samples were collected from different regions of western coast of Maharashtra, India (Figure 2) and were brought to our laboratory for further investigation.
Figure 2.
Map of western coast region of Maharashtra (INDIA).
2.2.2. Isolation of Marine Bacteria
The collected samples were serially diluted and plated over nutrient agar plates. The inoculated plates were incubated at 28 ± 2°C for 24–48 h. After incubation, the selected bacterial colonies were purified and subcultured on nutrient agar medium for further investigation. All the marine isolates were preserved in glycerol stocks under (−) 20°C.
2.3. Primary Screening of Marine Bacterial Isolates by H2O2-Plate Assay
About 100 isolated marine strains were randomly selected and screened for PUFA production (Figure 3). All the selected marine cultures were cultivated in Luria-Bertani (LB) medium (1% Tryptone, 0.5% Yeast Extract, 1% NaCl) at 28 ± 2°C for 24 h, 180 rpm. To this medium, 0.5% NaCl was added for proper growth of marine isolates rather than using artificial seawater [11]. To find out response of bacteria to H2O2, bacterial culture reaching an optical density at 660 nm of 1.0 was used and spreaded over plate containing LB medium and sodium azide (NaN3, 1 mM). On a surface filter paper discs of diameter 5 mm were placed and ten microliter aliquots of solution containing different concentration of H2O2 (0.01, 0.5, 1.0% prepared from 30% stock solution) were added on filter disc. Plates were then incubated at 28 ± 2°C for 24 h. Zone of inhibition was observed and further confirmation of PUFA producers was done by GCMS.
Figure 3.
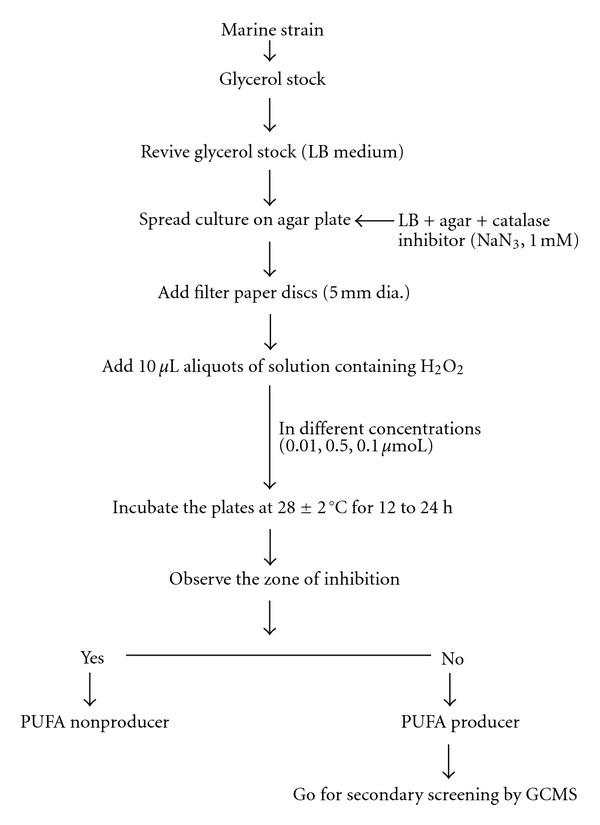
Protocol for primary screening of marine isolates.
2.4. Secondary Screening and Confirmation
2.4.1. Media and Cultivation Conditions
Amongst all screened marine bacterial cultures, the positive strains were selected on the basis of no zone of inhibition (PUFA producers) and used further for secondary screening and confirmation for the production of PUFA. A loopful of the bacterial culture grown on previously preserved LB medium slants was transferred into 20 mL of LB broth in a 100 mL Erlenmyer flask and left on a shaker at 28 ± 2°C for 24 h at 180 rpm. One mL of culture was transferred into 50 mL fresh sterilized LB broth and left on shaker at 28 ± 2°C for 24 h at 180 rpm. Cell biomass was harvested from fermented media by centrifugation at 8000 rpm at 25 ± 2°C for 10 min. The biomass was washed thoroughly with distilled water and was finally dried at 50°C overnight.
