Abstract
This study was designed to characterize binding of a DNA aptamer to breast cancer cells and to test whether that aptamer could be used to kill target cells in vitro as part of an aptamer-C1q protein conjugate by coupling to the classic complement cascade. A biotinylated DNA aptamer designated MUC1-5TR-1 was shown to decorate the plasma membranes of human breast adenocarcinoma (MCF7) cells via fluorescence confocal microscopy. Biotinylated aptamer binding successfully initiated the classical complement pathway leading to complement fixation on the target cells via a streptavidin-C1q conjugate as previously reported. Förster Resonance Energy Transfer (FRET) measurements demonstrated membrane depolarization upon aptamer binding, providing indirect evidence of membrane attack complex (MAC) formation as a result of aptamer binding. Transmission electron microscopy (TEM) and immunogold labeling confirmed that aptamer-mediated complement fixation results in MAC formation on the plasma membrane, leading to osmotic swelling and cell death. This approach may provide a much less toxic and more precisely targeted “antibody-like” treatment for cancers by coupling to the patient's innate immune system in much the same way as more expensive humanized monoclonal antibodies.
Introduction
Aptamers are short (typically less than 100 bases in length) DNA or RNA oligonucleotides, which fold to bind specifically to diverse targets against which they are selected from random libraries based on affinity for a given target. Aptamers can be used as surrogates for antibodies in vitro (JAYASENA, 1999). In vivo, aptamers appear to be significantly less immunogenic than allogeneic or humanized antibodies (Pendergast et al., 2005; Bouchard et al., 2010), making them preferable to traditional chemotherapeutic or monoclonal antibody-based therapeutics, which can still exhibit residual immunogenicity and toxicity in the grafted complementarity determining regions sections of fully “humanized” antibodies (Harding et al., 2010). Aptamers can linger for significant periods in vivo to enhance their pharmacokinetics, if stabilized for use in serum to resist nucleases (JAYASENA, 1999; Dougan et al., 2000; Pendergast et al., 2005). Adding weight to aptamers in the form of polyethylene glycol or protein conjugation also enhances aptamer half-life in serum by slowing clearance through the kidneys and other organs (Dougan et al., 2000).
The potential advantages of using aptamers in place of antibodies have been reviewed extensively (JAYASENA, 1999; Pendergast et al., 2005; Bouchard et al., 2010). Aptamers have been used effectively for detection of specific tumor markers (Ferreira et al., 2006; Ferreira et al., 2008; Li et al., 2009). Aptamers have also been used for targeted delivery of various chemotherapeutic agents to cancer cells, which can, in some cases, take up aptamers carrying toxic payloads via endocytosis (Chu et al., 2006; Ferreira et al., 2009; Meng et al., 2012).
Most relevant to the present report is recent success involving aptamer-complement protein (C1q) bioconjugates to activate the classical complement pathway and lyse Gram negative bacteria (Bruno et al., 2008) and human breast adenocarcinoma (MCF7) cells (BRUNO, 2010). The classical pathway of complement activation results in the formation of membrane attack complexes (MAC) composed of the C5b, C6-C8, and a varying number of pore forming C9 proteins. The resultant pores in the plasma membrane allow passage of water, resulting in osmotic lysis of the target cell (Halulinen and Meri, 1998; Wang and Weiner, 2008).
The present study verifies and advances the seminal work of Bruno in aptamer-mediated complement activation targeted to MCF7 cells (BRUNO, 2010) by providing additional evidence of complement protein C9 deposition on target cells. In the present study, the aptamer-biotin-streptavidin-C1q conjugate mechanism of complement activation and ultimate cell lysis is validated by transmission electron microscopic (TEM) analysis of the effects of aptamer-complement treatment on target cell size as well as images of putative MAC-induced pores. The TEM evidence also correlates well with immunofluorescence microscopic evidence of MAC formation on the surface of MCF7 cells and Förster resonance energy transfer (FRET) evidence of membrane depolarization indicative of cell permeation and the kinetics thereof.
Materials and Methods
Aptamer-complement bioconjugate formation
The MUC1-5TR-1 aptamer, developed by the Ferreira et al. (2008), was chosen for this study due to its low dissociation constant of 47.3 nM and its previously reported ability to bind and kill MCF7 cells when coupled to the complement cascade (BRUNO, 2010). Aptamers were synthesized with 5′-biotin or 5′-Alexa Fluor 546 (AF546) modifications by Integrated DNA Technologies (IDT) using the MUC1-5TR-1 aptamer sequence: 5′-GGGAGACAAGAATAAACGCTCAAGAAGTGAAAATGACAGAACACAACATTCGACAGGAGGCTCACAACAGGC-3′.
