Abstract
Purpose
Abnormalities in the constitutive and IFN-γ–inducible HLA class I surface antigen expression of tumor cells is often associated with an impaired expression of components of the antigen processing machinery (APM). Hence, we analyzed whether there exists a link between the IFN-γ signaling pathway, constitutive HLA class I APM component expression, and IFN-γ resistance.
Experimental Design
The basal and IFN-γ–inducible expression profiles of HLA class I APM and IFN-γ signal transduction cascade components were assessed in melanoma cells by real-time PCR (RT-PCR), Western blot analysis and/or flow cytometry, the integrity of the Janus activated kinase (JAK) 2 locus by comparative genomic hybridization. JAK2 was transiently overexpressed in JAK2− cells. The effect of IFN-γ on the cell growth was assessed by XTT [2,3-bis(2-methoxy-4-nitro-S-sulfophenynl)-H-tetrazolium-5-carboxanilide inner salt] assay.
Results
The analysis of 8 melanoma cell lines linked the IFN-γ unresponsiveness of Colo 857 cells determined by lack of inducibility of HLA class I surface expression on IFN-γ treatment to a deletion of JAK2 on chromosome 9, whereas other IFN-γ signaling pathway components were not affected. In addition, the constitutive HLA class I APM component expression levels were significantly reduced in JAK2− cells. Furthermore, JAK2-deficient cells were also resistant to the antiproliferative effect of IFN-γ. Transfection of wild-type JAK2 into JAK2− Colo 857 not only increased the basal APM expression but also restored their IFN-γ sensitivity.
Conclusions
Impaired JAK2 expression in melanoma cells leads to reduced basal expression of MHC class I APM components and impairs their IFN-γ inducibility, suggesting that malfunctional IFN-γ signaling might cause HLA class I abnormalities.
Introduction
Type I and type II IFNs represent multifunctional cytokines, which exert pleiotropic activities including antiproliferative, immunomodulatory, anti-inflammatory, apoptosis-inducing, and stress-mediated effects. In addition, they are involved in shaping the adaptive and innate immunity including tumor immunosurveillance (1–6). IFN-γ binds to the heterodimeric IFN-γ receptor 1 (R1) and 2 (R2) and activates a downstream signal transduction cascade leading to the transcriptional activation of IFN-stimulated genes (ISG). They include the receptor-associated Janus associated kinases JAK1 and JAK2, the signal transducers and activators of transcription STAT, suppressors of cytokine signaling (SOCS), and protein inhibitors of activated STATs that ensure the execution of the IFN-γ effects (7, 8). In addition to transducing signals from many ligands, STAT proteins can act as transcription factors. Phosphorylated STAT1 (pSTAT1) can interact with the IFN-γ activation site (GAS) or in combination with the IFN-γ–stimulated gene factor 3 (ISGF3), thereby transactivating a number of genes such as members of the IFN regulatory factor (IRF) family.
A prerequisite for the IFN-γ–mediated enhanced expression of the HLA class I antigen processing machinery (APM) components, such as the IFN-γ–inducible low-molecular-weight proteins LMP2, LMP7, and LMP10, the transporter associated with antigen processing (TAP), tapasin, β2-microglobulin (β2-m), and HLA class I heavy chain (HC), is the presence of GAS and IFN-sensitive response elements (ISRE) in their promoters. Although the IFN-γ– mediated binding of IRF1 or STAT1 to the interferon consensus sequence (ICS)-GAS is sufficient for the IFN-γ inducibility of many genes, LMP2 transcription requires the presence and binding of both factors (9). Furthermore, DNA-bound IRF2 and activation of STAT1 are crucial for the modulation of other HLA class I APM components by IFN-γ (10).
Some tumor cells have lost their susceptibility to modulation by IFN-γ; as a result, HLA class I and class II antigens are not upregulated when cells are exposed to IFN-γ (11, 12). This abnormality is likely to have a negative impact on the interactions of tumor cells with host immune system and provide them with an escape mechanism. The molecular mechanisms causing IFN-γ resistance have been investigated only to a limited extent, although this information might have an important impact on the development of targeted therapies. So far, IFN-γ–responsive genes have been shown to be frequently downregulated in tumor cells because of impaired IRF1 expression as well as defective transcriptional and posttranscriptional regulation of components of the IFN-γ signal transduction cascade. In addition, to the best of our knowledge, loss of the IFN-γ–mediated upregulation of TAP in one renal cell carcinoma (RCC) cell line has been found to be associated with the lack of IRF1 and STAT1 binding activities as well as of JAK1, JAK2, and STAT1 phosphorylation upon incubation with IFN-γ (13). Although JAK1 and/or JAK2 gene transfer could not restore the IFN-γ–mediated phosphorylation in this RCC cell line, their overexpression increased constitutive LMP2 and TAP1 expression independent of IFN-γ (13). Furthermore, an impaired STAT1 phosphorylation was accompanied by loss of IFN-γ–mediated HLA class I upregulation in melanoma and colorectal carcinoma cell lines (14). The purpose of this study was to determine the mechanisms of IFN-γ unresponsiveness of melanoma cells regarding the HLA class I upregulation as well as the role of the IFN-γ signal cascade for HLA class I APM component expression. Our results show loss of JAK2 expression in 1 of 8 melanoma cell lines, which associated with a lack of IFN-γ inducibility of HLA class I surface antigens and with a low constitutive HLA class I APM component expression. These defects could be corrected by JAK2 transfection; vice versa, JAK2-specific short hairpin RNA and the pharmacologic inhibitor AG490 inversely impairs constitutive APM component expression in JAK2-positive cells, which is associated with reduced HLA class I surface expression.
Material and Methods
Tissue culture
Eight human melanoma cell lines, which have already been described elsewhere or were obtained from the European tumor cell line database (ESTDAB project; see www.ebi.ac.uk/ipd/estdab) were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS; PAA Laboratories), 2% glutamine (Lonza Cologne AG), and 1% penicillin, and streptomycin (PAA Laboratories) in a humified atmosphere with 5% CO2.
Antibodies used
The low-molecular-weight polypeptide LMP2, LMP7, and LMP10-specific mouse monoclonal antibody (mAb) SY-1, HB-2, and TO-7, respectively (15), the TAP1-specific mAb NOB-1 (16), the TAP2-specific mAb NOB-2 (16), the tapasin-specific mAb TO-3 (17), and the HLA class I HC-specific mAb HC-10 (18) were developed and characterized as described. All of these are IgG1 mAbs with the exception of mAb HC-10, which is an IgG2a mAb. In addition, following antibodies were purchased, which were directed against the IFN-γR1 (clone 92101; R&D Systems), IFN-γR2 (clone MMHGR-2; Abcam), HLA-ABC (clone B9.12.1; Beckman Coulter), and HLA class II antigens (clone Tü39; Becton Dickinson, BD). The antibodies directed against the unphosphorylated and phosphorylated IFN signal transduction pathway components JAK1 (clone 6G4), pJAK1 (Tyr1022/1023), JAK2 (clone 24B11), STAT1 (clone 42H3), and pSTAT1 (clone 58D6) were all obtained from Cell Signaling [New England Biolabs GmbH)]. The fluorescein isothiocyanate (FITC)-conjugated IgG2a antibody (Beckman Coulter) served as a control in flow cytometry. The anti-GAPDH (clone 14C10; Cell Signaling) and the anti-β-actin (Abcam) mAbs served as loading controls, whereas the horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse IgGs were used as detection antibody in Western blot analysis.