2.4.2. Direct Lipid Extraction and Esterification of Fatty Acids
A rapid direct extraction and esterification method was developed and applied in this study. Extraction of lipids from biomass was performed according to the modified procedure of Hoshi et al. [12]. Lipids were extracted for 3-4 h with a 3 times volume of chloroform/methanol (2 : 1, v/v) containing 15 mg BHT for prevention of oxidation of PUFAs [13]. Esterification was done by adding 0.2 mL of 20 mM cupric acetate monohydrate in methanol and 1 mL of 0.5 N HC1 in methanol was added, and the mixture was left for the specified time (2-3 h) at room temperature or at 28 ± 2°C. The reaction was stopped with the addition of 0.4 mL of water. The lower chloroform layer was pooled and then evaporated and was concentrated by rotary evaporator (Buchi Rotavapor) at 35°C. Finally the fatty acid methyl esters (FAME) were dissolved in hexane (1 mL), filtered through Whatman No. 1 filter paper, and analysed by GCMS.
2.4.3. Analysis PUFA by GCMS
Analysis of the FAME was performed by GCMS using slightly modified procedure [14]. All compounds were identified by comparison of their retention times with those of known standards and confirmed by GCMS using Varian 220-MS ion trap mass spectrometer (Varian, Inc. Walnut Creek, CA) connected to Varian 450-GC equipped with CP-SIL 88 capillary column (25 m × 0.25 mm i.d. × 0.39 mm OD, Varian). The injector was maintained at 250°C, and the column oven was programmed to increase from 160 to 220°C at 7°C min and then maintained at 220°C for 10 min. Split ratio was adjusted to 1 : 20. Helium was used as the carrier gas and flow rate was maintained at 1 mL/min. GCMS was operated at an ionization voltage of 70 eV and trap temperature at 220°C with mass range of 40–350 atomic mass units.
3. Results and Discussion
3.1. Primary Screening of Marine Bacterial Isolates by H2O2-Plate Assay
In H2O2-plate assay method, the cells which are susceptible to externally-added H2O2 cannot able to grow suitably and thus shows a zone of inhibition which is dependant on added concentration of H2O2 on filter paper disc. The diameter of zone of inhibition is directly proportional to the concentration of added H2O2. The contradictory situation was observed for bacterial cells which produce PUFA. These cells able to grow in presence of added H2O2 on filter paper disc. As shown in Figure 4(a) the bacteria were grown even in presence of H2O2, due to the membrane-shielding effects of PUFAs. In most cases, PUFAs are among the molecules most vulnerable to oxygen and ROS, Okuyama et al. [10]. In Figure 4(b) the bacterial cells which were PUFA deficient or nonproducers were hampered by H2O2 and hence could not able to grow where zone of inhibition was directly proportional to the concentration of H2O2 added. Higher the concentration of H2O2 more will be the zone of inhibition.
Figure 4.
H2O2-plate assay.
To confirm that, growth of bacteria in presence of H2O2 is in reality mainly due to presence of PUFA; NaN3 was added in to the media which is a very powerful inhibitor of catalase. If microorganism is producing catalase enzyme, NaN3 inhibits catalase enzyme [15] which helps in interfering and promotes the actual interpretation of plate assay. About 1 mM concentration of NaN3 was used during experimental study. The concentration should be an adequate amount to act as catalase inhibitor at the same time should not be antimicrobial in nature. The concentration of NaN3 was decided from reported studied by Teixeira and Mota [16]. Out of selected 100 strains, 26 strains were found to give false positive results (Tables 1a and 1b). Out of 26, 10 strains were selected which gave false positive results at all H2O2 concentrations used during plate assay method. They were screened for further secondary analysis and confirmation.
Table 1.
Screening of marine strains using H2O2-plate assay method.