Cell culture and experimental treatments
MCF7 cells, derived from a human breast adenocarcinoma, were purchased from and authenticated by American Type Culture Collection. Cells were used within 6 months of initial thawing and seeding of frozen cells. MCF7 cells were cultured in RPMI-1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 0.1% bovine insulin (Sigma Aldrich), 100 U/mL of penicillin and streptomycin and were incubated at 37°C in a 95% air, 5% CO2 humid atmosphere.
MCF7 cells were grown on square glass coverslips in culture dishes to an 80%–90% confluent layer. Cells were washed twice in 1× binding buffer (1× BB; 0.5 M NaCl, 10 mM Tris-HCl, and 1 mM MgCl2, in nuclease-free water, pH 7.5–7.6) to remove shed MUC1 (Chan et al., 1999; BRUNO, 2010), then exposed to 2.4 mL of fresh filter-sterilized culture media and 200 μL of MUC1-5TR-1 5′ biotinylated aptamer (1.5 mg/mL) for 5 minutes. This step was followed in the full test group by addition of 200 μL of streptavidin-C1q (∼ 0.58 mg/mL) for 10 minutes, followed by 200 μL of human serum complement proteins (HSCP; Sigma Chemical Co., 30 U/mL) for 3 hours. The complement control group received only 200 μL of HSCP and equal volumes of 1× BB to compensate for the biotinylated aptamer and streptavidin-C1q conjugate volumes, while the negative control group received only equal volumes of 1× BB. All treatment groups thusly had a total volume of 3 mL and were incubated at 37°C with 95% air, 5% CO2 humid atmosphere.
Immunofluorescence laser scanning confocal microscopy and immunogold TEM
Following various aptamer and complement treatments, the cells were washed twice in 1× BB for 5 minutes each and then fixed in methanol at −20°C for 1 minute. After drying, cells were washed 3 times in 1× BB for 10 minutes per wash, then blocked with 20% nonfat dry milk for 2 hours at room temperature. After 3 more washes in 1× BB at 10 minutes per wash, cells were incubated in 1:20 monoclonal mouse anti-human C9 antibody (United States Biological, Inc.) overnight at 4°C. After 3 more 10-minute washes with 1× BB, cells were blocked with 20% nonfat dry milk for 2 hours at room temperature, followed by another three 10-minute washes with 1× BB. The cells were then incubated with a 1:300 anti-mouse secondary antibody tagged with Cy5 (Invitrogen) for 2 hours at room temperature. Cells were again washed 3 times in 1× BB for 10 minutes per wash, incubated for 20 minutes with a 1:2,000 Hoechst nuclear stain, washed again, and imaged as described below.
For TEM, suitable ultrathin sections were processed for immunogold labeling by floating grids with their section sides facing down on drops of 20% nonfat powdered milk in 1× BB for 2 hours. The sections were washed 3 times for 15 minutes per wash with 1× BB and floated on a drop of solution containing anti-complement C9 monoclonal antibody (U.S. Biological, Inc.) at a 1:100 dilution overnight at 4°C in a moist chamber to prevent drying. The grids were then washed and floated for 2 hours on drops of 1× BB containing a 1:20 dilution of anti-mouse immunoglobulin G conjugated to 10 nm colloidal gold (Sigma Chemical Co.). After washing and allowing the grids to dry, the sections were re-imaged and suitable areas were photographed using a Gatan SC1000 ORIUS® CCD TEM digital camera and edited using Gatan's Digital Micrograph ™ and Adobe Photoshop 12.0.
FRET analysis
The voltage sensitive probes dimyristoylphosphatidylethanolamine (CC2-DMPE) as donor and bis-(1,3-diethylthiobarbituric acid)trimethine oxonol (DiSBAC2(3)) as acceptor were used to signal membrane depolarization. When the membrane depolarizes, the acceptor molecule moves across the cell membrane placing it too far from the donor molecule for FRET to occur, resulting in a decrease in acceptor signal. Both probes were from Invitrogen Corp. The CC2-DMPE donor was loaded at a concentration of ∼10 μM for 30 minutes in the dark at ∼25°C, and after a brief wash in culture medium, the DiSBAC2(3) acceptor was loaded at 10 μM, along with 250 μL of biotinylated aptamer (1.5 mg/mL) and 200 μL streptavidin-C1q conjugate. The dish containing the loaded MCF7 cells was placed on a 37°C stage of the FV1000 Laser Scanning Confocal Microscope (LSCM) and was excited only at the wavelength (405 nm) appropriate for the donor (CC2-DMPE). Images were acquired by collection of emitted light at 465 nm, characteristic emission of CC2-DMPE when uncoupled from DiSBAC2(3) and 580 nm, the characteristic emission of FRET-coupled CC2-DMPE - DiSBAC2(3).