Cytokines and pharmacologic agents
Recombinant human IFN-γ, IFN-α, and TNF-α were purchased from Pan Biotech.
Flow cytometry
The expression of IFN-γR and HLA class I and class II surface antigens was assessed by direct immunofluorescence. For determination of surface expression, 1 × 105 cells were trypsinized, washed with PBS containing 1% FCS, and consecutively incubated with FITC-conjugated respective antibodies for 30 minutes at 4−C in the dark. After 2 washes with PBS, flow cytometry was carried out using the BD FACSCalibur flow cytometer (Becton Dickinson). The results were expressed as mean specific fluorescence intensity (MFI) − SD of the values obtained in 3 independent experiments. Staining with an FITC-conjugated respective IgG antibodies served as the negative control.
Growth properties
A total of 3 × 103 cells were plated in triplicate in the cavities of a 96-well plate overnight before supplementation of different concentrations of IFN-γ (200–1,600 units). After 48 hours, cell viability was measured by standard XTT [2,3-bis(2-methoxy-4-nitro-S-sulfophenynl)-H-tetrazolium-5-carboxanilide inner salt] method. Results are expressed as relative growth in comparison with untreated cells of 3 independent experiments.
Semiquantitative RT-PCR and quantitative RT-PCR analysis
For quantitative RT-PCR (qRT-PCR), total cellular RNA was extracted using the RNeasy Mini Kit (Macherey and Nagel), followed by digestion with DNase I (Invitrogen). cDNA was synthesized from 2 µg of total RNA employing the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas), according to the manufacturer’s instructions.
Comparative quantification of gene expression was carried out by qRT-PCR on a Rotor Gene 6000 system (Corbett Research), using the quantitative SYBR green kit (Invitrogen GmbH) and the target-specific primers listed in Table 1. Amplifications were carried out by an initial hold at 50−C for 2 minutes, followed by denaturation at 95−C for 2 minutes. After 40 cycles with denaturation at 95−C for 15 seconds and annealing at 60−C for 30 seconds, the melting steps occurred, starting at 60−C and increasing to 99−C by 1−C steps. For STAT1, the melting step started at 55−C, rising to 99−C. The melting curve analysis was provided at the end of each run to control PCR specificity. Results of the qRT-PCR data were expressed as relative mRNA expression quantified with the Rotor Gene analysis software and normalized to the averaged glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and peptide prolyl isomerase A (PPIA) transcription levels.
Table 1.
Primers used for RT-PCR analysis
| Application | Gene name | Sequence | Tm |
|---|---|---|---|
| IFN-γ signaling | |||
| rev: 3′-tcc agt gag ctg gca tca ag-5′ | JAK1 | fw: 5′-gca cca tca ccg ttg atg ac-3′ | 60°C |
| rev: 3′-att acg ccg acc agc act gt-5′ | JAK2 | fw: 5′-tgt gga gat gtg ccg gta tg-3′ | 60°C |
| rev: 3′-gcc agg tac tgt ctg att-5′ | STAT1 | fw: 5′-atc ctc gag agc tgt cta-3′ | 55°C |
| Antigen processing | |||
| rev: 3′-tgc tgc atc cac ata acc at-5′ | LMP2 | fw: 5′-tgt gca ctc tct ggt tca gc-3′ | 60°C |
| rev: 3′-cag ccc cac agc agt aga tt-5′ | LMP10 | fw: 5′-ggg ctt ctc ctt cga gaa ct-3′ | 60°C |
| rev: 3′-tgg gtg aac tgc atc tgg ta-5′ | TAP1 | fw: 5′-gga atc tct ggc aaa gtc ca-3′ | 60°C |
| rev: 3′-ttc atc cag cag cac ctg tc-5′ | TAP2 | fw: 5′-cca aga cgt ctc ctt tgc at-3′ | 60°C |
| rev: 3′-acc tgt cct tgc agg tat gg-5′ | Tapasin | fw: 5′-tgg gta agg gac atc tgc tc-3′ | 60°C |
| rev: 3′-ggt ggc ctc atg gtc aga ga-5′ | HLA-ABC | fw: 5′-gcc tac gac ggc aag gat tac-3′ | 60°C |
| Locus specificity | |||
| rev: 3′-tgt tgc atg ggt tgt tgt ct-5′ | RFX3 | fw: 5′-aaa ctg gac cca gtc aat gc-3′ | 57°C |
| rev: 3′-atg ggc ttc aag acc ttc ct-5′ | RCL1 | fw: 5′-tct tct ttg ctt ggc tcc at-3′ | 57°C |
| rev: 3′-cag att ccc acc tga gca tt-5′ | GLDC | fw: 5′-tcg atg cag ttc acc tca ag-3′ | 57°C |
| rev: 3′-ggc cct cct tat ttc agt cc-5′ | BNC | fw: 5′-gtc aag cat gcc tgt gaa ga-3′ | 59°C |
| Control | |||
| rev: 3′-ctg aag gcc atg caa gtg ag-5′ | GAPDH | fw: 5′-cct gca cca cca act gct ta-3′ | 60°C |
| rev: 3′-cag tca gca atg gtg atc ttc t-5′ | PPIA | fw: 5′-cca tct atg ggg aga aat ttg a-3′ | 60°C |
| rev: 3′-gaa gca ttt gcg gtg gac gat-5′ | β-Actin | fw: 5′-tcc tgt ggc atc cac gaa act-3′ | 60°C |
Abbreviations: fw, forward; rev, reverse.
For semiquantitative RT-PCR, cDNA was synthesized from 500 ng of total RNA employing the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturers’ instructions. Amplification was carried out in a final volume of 25 µL supplemented with 1.25 units of Taq polymerase, MgCl2 (Invitrogen), and 0.2 mmol/L dNTP mix (PeqLab). Reaction conditions for the initial denaturation step were 95−C for 2 minutes, followed by denaturation at 95−C for 30 seconds. The annealing occurred at 60−C for 30 seconds, and the extension was done at 72−C for 30 seconds. After 35 cycles, a final extension was done at 72−C for 5 minutes.