(a)
| No. | Strain | H2O2 @ (%) | PUFA* | No. | Strain | H2O2 @ (%) | PUFA* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.1 | 0.5 | 1.0 | +ve/−ve | 0.1 | 0.5 | 1.0 | +ve/−ve | ||||
| 1 | JAY B 8 | − | − | − | −ve | 26 | TH B 9 | − | − | − | −ve |
| 2 | TAV B 31 | − | − | − | −ve | 27 | DI BB 152 | − | − | + | +ve |
| 3 | PA BB 8 | − | − | − | −ve | 28 | TH BB 21 | + | ++ | ++ | +ve |
| 4 | TH BB 31 | − | − | − | −ve | 29 | ALI B 71 | − | − | − | −ve |
| 5 | DIG BB 113 | − | − | − | −ve | 30 | R BB 11 | + | + | − | +ve |
| 6 | TH BB 22 | − | − | − | −ve | 31 | BB 82 | − | − | − | −ve |
| 7 | PA BB 3 | − | − | − | −ve | 32 | JAY B 12 | − | − | − | −ve |
| 8 | TH BB 15 | − | − | − | −ve | 33 | TH BB 22 | − | − | − | −ve |
| 9 | P BB 13 | − | − | − | −ve | 34 | MAN BB 358 | − | − | − | −ve |
| 10 | DIG BB 4 | + | + | + | +ve | 35 | P BB 9 | − | − | − | −ve |
| 11 | M BB 318 | − | + | ++ | +ve | 36 | TH 33 | − | − | − | −ve |
| 12 | DI BB 146 | − | − | + | +ve | 37 | R 23 | − | − | − | −ve |
| 13 | GP B 2 | − | + | ++ | +ve | 38 | RAT B 18 | − | − | − | −ve |
| 14 | AH B 53 | − | + | ++ | +ve | 39 | RA 21 | − | − | − | −ve |
| 15 | MD BB 345 | + | + | ++ | +ve | 40 | MD BB 325 | − | − | − | −ve |
| 16 | PA BB 8 | − | + | ++ | +ve | 41 | SHI BB 2 | − | − | − | −ve |
| 17 | TH BB 5 | − | + | + | +ve | 42 | PA B 12 | − | − | − | −ve |
| 18 | MAN BB 200 | − | − | − | −ve | 43 | PA BB 4 | − | − | − | −ve |
| 19 | MAN BB 361 | − | − | − | −ve | 44 | TH BB 27 | − | − | − | −ve |
| 20 | AL BB 156 | − | + | ++ | +ve | 45 | TH BB 25 | − | − | − | −ve |
| 21 | TH BB 20 | − | + | ++ | +ve | 46 | BB 227 | + | + | ++ | +ve |
| 22 | TAV B 27 | + | + | ++ | +ve | 47 | TH BB 30 | − | − | − | −ve |
| 23 | ALI B 51 | + | + | ++ | +ve | 48 | MAN 10 | − | − | − | −ve |
| 24 | BB 232 | − | + | + | +ve | 49 | BB 72 | − | − | − | −ve |
| 25 | MAN 353 | − | − | + | +ve | 50 | R 37 | − | − | − | −ve |
@describes zone of inhibition due to presence H2O2; +/++ describes growth of microorganisms or no zone of inhibition due to presence of PUFA.
*PUFA +ve denotes PUFA producer and −ve denotes PUFA nonproducer.
Note: highlighted strains were further screened by secondary analysis.
(b)
| No. | Strain | H2O2 @ (%) | PUFA* | No. | Strain | H2O2 @ (%) | PUFA* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.1 | 0.5 | 1.0 | +ve/−ve | 0.1 | 0.5 | 1.0 | +ve/−ve | ||||
| 51 | TH BB 19 | ++ | ++ | + | +ve | 76 | MD BB 312 | − | − | − | −ve |
| 52 | BB 83 | + | + | + | +ve | 77 | MAN BB 63 | − | − | − | −ve |
| 53 | RAT B 3 | − | − | − | −ve | 78 | TH BB 28 | − | − | − | −ve |
| 54 | AK BB 189 | − | − | − | −ve | 79 | JAY B 38 | − | − | − | −ve |
| 55 | BB 201 | − | − | − | −ve | 80 | BB 20 | + | − | − | +ve |
| 56 | MAN 352 | − | − | − | −ve | 81 | P BB 22 | − | − | − | −ve |
| 57 | TH 33 | + | − | − | +ve | 82 | RAT B 85 | − | − | − | −ve |
| 58 | BB 198 | ++ | + | + | +ve | 83 | SHI BB 2 | − | − | − | −ve |
| 59 | R 13 | − | − | − | −ve | 84 | PA BB 33 | − | − | − | −ve |