Imaging was performed using a 256×256 pixel scan at a single 500-nm-thick focal plane that included the nucleus of most cells within the field of view. The dwell time was 20 μseconds/pixel, and laser intensity was 3% of maximum (17 mW) to minimize cell damage from photo-oxidation. Images were collected once per minute for 16 minutes and HSCP was added at the 7-minute mark. FRET results were determined by measuring pixel intensities from the MCF7 cell membranes simultaneously at both the donor and acceptor characteristic emission wavelengths. This measurement was accomplished and automated using ImageJ (National Institutes of Health; rsbweb.nih.gov/ij) by writing a macro that selected only the cell membranes as the region of interest and then reported pixel intensity per unit area at each of the respective wavelengths (as described in Weigum et al., 2010). Images were imported into ImageJ and fluorescence from the periphery of every cell in the field of view was measured at each time point. The experiment was repeated three times. Statistical analysis of the acceptor FRET response was done by comparing the pixel intensities/μm2 before and after addition of HSCP by single factor analysis of variance followed by the post-hoc Tukey honestly significant difference test for differences between means.
LSCM image preparation
Acquisition and primary processing of LSCM images were performed using the Olympus FV1000 system and software. Imaging was accomplished using an Olympus 20× water immersion lens, NA=0.95. For live cells, imaging was accomplished as described above for FRET analyses. Cells on coverslips were mounted in glycerol, cell side down, on slides and imaged with the same lens immersed in water. Image collection parameters were calibrated on the experimental treatment group of each experiment and then held constant for variations and control observations (e.g., Figs. 1 and 2). Formatting and labeling for publication was done using Adobe Photoshop CS5.1. Brightness and contrast were enhanced globally in illustrations showing fluorescence to make it more obvious in print media. Presentation of these images conforms to accepted guidelines for the ethical use of digital technology (CROMEY, 2010).
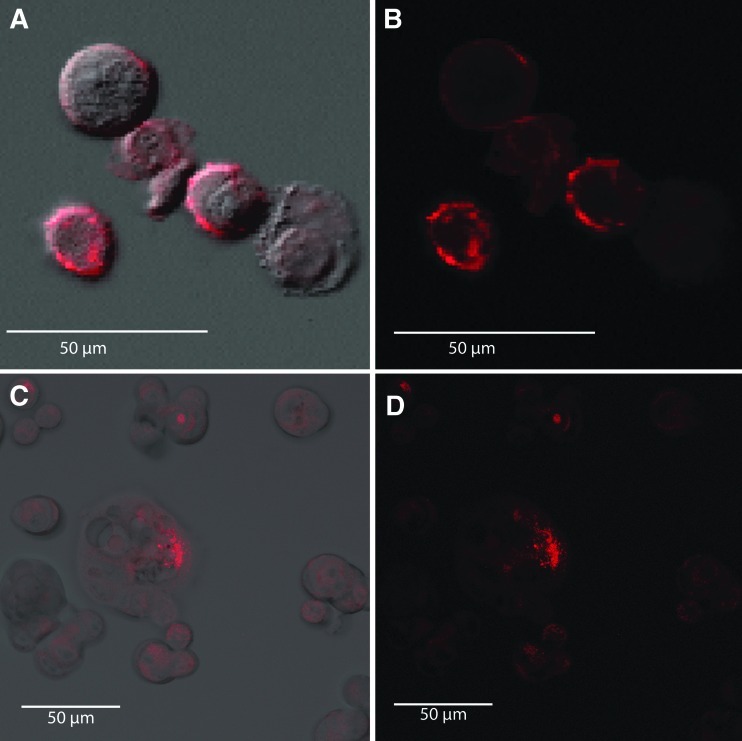
FIG. 1.
Live human breast adenocarcinoma (MCF7) cells treated with the AF546-labeled aptamer or 5′-biotinylated aptamer plus streptavidin-AF546. (A) merged fluorescence and differential interference contrast (DIC) microscopy; (B) shows only fluorescence microscopy of AF546-labeled aptamer cell membrane decoration within 5 minutes of aptamer addition. (C) and (D) live MCF7 cells treated with biotinylated aptamer followed by AF546-labeled streptavidin and demonstrating aptamer-dependent labeling of MCF7 cell membranes (C merged fluorescence and DIC; D, fluorescence only). Each image is a z-stack projection of ten 500-nm-thick optical sections.