Western blot analyses
Proteins (50 µg per lane) extracted from melanoma cells were separated in a 8% to 15% SDS-PAGE gel, depending on the protein size, and transferred to nitrocellulose membranes (Schleicher & Schuell), which were probed with primary mAbs directed against the major HLA class I APM components and molecules of the IFN-γ signal transduction pathway. Following an overnight incubation at 4−C, membranes were incubated for 4 hours with the appropriate HRP-conjugated secondary antibodies as recently described (6). Proteins were detected using a Lumilight detection kit (Roche Diagnostics). Staining of the blots with the anti-β-actin or anti-GAPDH mAbs served as loading controls.
Array comparative genomic hybridization
Genomic DNA was extracted from cultured Colo 857 melanoma cells (test sample) or normal donor peripheral blood mononuclear cells (PBMC; reference sample), using the Qiagen Mini Kit). DNA labeling was conducted using a BioPrime array comparative genomic hybridization (aCGH) genomic labeling kit (Invitrogen). Test and reference DNAs (4 µg of each) were labeled with Cy5 and Cy3, respectively, and cohybridized to the cDNA clone microarray printed at the Infectious Disease and Immunogenetics Section, Department of Transfusion Medicine, Clinical Center, NIH, at 65−C overnight with a configuration of 32 − 24 − 23 spots and contained 17,500 cDNAs (19). Microarray slides were scanned at 10-µm resolution on a GenePix 4000 scanner (Axon Instruments) to obtain maximal signal intensities with less than 0.1% probe saturation.
Genomic PCR
Genomic DNA from melanoma cells was extracted using the Qiagen Mini Kit and genomic PCR was carried out as recently described (20). The forward and reverse primers used for JAK2 amplification were 5′-cat tcc ctt ggg aaa tct ga-3′ and 3′-tgc atg tga aaa cac aca cg-5′, respectively.
Transfection
The JAK2 cDNA was amplified using JAK2-specific primers: forward: 5′-aaa atc gat atg gga atg gcc tgc ctt ac-3′ and reverse: 3′- ttt gcg gcc gct cat cca gcc atg tta tcc c-5′. The 3,339-bp amplification product was directly cloned into the multiple cloning site of the pCMV-IRES expression vector as previously described (21). JAK2-negative cells were stably transfected with the JAK2 expression vector or the control vector (mock), using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. Transfectants were selected in 800 µg/mL G418, and neoR clones were cultivated and further analyzed.
Data analysis of comparative genomic hybridization (aCGH)
The fluorescence intensity and ratio data for Cy5 and Cy3 were transformed into a log base 2 scale and normalized against the median over the entire array using BRBArray Tools developed by the Biometric Research Branch, National Cancer Institute (22). Further data preprocessing was done using Web tool prep (http://prep.bioinfo.cnio.es/). Duplicate data points were merged by taking the average over the UniGene cluster IDs. Missing data were imputed using K-nearest neighbor imputation method with K = 15. Gene location was extracted according to UniGene cluster IDs from Ensemble and University of California at Santa Cruz by using ID converter (http://idconverter.bioinfo.cnio.es/). The preprocessed aCGH data were then segmented using the circular binary segmentation method implemented in ADaCGH (http://adacgh.bioinfo.cnio.es/) to detect regions with abnormal DNA copy number. The variables were set as defaults. The significance level for the test to accept change points was set to be 0.01 under 10,000 permutations. The cutoff for gain/loss calls was F0.12 on the log base 2 scale (22). In PBMCs from normal volunteers, this threshold was higher than the 99th percentile of data obtained from autosomes, excluding chromosome X ratios that fell under the 15th percentile. This threshold was then applied to pick up the gain/loss regions in the Colo 857 cell line. To ensure the reproducibility of the array data, the array experiments were repeated twice by using DNA isolated from 2 different passages.
Results
Impaired constitutive HLA class I APM component expression in IFN-γ–resistant melanoma cells
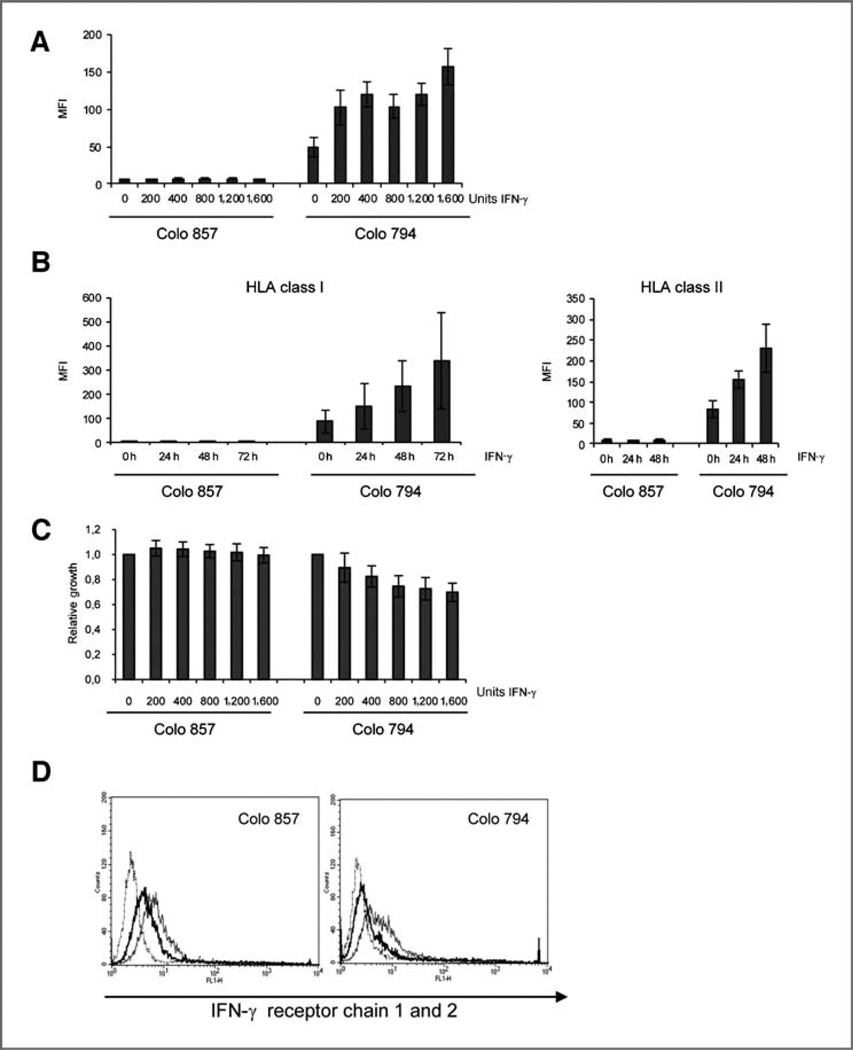
Flow cytometric analysis of 8 untreated or IFN-γ–treated melanoma cell lines using the HLA class I antigen-specific mAb B9.12.1 (Fig. 1A and C, Supplementary Fig. S1) or the HLA class II-antigen-specific mAb Tü39 (Fig. 1C, Supplementary Fig. S1) showed a marked variability in the IFNγ–mediated modulation of both HLA antigen classes. The different melanoma cell lines heterogeneously responded in a dose- and time-dependent manner to IFN-γ treatment, ranging from lack of to low to strong IFN-γ–mediated upregulation of HLA class I (Supplementary Fig. S1A) and class II surface antigens (Supplementary Fig. S1B). The representative results shown in Supplementary Figure S1 show that 4 of 8 melanoma cell lines tested exhibited a 2- to 3-fold upregulation of both HLA class I and class II surface antigens, whereas the remaining 4 failed to upregulate HLA class II antigens. The melanoma cell line Colo 857 was completely resistant to IFN-γ treatment, lacking IFN-γ–mediated upregulation of both HLA class I and class II surface antigens (Fig. 1A and B) as well as responsiveness to the antiproliferative effect of IFN-γ (Fig. 1C). The resistance of Colo 857 cells was selective for IFN-γ because HLA class I surface expression was induced in these cells in a dose- and time-dependent manner by IFN-α as well as by TNF-α, although the degree of upregulation varied between both cytokines (Supplementary Fig. S2). Because the IFN-γ receptor was expressed in the IFN-γ– resistant Colo 857 cells to levels comparable with the IFN-γ–sensitive control cell line Colo 794 (Fig. 1D), the IFN-γ resistance seemed to be due to defects in the IFN-γ signal transduction pathway rather than at the receptor level.