| 60 | HAR BB 118 | − | − | − | −ve | 85 | MD BB 122 | − | − | − | −ve |
| 61 | RO BB 24 | + | − | − | +ve | 86 | MAN BB 229 | + | − | − | +ve |
| 62 | TH BB 24 | − | − | − | −ve | 87 | TH BB 82 | − | − | − | −ve |
| 63 | SRK 6 | − | − | − | −ve | 88 | DIG BB 122 | − | − | − | −ve |
| 64 | BB 245 | − | − | − | −ve | 89 | G BB 89 | − | − | − | −ve |
| 65 | BB 86 | − | − | − | −ve | 90 | MAN 63 | − | − | − | −ve |
| 66 | G BB 249 | − | − | − | −ve | 91 | AL BB 148 | − | − | − | −ve |
| 67 | DIG BB 108 | − | − | − | −ve | 92 | TAV B 47 | − | − | − | −ve |
| 68 | MD BB 338 | − | − | − | −ve | 93 | R 39 | − | − | − | −ve |
| 69 | TH BB 166 | − | − | − | −ve | 94 | BB 289 | + | − | − | +ve |
| 70 | MAN BB 364 | − | − | − | −ve | 95 | JAY B 23 | − | − | − | −ve |
| 71 | SRK 23 | − | − | − | −ve | 96 | DI BB 278 | − | − | − | −ve |
| 72 | TH BB 83 | − | − | − | −ve | 97 | TH B 34 | − | − | − | −ve |
| 73 | BB 39 | − | − | − | −ve | 98 | AK BB 148 | − | − | − | −ve |
| 74 | TAV B 29 | − | − | − | −ve | 99 | HAR BB 72 | − | − | − | −ve |
| 75 | SHE BB 29 | − | − | − | −ve | 100 | RO BB 41 | − | − | − | −ve |
@describes zone of inhibition due to presence H2O2; +/++ describes growth of microorganisms or no zone of inhibition due to presence of PUFA.
*PUFA +ve denotes PUFA producer and −ve denotes PUFA nonproducer.
Note: highlighted strains were further screened by secondary analysis.
EPA- and DHA-expressing bacteria were reported to be more resistant to exogenous H2O2 [10]. But there were no investigations whether other long chain (LC) PUFAs than EPA and DHA have similar effects. The bacterial cell gets protected by the effect of EPA that has reported earlier [10]. The membrane-shielding effects of n-3 LC-PUFAs have been shown only for bacterial cells producing EPA [17, 18]. From the study we have observed that other than EPA and DHA other PUFA might be responsible for the same protecting effect of exogenous H2O2 and our hypothesis was well supported with studied carried out by Okuyama et al. [10].
As shown in Figure 5, the fatty acid profile obtained from the bacterial culture which was able to grow in presence of H2O2 shows mainly AA production along with EPA. Hence, from this study not only EPA and DHA but also other PUFAs like AA act as the shield molecules against such oxidative challenges exogenously and endogenously raised in marine environments. There are some reports on protective effect of DHA against the external hydrogen peroxide [19]. It was reported that PUFAs including n-3 PUFAs are the molecules which are most susceptible to oxygen and reactive oxygen species (ROS). But from our study, it was observed that not only n-3 PUFAs are susceptible to oxygen and reactive oxygen species but also n-6 PUFAs like AA may behave in same way [20].
Figure 5.
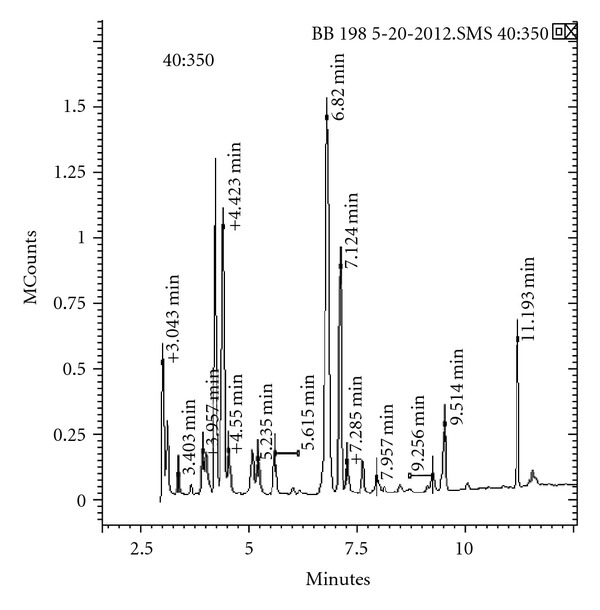
Gas chromatogram showing fatty acid profile of a selected marine isolate after primary screening.