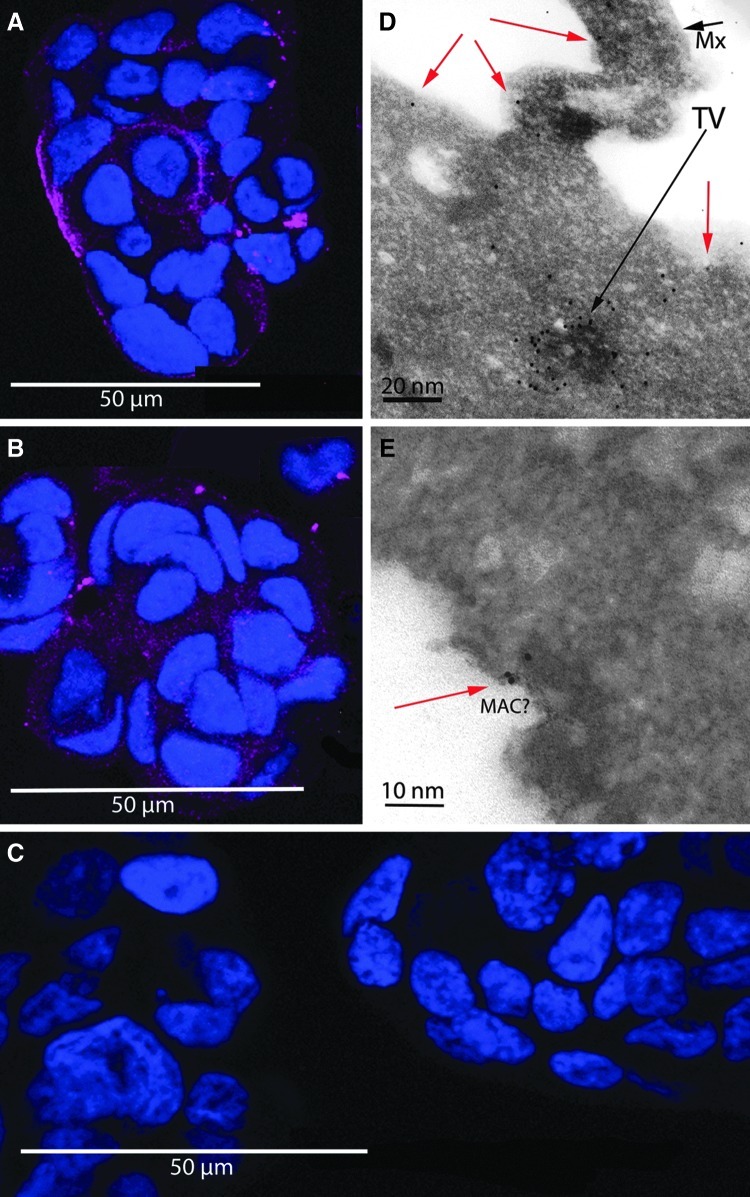
FIG. 2.
Live MCF7 cells exposed to biotinylated aptamers, streptavidin-C1q conjugate, and human serum complement proteins (HSCP). After fixation, cells were labeled with an anti-human C9 antibody and a Cy5-conjugated secondary (magenta) then counterstained with Hoechst nuclear stain (blue). Aptamer-biotin-streptavidin-C1q plus HSCP-treated cells were strongly decorated (A), while the complement control group, in which cells were not exposed to streptavidin-C1q conjugate but did experience HSCP exposure, was only moderately decorated (B). The negative control group, in which cells were not exposed to either streptavidin-C1q or HSCP, appeared devoid of anti-C9 binding (C). Each of the A, B, and, C image panels is a z-stack projection of ten 500-nm-thick optical sections. Panel D shows a transmission electron microscopy image of an immunogold anti-C9 stained ultrathin section of an MCF7 cell treated as described for panel A. The red arrows indicate gold particles on or near the cell surface. The black arrow labeled “TV” indicates a heavily anti-C9 labeled structure in the cytoplasm, possibly a transport vesicle (TV). The black arrow labeled “Mx” indicates a mucin exocytotic structure, characteristic of MCF7 cells. In panel E, the edge portion of an MCF7 cell, treated and stained as in panel D, containing a possible MAC complex is shown. The red arrow points to immunogold anti-C9 labeling in close proximity to the putative membrane attack complex pore as would be expected for authentic MAC pores.