Figure 1.
Constitutive and cytokine-mediated regulation of HLA class I and class II surface expression. A, constitutive and cytokine-mediated dose-dependent upregulation of HLA class I surface antigen expression. B, constitutive and cytokine-mediated time-dependent upregulation of HLA class I and HLA class II antigen surface expression. The cells were either left untreated or treated with 200 units of IFN-γ for the time points indicated. Flow cytometry was carried out using FITC-conjugated HLA class I- and class II-specific mAb. Results are expressed as MFI − SD. The experiments were carried out at least 3 times. C, antiproliferative effect of IFN-γ in Colo 794. Cells were treated with the indicated concentrations of IFN-γ for 48 hours and the number of viable cells was quantified by XTT measurements. Results are expressed as relative growth to untreated cells. D, presence of IFN-γR on the cell surface expression. The IFN-γR expression was determined by flow cytometry by using IFN-γR chain-specific mAbs. The results are presented as histograms. The dotted line represents the IgG1 isotype control, the thin line the IFN-γR1, and the thick line the IFN-γR2.
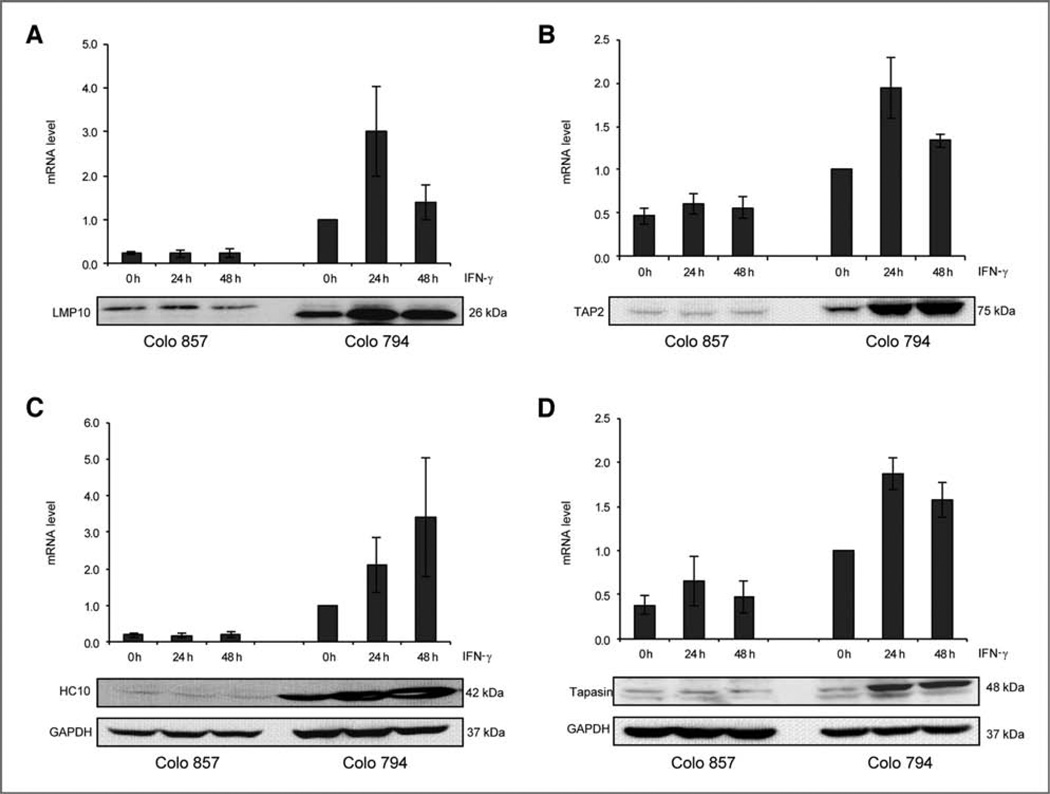
To investigate whether the loss of IFN-γ inducibility of HLA class I surface antigens was associated with altered expression levels of HLA class I APM components, constitutive and IFN-γ inducible LMP10, TAP2, tapasin, HLA class I HC (Fig. 2), LMP2, TAP1, and β2-m (data not shown) mRNA and protein expression levels were monitored by qRT-PCR and Western blot analysis. With the exception of β2-m, the constitutive expression pattern of these molecules was lower and not inducible in IFN-γ–resistant Colo 857 cells than that in IFN-γ–sensitive Colo 794 melanoma cells (Fig. 2). In contrast, IFN-α treatment increased APM component transcription levels and protein expression in both Colo 857 and Colo 794 cells (data not shown).
Figure 2.
IFN-γ resistance of melanoma cells associated with low levels of HLA class I APM component expression. mRNA and protein expression patterns of HLA class I APM components in melanoma cells either left untreated or treated for 24 to 48 hours with IFN-γ were determined by qRT-PCR and Western blot analysis as described in Materials and Methods by using APM-specific primers and antibodies, respectively. A, LMP10; B, TAP2; C, HLA class I HC; D, tapasin. At least 3 independent experiments were carried out. The results are either expressed as mean of the values obtained in 3 independent experiments (qRT-PCR) or shown by a representative Western blot.