3.2. Secondary Screening and Confirmation
From few selected false positive strains all strains were found to give remarkable response when their lipid extract was injected into GCMS. The mass spectra of all selected strains were found to produce different kind of fatty acids. Some of them were found to produce AA and EPA which are very essential PUFAs from nutritional point of view. The fatty acid profile of 10 selected strains was shown in Table 2 that provides the information about different fatty acids produced by selected marine bacteria after primary screening using H2O2-plate assay. Hence this method is a qualitative estimation of PUFAs produced from microorganisms.
Table 2.
Fatty acid profile of marine isolate produced after fermentation in secondary screening.
| Fatty acid methyl esters (MUFA + PUFA) | Selected strains for secondary screening and their fatty acid profile | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M BB 318 | BB 83 | ALI B 51 | BB 227 | MD BB 345 | BB 198 | DIG BB 4 | TAV B 27 | TH BB 19 | TH BB 21 | |
| Pentadecanoic acid | + | + | + | + | + | + | − | − | − | − |
| Hexadecanoic acid | + | + | + | + | + | + | + | + | + | + |
| 14-methylhexadecanoate | + | − | + | + | + | + | − | − | − | − |
| Margaric acid | + | + | + | + | + | + | − | − | − | − |
| Oleic acid | + | + | + | + | + | + | − | + | + | + |
| 16-Octadecenoic acid | − | − | − | − | − | − | + | − | − | − |
| 7,10-Octadecadienoic acid | − | − | − | − | − | − | + | − | − | − |
| Linoleic acid | + | + | + | + | + | + | − | + | + | + |
| 7,10,13-Hexadecatrienoic acid | − | − | − | − | − | − | + | − | − | − |
| γ-Linolenic acid | − | − | − | − | − | + | − | − | − | − |
| Linolenic acid | + | + | + | + | + | + | − | + | + | + |
| Eicosanoic acid | + | − | − | − | − | − | − | − | − | − |
| Arachidonic acid | − | − | − | − | − | + | − | − | − | − |
| Eicosapentaenoic acid | − | − | − | − | − | + | − | − | − | − |
Note: MUFA- monounsaturated fatty acid.
4. Conclusion
Thus, the present investigation has clearly revealed the presence of PUFAs in marine bacteria in the marine samples by newly developed novel simplified and rapid plate culture method for screening of PUFAs-producing marine bacteria; collected from different regions of western coast of Maharashtra, India. GCMS analysis studies confirmed the actual production of PUFA in various selected marine bacteria after primary screening. In order to minimise the time required for analysis as well as economic loss, this method gives the suitable solution for large number of samples screening which are abundant in the marine environment.
Acknowledgment
Authors are thankful to the University Grants Commission, Government of India, India, for providing financial assistance during the course of this research.