Transmission Electron Microscopy
Transmission electron microscopy (TEM) was used to search for MAC pores, and images obtained were evaluated to analyze differences in cell swelling and MAC formation between treatment and control groups. Cells were detached from their dishes using trypsin-EDTA, pelleted by centrifugation at 14,000 rpm in a microfuge for 1 minute, and then resuspended in 3 mL of filter-sterilized complete culture medium containing FBS. All experimental treatments and controls were conducted as described previously.
Cells were then pelleted in 3% glutaraldehyde fixative in 0.05 M cacodylate buffer, pH 7.5, and fixed overnight at 4°C. Cells were washed 3 times for 15 minutes each in 0.05 M cacodylate buffer by centrifugal pelleting again without resuspension. Cells were then fixed with 1% OsO4 in 0.1 M cacodylate buffer, pH 7.5 for 3 hours and washed twice in 1× BB for 15 minutes each and once more overnight. Cells were dehydrated with two 30-minute washes of 70% ethanol. Cell pellets were embedded in London Resin White (LR White; Electron Microscopy Sciences) and 70% ethanol for 1 hour, followed by three 30-minute washes in 100% LR White followed by polymerization for 48 hours at 60°C.
Samples were cut into 70-nm sections using a Leica Reichert Ultracut S ultramicrotome with a Diatome diamond knife, and collected on 200 mesh, hexagonal nickel grids. Imaging was performed using a JEOL JEM-1200EX II TEM system. Images were captured by a Gatan SC1000 ORIUS® CCD TEM digital camera. Image editing was done using Gatans' Digital Micrograph™ and Adobe Photoshop 12.0.
To avoid bias in image acquisition and statistical analysis, a double-blind experiment was set up by re-labeling groups for image acquisition, and re-labeling again for analysis of the data. Care was taken to avoid counting any cell twice. A standard area was designated to use for cell counting across all grids, and only cells with a single visible nucleus were counted. To determine if the cell was swollen, low magnification images (7.5K, 10K, or 12K) were used. Swollen cells were recognized by criteria described by Bittar et al. (1975), Koke et al. (1976), and Folts et al. (1978) and were easily discernible by the low density of the cytoplasm and overall rounded morphology resulting in the typical low surface to volume ratio characteristic of swollen cells. Ambiguous cells were counted as not swollen. A single 200,000× image was taken of the membrane at 0°, 90°, 180°, and 270° angles from the nucleus to check for membrane breaks. Twenty cells from each experimental group were analyzed.
Results
LSCM visualization of aptamer-AF546 and aptamer-biotin-streptavidin-AF546 binding
To assess the characteristics of individual components of our experimental system, LSCM was used to detect AF546-labeled MUC1-5TR-1 aptamer and biotinylated aptamer plus AF546-streptavidin conjugate binding to MCF7 cells. First, live MCF7 cells were treated with the AF546-labeled aptamer, which decorated the membranes of cells within 5 minutes of addition (Fig. 1A, B). Live MCF7 cells were also treated with 5′-biotinylated aptamer followed by AF546-labeled streptavidin, which similarly decorated MCF7 cell membranes (Fig. 1C, D). Observations were made with and without the aid of differential interference contrast (DIC) microscopy (Fig. 1A, C). No AF546-streptavidin decoration was seen on MCF7 cells that were not first treated with biotinylated aptamer (image not shown).
Immunofluorescence and immunogold analysis of C9 deposition
Once the assembly of the aptamer-biotin-streptavidin complex on cell surfaces had been confirmed in culture, we next tested the system's ability to initiate the classical complement pathway using a streptavidin-C1q conjugate in place of the streptavidin-AF546 conjugate. Live MCF7 cells were exposed to biotinylated MUC1 aptamer, streptavidin-C1q conjugate, and HSCP. After fixation, cells were labeled with an anti-human C9 antibody and a Cy5-conjugated secondary antibody. Aptamer-biotin-streptavidin-C1q plus HSCP-treated cells were strongly decorated (Fig. 2A) while the complement control group, in which cells were not exposed to streptavidin-C1q conjugate, but did experience HSCP exposure, was only moderately decorated (Fig. 2B). The negative control group, in which cells were not exposed to either streptavidin-C1q or HSCP, was minimally decorated (Fig. 2C). TEM imaging of immunogold anti-C9 stained ultrathin sections of MCF7 cells treated experimentally confirmed anti-C9 labeling consistent with membrane localization of C9 (Fig. 2D) and may have revealed MAC profiles (Fig. 2E). These results were consistent with the hypothesis that aptamer-biotin-streptavidin-C1q recruited C9 to the cell surface and greatly enhanced MAC formation over the levels of MAC formation due to nonspecific binding of C9 from HSCP treatment.