Lack of IFN-γ sensitivity due to defects in the IFN-γ signal cascade
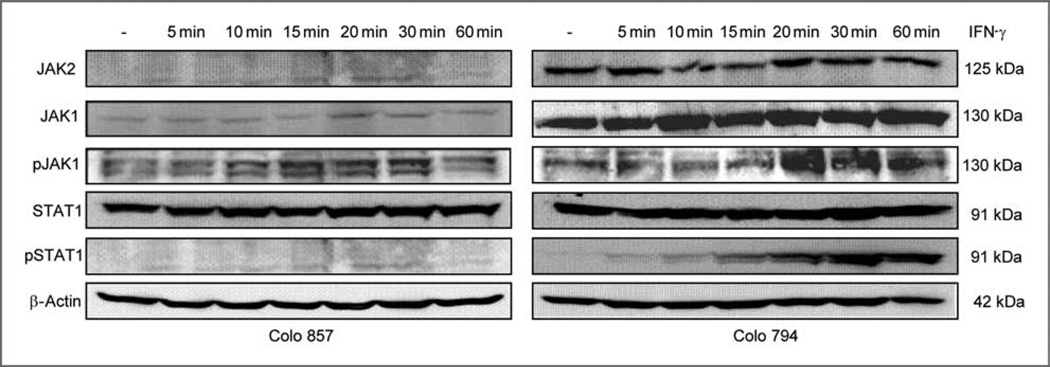
To determine whether the resistance of Colo 857 cells to IFN-γ is due to an impaired IFN-γ signal transduction, constitutive and IFN-γ–inducible transcription of the major signal transduction molecules including JAK1, JAK2, and STAT1 were investigated. In contrast to the IFN-γ–sensitive cell line Colo 794, RT-PCR revealed a lack of constitutive and IFN-γ–inducible JAK2 mRNA expression in Colo 857 cells, whereas the mRNA of JAK1 and STAT1 was expressed in these cells (data not shown). With the exception of JAK1, the signal transduction components were not upregulated by IFN-γ (data not shown). These data were further confirmed by Western blot analysis, which showed JAK2 protein expression in Colo 794 and all other melanoma cells analyzed (data not shown) but not in Colo 857 cells despite their constitutive expression of JAK1 and STAT1 proteins (Fig. 3). The functionality of the IFN-γ signaling components was determined using antibodies specifically directed against the selected phosphorylated counterparts JAK1 and STAT1. In Colo 794 cells, an increased phosphorylation of JAK1 and STAT1 was observed after IFN-γ treatment. In contrast, phosphorylation of JAK1 and STAT1 was not detected in Colo 857 cells (Fig. 3). This defect is selective for IFN-γ, as IFN-α did induce STAT1 phosphorylation (data not shown). Thus, the impaired phosphorylation of signal cascade members by IFN-γ treatment reflects the loss of JAK2 expression.
Figure 3.
Association of impaired JAK2 expression in Colo 857 cells with impaired STAT1 phosphorylation. Melanoma cells were either left untreated or treated with IFN-γ for various time points before being harvested for protein extraction. Western blot analysis was carried out using unphosphorylated and phosphorylated JAK/STAT pathway component-specific antibodies. Staining of the Western blot with an anti-GAPDH- or β-actin–specific antibody served as controls.
Molecular mechanisms underlying deficient JAK2 expression
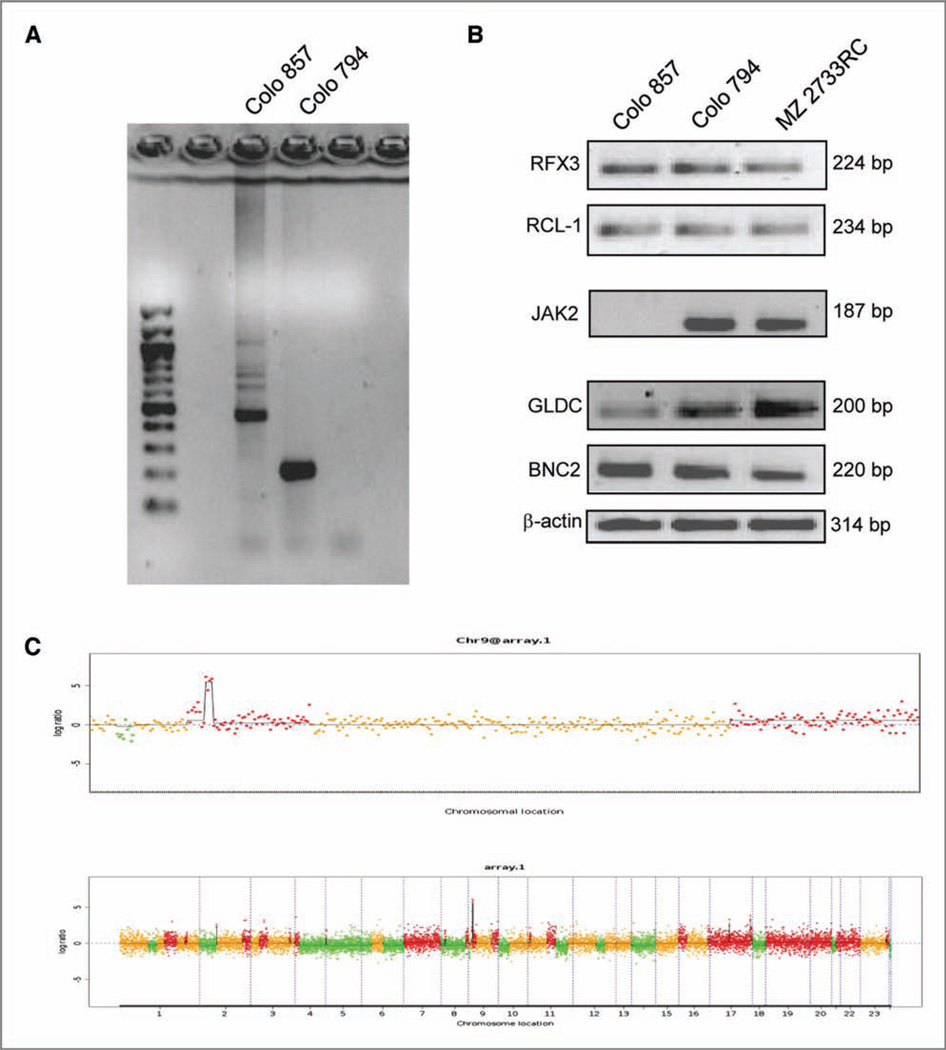
To define the molecular mechanisms involved in the lack of JAK2 mRNA and protein expression in Colo 857 cells, JAK2-specific genomic PCR was carried out. Despite the presence of JAK2 amplification products in the Colo 794 control cells, no JAK2-specific genomic PCR product was obtained in Colo 857, suggesting a structural abnormality of JAK2 in these cells (Fig. 4A). In contrast, RT-PCR analyses of 2 genes flanking upstream (RFX3 and RCL1) or downstream (GLDC and BNC2) the JAK2 locus showed amplification products in both the JAK2− Colo 857 and the JAK2+ Colo 794 cells (Fig. 4B). To confirm these data, CGH of the JAK2− Colo 857 cell line was done (Fig. 4C) using PBMCs as a control. In addition to multilocus gene amplifications and deletions across the whole genome, a deletion on chromosome 9 from positions 24,466 to 22,022,985 containing the JAK2 gene was found in Colo 857 cells (Fig. 4C). These results indicate that the lack of JAK2 expression in this melanoma cell line is caused by a genomic deletion.