References
- 1.Drevon CA, Baksaas I, Krokan HE. Omega-3 Fatty Acids: metabolism and Biological Effects. Basel, Switzerland: Birkhäuser; 1993. [Google Scholar]
- 2.Simopoulos AP, Leaf A, Salem N. Essentiality of and recommended dietary intakes for omega-6 and omega-3 fatty acids. Annals of Nutrition and Metabolism. 1999;43(2):127–130. doi: 10.1159/000012777. [DOI] [PubMed] [Google Scholar]
- 3.Crawford MA. Placental delivery of arachidonic and docosahexaenoic acids: implications for the lipid nutrition of preterm infants. American Journal of Clinical Nutrition. 2000;71(1):275S–284S. doi: 10.1093/ajcn/71.1.275S. [DOI] [PubMed] [Google Scholar]
- 4.Bergstrom S, Danielsson H, Samuelsson B. The enzymatic formation of prostaglandin E2 from arachidonic acid. Biochimica et Biophysica Acta. 1964;90:207–210. doi: 10.1016/0304-4165(64)90145-x. [DOI] [PubMed] [Google Scholar]
- 5.Van Dorp DA, Beerthuis RK, Nugteren DH, Vonkeman H. The biosynthesis of prostaglandins. Biochimica et Biophysica Acta. 1964;90(1):204–207. doi: 10.1016/0304-4165(64)90144-8. [DOI] [PubMed] [Google Scholar]
- 6.Jakschik BA, Sams AR, Sprecher H, Needleman P. Fatty acid structural requirements for leukotriene biosynthesis. Prostaglandins. 1980;20(2):401–410. doi: 10.1016/s0090-6980(80)80057-8. [DOI] [PubMed] [Google Scholar]
- 7.Okuyama H, Kobayashi T, Watanabe S. Dietary fatty acids—The N-6/N-3 balance and chronic elderly diseases. Excess linoleic acid and relative N-3 deficiency syndrome seen in Japan. Progress in Lipid Research. 1996;35(4):409–457. doi: 10.1016/s0163-7827(96)00012-4. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi J. Bioactive products from marine micro-and macro-organisms. Pure and Applied Chemistry. 1999;71(6):1123–1126. [Google Scholar]
- 9.Bowles RD, Hunt AE, Bremer GB, Duchars MG, Eaton RA. Long-chain n-3 polyunsaturated fatty acid production by members of the marine protistan group the thraustochytrids: screening of isolates and optimisation of docosahexaenoic acid production. Journal of Biotechnology. 1999;70(1–3):193–202. [Google Scholar]
- 10.Okuyama H, Orikasa Y, Nishida T. Significance of antioxidative functions of eicosapentaenoic and docosahexaenoic acids in marine microorganisms. Applied and Environmental Microbiology. 2008;74(3):570–574. doi: 10.1128/AEM.02256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliver JD, Colwell RR. Extractable lipids of gram negative marine bacteria: fatty acid composition. International Journal of Systematic Bacteriology. 1973;23(4):442–458. doi: 10.1128/jb.114.3.897-908.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoshi M, Williams M, Kishimoto Y. Esterification of fatty acids at room temperature by chloroform methanolic HCl cupric acetate. Journal of Lipid Research. 1973;14(5):599–601. [PubMed] [Google Scholar]
- 13.Cheng MH, Walker TH, Hulbert GJ, Raman DR. Fungal production of eicosapentaenoic and arachidonic acids from industrial waste streams and crude soybean oil. Bioresource Technology. 1999;67(2):101–110. [Google Scholar]
- 14.Song X, Zhang X, Kuang C, Zhu L, Guo N. Optimization of fermentation parameters for the biomass and DHA production of Schizochytrium limacinum OUC88 using response surface methodology. Process Biochemistry. 2007;42(10):1391–1397. [Google Scholar]
- 15.Lichstein HC, Soule MH. Studies of the effect of sodium azide on microbic growth and respiration. II. The action of sodiuim azide on bacterial catalase. Journal of Bacteriology. 1944;47(3):231–238. doi: 10.1128/jb.47.3.231-238.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teixeira JA, Mota M. Determination of catalase activity and its inhibition by a simple manometric method. Biochemical Education. 1992;20(3):174–175. [Google Scholar]
- 17.Nishida T, Orikasa Y, Watanabe K, Okuyama H. The cell membrane-shielding function of eicosapentaenoic acid for Escherichia coli against exogenously added hydrogen peroxide. FEBS Letters. 2006;580(28-29):6690–6694. doi: 10.1016/j.febslet.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Nishida T, Morita N, Yano Y, Orikasa Y, Okuyama H. The antioxidative function of eicosapentaenoic acid in a marine bacterium, Shewanella marinintestina IK-1. FEBS Letters. 2007;581(22):4212–4216. doi: 10.1016/j.febslet.2007.07.065. [DOI] [PubMed] [Google Scholar]
- 19.Bechoua S, Dubois M, Dominguez Z, et al. Protective effect of docosahexaenoic acid against hydrogen peroxide-induced oxidative stress in human lymphocytes. Biochemical Pharmacology. 1999;57(9):1021–1030. doi: 10.1016/s0006-2952(99)00012-x. [DOI] [PubMed] [Google Scholar]
- 20.Halliwell B, Gutteridge JM. Free Radicals in Biology and Medicine. 3rd edition. Oxford, UK: Oxford University Press; 1998. [Google Scholar]