Förster resonance energy transfer analysis
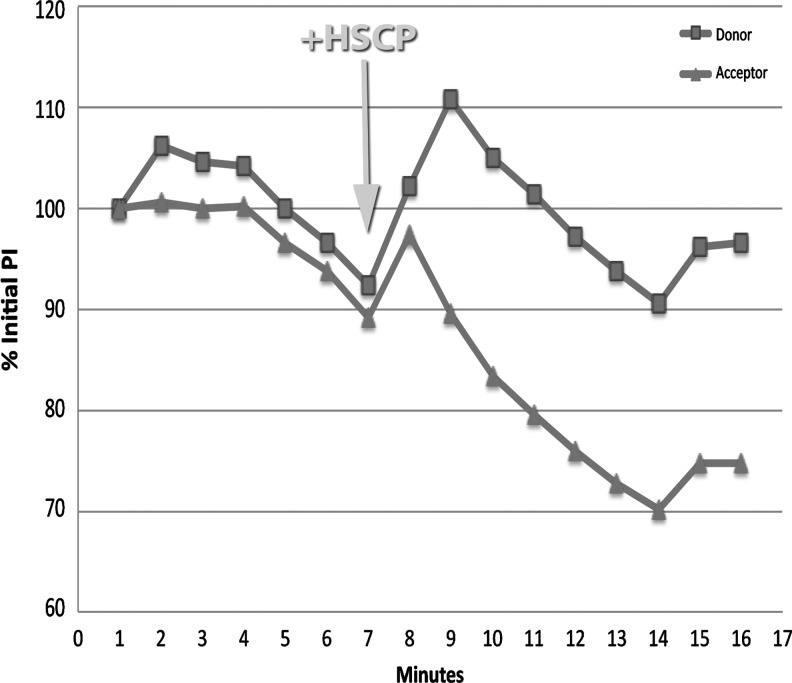
Addition of HSCP to cells treated with biotinylated aptamer and streptavidin-conjugated C1q caused an immediate (<1 minute) FRET response. There was an initial increase in both donor and acceptor fluorescence, but after 1 minute, FRET radiation from the acceptor declined dramatically and continued to decline for 6 minutes, becoming stable for the remaining 3 minutes of observation. Comparison of acceptor fluorescence before and after addition of HSCP showed a substantial and significant decrease (P=0.00013; Fig. 3).
FIG. 3.
Graph showing the effects on Förster resonance energy transfer (FRET) due to addition of HSCP to cells treated with biotinylated aptamer and streptavidin-conjugated C1q. The cells were preloaded with donor and acceptor molecules that FRET-pair across a polarized cell membrane as described in Methods. HSCP was added at minute 7 as indicated by the arrow. There was an initial increase in both donor and acceptor fluorescence intensity (% of initial pixel intensity on the y-axis), but 1 minute after HSCP addition, FRET radiation from the acceptor declined dramatically and continued to decline for 6 minutes, becoming stable for the remaining 3 minutes of observation. Comparison of acceptor fluorescence before and after addition of HSCP showed a substantial and significant decrease (P=0.00013).
Morphological analysis of MAC formation and cell swelling by TEM
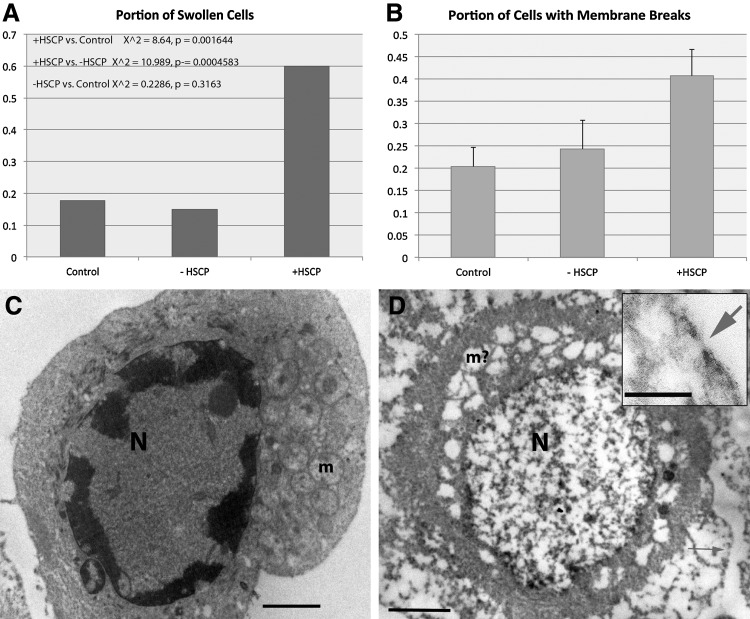
MCF7 cells fixed after treatment with or without HSCP were imaged using TEM. Two categorical variables were collected for each cell. Each cell was classified as either swollen (representative of potential osmotic lysis) or not swollen and additionally classified as having small breaks in the plasma membrane (potential MACs) or no breaks in the plasma membrane. The first analysis was to determine if there was an association between these 2 variables within each group: full treatment, HSCP only control, and an untreated negative control. A Fisher's Exact Test was also used to analyze the significance of the association between swollen cells and membrane breaks, and only the full treatment group was found to have significantly associated variables (p<0.005). An overall comparison of all the cells from each group using a Fisher's Exact Test was also used and the degree of association between swollen cells and membrane breaks was also found to be significant (p<0.0001). Representative images and results are shown in Fig. 4.