Figure 4.
Genomic deletion of JAK2 in Colo 857 cells. A, genomic PCR using JAK2-specific primers showed no amplification product in Colo 857 but in Colo 794. B, locus-specific gene amplification (end point PCR) was carried out as described in Materials and Methods. The RCC cell line MZ 2733RC known to have no observed abnormalities in IFN-γ signaling pathway served as a control (C) CGH. The deletion of JAK2 was determined by 3 independent technologies as described in detail in Materials and Methods.
Restoration by JAK2 gene transfer of constitutive and IFN-γ–inducible HLA class I APM component expression by melanoma cells Colo 857
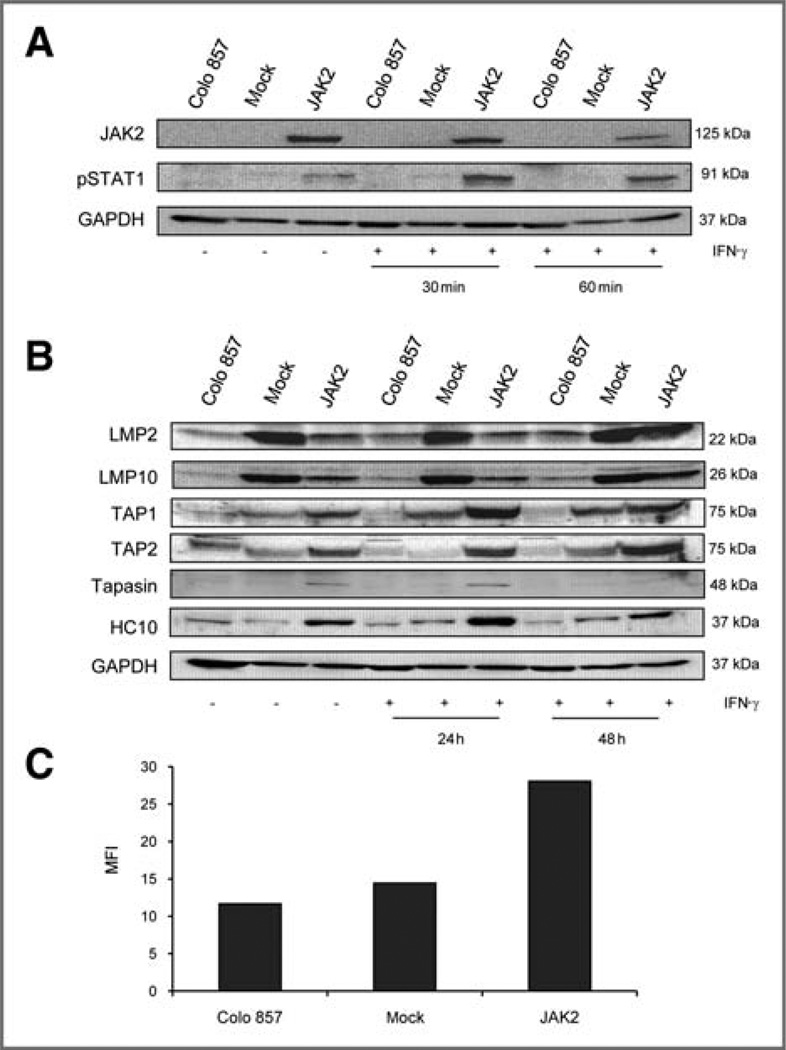
To determine whether the IFN-γ resistance and low levels of HLA class I APM molecules could be restored by JAK2 gene transfer, Colo 857 cells were transfected with a JAK2 expression vector and a control vector (mock) carrying only the neoR gene, respectively. The JAK2 transfectants, but not the mock-transfected control cells, did express high levels of JAK2 (Fig. 5A), which was associated with an increased phosphorylation of STAT1 after IFN-γ treatment (Fig. 5A). JAK2 overexpression in JAK2− cells was accompanied by an enhanced constitutive expression of APM components as representatively shown for TAP1, TAP2, tapasin, and HLA class I HC protein (Fig. 5B), which was accompanied by an increased HLA class I surface antigen expression (Fig. 5C). As expected, the JAK2-transfected cells acquired the susceptibility to modulation by IFN-γ treatment, as indicated by the upregulation of HLA class I APM component expression in these cells (Fig. 5B).
Figure 5.
Reconstitution of HLA class I APM component expression by Colo 857 cells following JAK2 gene transfer. A, JAK2 and pSTAT1 expression were determined in untreated and IFN-γ–treated Colo 857 cells transfected with the JAK2 expression vector as described in Materials and Methods. The results showed restoration of JAK2 expression and pSTAT1 upregulation by JAK2 gene transfer. B, constitutive and IFN-γ–inducible upregulation of APM component expression in mock-transfected and JAK2-transfected Colo 857 cells was carried out as described in Materials and Methods by using APM-specific mAbs. C, restoration of constitutive HLA class I surface expression in Colo 857 cells. Flow cytometry of untransfected, mock, and JAK2-overexpressing cells was carried out as described using an anti-HLA class I–specific mAb. The results are expressed as MFI with subtraction of the isotype control.
Discussion
The physiologic relevance of the IFN-γ–dependent JAK/STAT pathway was characterized by the functional analysis of JAK/STAT knockout mice and has been linked to anti-tumor responses (23). IFN-γ can directly act on tumor cells by exerting antiproliferative, proapoptotic, and antiangiogenic effects (24). STATs and JAKs are thought to play a role in promoting these IFN-γ effects on tumor cells, and defects of the JAK/STAT signal transduction intermediates have been associated with an IFN-γ–resistant phenotype in lung carcinoma and melanoma cells (3, 25, 26). This might provide tumor cells with a selective growth advantage. Indeed, more than 30% of human tumors exhibited unresponsiveness or reduced sensitivity to IFN-γ associated with tumor progression. The variable IFN-γ responsiveness of melanoma cells could be (i) associated with a lower capacity of IFN-γ to induce JAK/STAT (12, 27) or (ii) mediated by downstream components or an additional signaling pathway (28) or (iii) due to lack of STAT1 phosphorylation and epigenetic silencing of the IRF1 transactivation (14).
Although abnormalities of HLA class I APM components represent one major mechanism of tumor cells to evade immune surveillance, there exist only limited information about the role of deficient IFN signal transduction in the regulation of these immunomodulatory molecules. Therefore, the constitutive and IFN-γ–inducible expression pattern of several components of the HLA class I APM and the IFN signal transduction pathway was determined in a number of melanoma cells. With the exception of Colo 857 cells, the other 7 melanoma cell lines analyzed constitutively expressed heterogeneous levels of JAK2 whereas the other IFN signal transduction molecules were constitutively expressed and upregulated by IFN-γ in these tumor cells. Although genetic abnormalities of the IFN-γ signal pathway such as mutations, deletions, and recombinations have been described in tumors of distinct origin (29), no structural alterations in these molecules have been yet reported in melanoma. In this context, it is noteworthy that resistance of melanoma cells to IFN-α is due to multiple defects in the type I IFN signal transduction pathway including lack of Tyk2 (tyrosine kinase 2; ref. 30).
Our results showed that loss of JAK2 expression in the melanoma cell line Colo 857 was caused by a deletion of the JAK2 gene, which is accompanied by a defective IFN-γ signaling and lack of the IFN-γ–mediated HLA class I inducibility. Even more important, impaired JAK2 expression significantly downregulated the constitutive mRNA and protein levels of HLA class I APM components despite a functional APM pathway, thereby providing a selective advantage to tumors. However, this was neither mediated by loss of the IFN-γ R, as Colo 857 cells express this receptor, nor by abnormalities of components of the HLA class I APM. The latter was confirmed by the induction of HLA class I APM molecules, which was accompanied by an upregulation of HLA class I surface expression in these cells on TNF-α and IFN-α treatment, respectively. In addition, JAK2 is required for IFN-γ–induced growth inhibition, as Colo 857 cells lacking JAK2 were not growth inhibited by IFN-γ in contrast to JAK2+ melanoma cells such as Colo 794. This loss of growth-restraining functions might influence tumor progression of JAK2− cells, which will be investigated in future studies.
To confirm the importance of a functional IFN-γ signaling for constitutive HLA class I APM component expression, JAK2 expression was restored in the JAK2− melanoma cells Colo 857 by stable transfection by using a JAK2-specific expression vector. JAK2 gene transfer into JAK2− Colo 857 cells increased the constitutive HLA class I APM component and surface antigen expression. Furthermore, functional JAK2 restored IFN-γ inducibility of HLA class I APM components. Thus, there exists a direct link between abnormalities of HLA class I antigen processing and presentation molecules and impaired JAK2 function. These might also result in reduced CTL sensitivity but increased susceptibility to natural killer cell–mediated lysis. Although a positive correlation between JAK2 and HLA class I antigens has been proven in this study for the first time, a recent publication has shown an improved patient survival when tumors expressed high MHC class I and STAT1 levels in association with a broad T-cell infiltrate (31). Furthermore, loss of STAT1 signaling has been shown to be associated with a higher incidence of tumors in mice (23, 32). These results strengthen our hypothesis of an important role of a functional IFN-γ signal cascade for the immunosurveillance of tumor cells.
Owing to the complexity of the IFN-γ signal transduction pathway, a comprehensive explanation how and at which level other elements of the IFN-γ system besides JAK2 and STAT1 modulate HLA class I APM component expression is still awaiting. Because JAK2 is a key regulator of IFN-γ responses and is induced by other growth factors and DNA damage, tumors acquiring resistance to IFN-γ by dysregulation or structural alterations of JAK2 might evade the immunosurveillance leading to tumor progression; vice versa, an impaired IFN-γ signaling in association with a reduced HLA class I APM component expression pattern suggests that defects in the IFN-γ cascade might play a crucial role in the malignant transformation process and might be involved in the frequent development of immune escape phenotypes caused by HLA class I APM component abnormalities. This is further supported by cDNA microarray analysis of different tumors showing an altered expression of IFN signaling molecules. In conclusion, the present study identified for the first time that a deletion of JAK2 in melanoma cells or inhibition of JAK2 signaling caused an impaired MHC class I APM component expression, suggesting that abnormalities of the JAK/STAT pathway might play an important role during development of melanoma. In addition, these results underscore the biological significance of JAK2 for IFN-γ treatment-independent immunosurveillance and provide evidence that defects in JAK2 might represent a novel immune escape mechanism and a potential target in combination with T-cell–based therapies. Further experiments are currently been carried out to validate these data in a large series of tumor lesions by using a tissue microarray and associate them with the survival of melanoma patients.
Supplementary Material
Translational Relevance.
Malignant transformation of cells is frequently associated with defects in HLA class I antigen processing machinery (APM), which plays a crucial role in the recognition of tumor cells by host immune system. The potential clinical relevance of these defects has stimulated interest in the molecular characterization of the underlying mechanisms, as this information may suggest targeted therapies to restore APM function. Multiple escape mechanisms have been identified and characterized. However, little information is available about defects in the IFN-γ signal transduction pathway in malignant cells despite the susceptibility of several APM components to modulation by IFN-γ. This study has identified for the first time loss or downregulation of JAK2 as a mechanism underlying HLA class I antigen downregulation in melanoma cells and lack of inducibility by IFN-γ. Furthermore, this information contributes to the design of strategies both to identify the mechanisms underlying defects in HLA class I antigen expression in malignant cells and to correct these defects.
Acknowledgments
Grant Support
This work was sponsored by DFG grant SE-581-9 and SE-581-11.1 and by PHS grants RO1CA104947 (to S. Ferrone), RO1CA110249 (to S. Ferrone), and PO1CA109688 (to S. Ferrone) awarded by the National Cancer Institute.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors have no conflicting financial interests.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
The article is part of the PhD thesis of Annedore Respa.