FIG. 4.
(A) Graph showing portion of swollen MCF7 cells as a function of no treatment (control), full aptamer treatment in the absence (−) and presence (+) of HSCP. Statistics were performed as described in Methods. (B) Graph showing the frequency of membrane breaks in the same categories as A. Error bars represent standard error of the mean. N=12 for each category. (C) MCF7 cell, −HSCP, categorized as not swollen. “N” indicates the nucleus, and “m” mitochondria. The scale bar indicates 25 μm. (D) Swollen MCF7 cell (+HSCP). “N” indicates the nucleus and “m?” indicates what may have been mitochondria. The arrows in D indicate membrane breaks. The scale bar in D indicates 25 μm, and in the inset, a 20-nm scale bar.
Discussion
We have demonstrated the ability of an aptamer-streptavidin-C1q bioconjugate to initiate the classical complement pathway, resulting in the formation of putative membrane attack complexes (MAC) on the surfaces of MCF7 cancer cells. That fact makes this work one of the few extant demonstrations of non-antibody (i.e., DNA aptamer-biotin-streptavidin-C1q complex) triggering of the classical complement cascade (Bruno et al., 2008; BRUNO, 2010). The interaction between MUC1-5TR-1 aptamer and the mucin 1 protein has demonstrated very high affinity (Kd=47.3 nM; Ferreira et al., 2008). In addition, mucin 1 is a dramatically overexpressed proteoglycan in breast cancers (Ferreira et al., 2006), making it a somewhat attractive target for aptamer or antibody binding and complement-mediated lysis in vivo, if non-target cells could remain relatively unaffected.
It is noteworthy that Ferreira et al. (2009) also discovered MUC1-5TR-1 aptamer internalization in MCF7 cells, which should work against complement-mediated lysis by removing at least some of the aptamer-C1q complexes from the cell surface. In another aptamer-target cell culture system which our group has studied (TLS11a aptamer and MEAR murine hepatoma cells, Meng et al., 2012), aptamer endocytosis appeared to be sufficient to essentially defeat complement activation in vitro, presumably because insufficient amounts of the aptamer-biotin-streptavidin-C1q remained on the cell surface long enough to significantly activate the complement pathway. However, the presently reported work and Bruno's original work (BRUNO, 2010) suggest that enough aptamer-biotin-streptavidin-C1q is present on the abundant mucin layer of MCF7 cells to induce cell lysis in the MCF7 system. Shedding of mucin into the culture medium or in vivo (Chan et al., 1999) as well as membrane complement regulatory proteins (mCRPs, e.g., CD59; Farkas et al., 2002; Donin et al., 2003; Yan et al., 2008) are also mechanisms by which complement-mediated lysis is limited by cancer cells and normal cells. Shedding is easily overcome in vitro by washing cells immediately prior to the addition of aptamer conjugate and HSCP to prevent aptamer blockade in solution.