References
- 1.Amadori M. The role of IFN-alpha as homeostatic agent in the inflammatory response: a balance between danger and response? J Interferon Cytokine Res. 2007;27:181–189. doi: 10.1089/jir.2006.0110. [DOI] [PubMed] [Google Scholar]
- 2.Lesinski GB, Trefry J, Brasdovich M, Kondadasula SV, Sackey K, Zimmerer JM, et al. Melanoma cells exhibit variable signal transducer and activator of transcription 1 phosphorylation and a reduced response to IFN-alpha compared with immune effector cells. Clin Cancer Res. 2007;13:5010–5019. doi: 10.1158/1078-0432.CCR-06-3092. [DOI] [PubMed] [Google Scholar]
- 3.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seliger B, Ruiz-Cabello F, Garrido F. IFN inducibility of major histocompatibility antigens in tumors. Adv Cancer Res. 2008;101:249–276. doi: 10.1016/S0065-230X(08)00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thyrell L, Erickson S, Zhivotovsky B, Pokrovskaja K, Sangfelt O, Castro J, et al. Mechanisms of Interferon-alpha induced apoptosis in malignant cells. Oncogene. 2002;21:1251–1262. doi: 10.1038/sj.onc.1205179. [DOI] [PubMed] [Google Scholar]
- 6.Vannucchi S, Chiantore MV, Mangino G, Percario ZA, Affabris E, Fiorucci G, et al. Perspectives in biomolecular therapeutic intervention in cancer: from the early to the new strategies with type I interferons. Curr Med Chem. 2007;14:667–679. doi: 10.2174/092986707780059616. [DOI] [PubMed] [Google Scholar]
- 7.Rane SG, Reddy ES. Janus kinases: components of multiple signaling pathways. Oncogene. 2000;19:56662–56679. doi: 10.1038/sj.onc.1203925. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, Jove R. The STATs of cancer: new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 9.Marqués L, Brucet M, Lloberas J, Celada A. STAT1 regulates lipopolysaccharide- and TNF-alpha-dependent expression of transporter associated with antigen p processing 1 and low molecular mass polypeptide 2 genes in macrophages by distinct mechanisms. J Immunol. 2004;173:1103–1110. doi: 10.4049/jimmunol.173.2.1103. [DOI] [PubMed] [Google Scholar]
- 10.Rouyez MC, Lestingi M, Charon M, Fichelson S, Buzyn A, Dusanter-Fourt I. IFN regulatory factor-2 cooperates with STAT1 to regulate transporter associated with antigen processing-1 promoter activity. J Immunol. 2005;174:3948–3958. doi: 10.4049/jimmunol.174.7.3948. [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez T, Méndez R, Del Campo A, Aptsiauri N, Martín J, Orozco G, et al. Patterns of constitutive and IFN-gamma inducible expression of HLA class II molecules in human melanoma cell lines. Immunogenetics. 2007;59:123–133. doi: 10.1007/s00251-006-0171-9. [DOI] [PubMed] [Google Scholar]
- 12.Wong LH, Krauer KG, Hatzinisiriou I, Estcourt MJ, Hersey P, Tam ND, et al. Interferon-resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48-ISGF3gamma. J Biol Chem. 1997;272:28779–28785. doi: 10.1074/jbc.272.45.28779. [DOI] [PubMed] [Google Scholar]
- 13.Dovhey SE, Ghosh NS, Wright KL. Loss of interferon-gamma inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line. Cancer Res. 2000;60:5789–5796. [PubMed] [Google Scholar]
- 14.Rodríguez T, Méndez R, Del Campo A, Jiménez P, Aptsiauri N, Garrido F, et al. Distinct mechanisms of loss of IFN-gamma mediated HLA class I inducibility in two melanoma cell lines. BMC Cancer. 2007;7:34. doi: 10.1186/1471-2407-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bandoh N, Ogino T, Cho HS, Hur SY, Shen J, Wang X, et al. Development and characterization of human constitutive proteasome and immunoproteasome subunit-specific monoclonal antibodies. Tissue Antigens. 2005;66:185–194. doi: 10.1111/j.1399-0039.2005.00462.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Campoli M, Cho HS, Ogino T, Bandoh N, Shen J, et al. A method to generate antigen-specific mAb capable of staining formalin- fixed, paraffin-embedded tissue sections. J Immunol Methods. 2005;299:139–151. doi: 10.1016/j.jim.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Ogino T, Wang X, Kato S, Miyokawa N, Harabuchi Y, Ferrone S. Endoplasmic reticulum chaperone-specific monoclonal antibodies for flow cytometry and immunohistochemical staining. Tissue Antigens. 2003;62:382–393. doi: 10.1034/j.1399-0039.2003.00114.x. [DOI] [PubMed] [Google Scholar]
- 18.Stam NJ, Spits H, Ploegh HL. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J Immunol. 1986;137:2299–2306. [PubMed] [Google Scholar]
- 19.Sabatino M, Zhao Y, Voiculescu S, Monaco A, Robbins P, Karai L, et al. Conservation of genetic alterations in recurrent melanoma supports the melanoma stem cell hypothesis. Cancer Res. 2008;68:122–131. doi: 10.1158/0008-5472.CAN-07-1939. [DOI] [PubMed] [Google Scholar]
- 20.Hess G, Rose P, Gamm H, Papadileris S, Huber C, Seliger B. Molecular analysis of the erythropoietin receptor system in patients with polycythaemia vera. Br J Haematol. 1994;88:794–802. doi: 10.1111/j.1365-2141.1994.tb05119.x. [DOI] [PubMed] [Google Scholar]
- 21.Jung D, Hilmes C, Knuth A, Jaeger E, Huber C, Seliger B. Gene transfer of the co-stimulatory molecules B7-1 and B7-2 enhances the immunogenicity of human renal cell carcinoma to a different extent. Scand J Immunol. 1999;50:242–249. doi: 10.1046/j.1365-3083.1999.00588.x. [DOI] [PubMed] [Google Scholar]
- 22.Simon R. Microarray-based expression profiling and informatics. Curr Opin Biotechnol. 2008;19:26–29. doi: 10.1016/j.copbio.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 25.Huang S, Bucana CD, Van Arsdall M, Fidler IJ. Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene. 2002;21:2504–2512. doi: 10.1038/sj.onc.1205341. [DOI] [PubMed] [Google Scholar]
- 26.Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. Interferon: cellular executioner or white knight? Curr Med Chem. 2007;14:1279–1289. doi: 10.2174/092986707780597907. [DOI] [PubMed] [Google Scholar]
- 27.Kreis S, Munz GA, Haan S, Heinrich PC, Behrmann I. Cell density dependent increase of constitutive signal transducers and activators of transcription 3 activity in melanoma cells is mediated by Janus kinases. Mol Cancer Res. 2007;5:1331–1341. doi: 10.1158/1541-7786.MCR-07-0317. [DOI] [PubMed] [Google Scholar]
- 28.Jackson DP, Watling D, Rogers NC, Banks RE, Kerr IM, Selby PJ, et al. The JAK/STAT pathway is not sufficient to sustain the antiproliferative response in an interferon-resistant human melanoma cell line. Melanoma Res. 2003;13:219–229. doi: 10.1097/00008390-200306000-00001. [DOI] [PubMed] [Google Scholar]
- 29.Xiang Z, Zhao Y, Mitaksov V, Fremont DH, Kasai Y, Molitoris A, et al. Identification of somatic JAK1 mutations in patients with acute myeloid leukemia. Blood. 2008;111:4809–4812. doi: 10.1182/blood-2007-05-090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pansky A, Hildebrand P, Fasler-Kan E, Baselgia L, Ketterer S, Beglinger C, et al. Defective JAK-STAT signal transduction pathway in melanoma cells resistant to growth inhibition by interferon-alpha. Int J Cancer. 2000;85:720–725. doi: 10.1002/(sici)1097-0215(20000301)85:5<720::aid-ijc20>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 31.Simpson J, Al-Attar A, Watson N, Scholefield JH, Ilyas M, Durrant LG. Intratumoral T cell infiltration, MHC class I and STAT1 as biomarkers of good prognosis in colorectal cancer. Gut. 2010;59:926–933. doi: 10.1136/gut.2009.194472. [DOI] [PubMed] [Google Scholar]
- 32.Levy DE, Gilliland DG. Divergent roles of STAT1 and STAT5 in malignancy as revealed by gene disruptions in mice. Oncogene. 2000;19:2505–2510. doi: 10.1038/sj.onc.1203480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.