The family of mCRPs normally keep complement fixation in check. Because the aptamer-C1q complex specifically activates and amplifies the classical pathway in a dose-dependent fashion (BRUNO, 2010), we expected it to largely overcome any regulatory protein's influence. Strong C9 recruitment to the MCF7 target cell membrane as evidenced by anti-C9 immunofluorescence labeling in our experimental group (Fig. 2A) supports the hypothesis that mCRPs can be overwhelmed. Other investigations aimed at specifically binding and inhibiting mCRPs (Hakulinen and Meri, 1998; Jurianz et al., 1999) demonstrated augmentation of antibody-activated complement-mediated cell killing rates. We are currently planning to overcome the effects of major mCRPs such as CD59 with Fab or F(ab′)2 antibody fragments (i.e., removal of the Fc tail to prevent complement activation due to surface-fixed antibodies on mCRPs) or new aptamers directed against these mCRPs in conjunction with anti-MUC 1 aptamers. The combined anti-mCRP and MUC 1 aptamer cocktail strategy should knock out most of the cancer cell's surface defenses and enable greater than 50% cell death, which was the functional limit that Bruno (2010) first reported for in vitro killing of MCF7 cells even with high doses of the aptamer-C1q plus HSCP when mCRPs were presumably active.
While we have found TEM evidence of putative MAC pore formation of the correct dimensions (∼15 nm deep and 10 nm wide) on the surface of aptamer-C1q complex plus HSCP-treated cells in close proximity to some anti-C9-colloidal gold particles (red arrow in Fig. 2E) as would be expected for real MAC pores, these lesions were not as abundant as might have been expected, but rather rare and relatively difficult to find. Part of the explanation for observing few MAC lesions might be related to the low probability of catching tiny pores amidst ultrathin sections by TEM. The paucity of MAC pores may also be partly due to the competing effect of aptamer internalization by endocytosis (Ferriera et al., 2009) as mucin is cycled into the target cells and takes bound aptamer complexes along with it.
Complement fixation on the target cell membrane positively feeds back on the pathway, causing more complement fixation on the cell surface. Thus, low levels of nonspecific HSCP binding to cells even in the absence of the aptamer-biotin-streptavidin-C1q bioconjugate, leads to some baseline level of cell mortality. Therefore, we expected the small amount of C9 immunolabeling with concomitant low mortality observed in our complement control group. The minimal baseline complement activation due only to HSCP treatment is also why we expected to see a response in our control group when using FRET to detect membrane depolarization. As the treatment group contained the aptamer bioconjugate for classical complement pathway activation, the stronger FRET response which we observed in the full treatment group met our expectations as well.
We found similar dead to live cell ratios as previously reported by Bruno (2010), but the results presented here suggest a somewhat higher dead to live cell ratio of approximately 60%. Further increases in the dead to live ratio might be achieved by a cocktail of aptamer-C1q complexes targeting different surface bound tumor markers individually or in concert with the MUC1-5TR-1 aptamer, along with inhibition of some mCRPs present on MCF7 cells. The mCRP CD59 seems to be a likely target, as it directly interferes with the formation of membrane attack complexes by blocking C9 recruitment when it binds to the C5b-8 complex (Farkas et al., 2002).
The broader implications of utilizing a nonimmunogenic nucleic acid aptamer conjugate to emulate antibodies as a means of activating the classical complement immune response and providing a targeted cancer treatment, should not be understated. Humanized monoclonal antibodies such as Herceptin are quite effective, but they are expensive (Barrett et al., 2006). Aptamer-C1q or Fc bioconjugatges could be used in place of humanized monoclonal antibodies that can exhibit adverse side effects despite being fully “humanized” (Harding et al., 2010). As an example, the U.S. Food and Drug Administration recently revoked approval for use of Avastin (bevacizumab) against breast cancer due to its potentially serious side effects. Aptamer-C1q or Fc bioconjugates which structurally mimic antibodies, may provide a means of specifically guiding the body's innate immune system to lyse and remove cancerous tumors in a natural way at much reduced cost if mass produced (Bruno and Crowell, 2008; Bruno and Miner, 2011). Furthermore, aptamers are easily and quickly selected, meaning personalized treatments could be rapidly and inexpensively developed from biopsies of a patient's specific cancer phenotype (Li et al., 2009), especially if the cancer mutates and expresses new surface markers.
Because mucin is expressed on a wide variety of cell types in vivo, it is not clear yet if the MUC1 aptamer family could ever be used clinically. New aptamers against highly cancer-specific molecules on the surface of target cells are needed if we are to unleash the powerful lytic effects of the aptamer-C1q or aptamer-Fc conjugate-directed complement cascade in vivo with confidence in the specificity of this new brand of “magic bullet.”
The present study provided an important validation of Bruno's initial work (BRUNO, 2010) and solidified the preliminary finding of aptamer-C1q-mediated complement activation with confocal and TEM evidence. However, these same studies suggest that more information on aptamer specificity and binding kinetics as well as the compensatory effects of mCRPs and endocytosis is needed before aptamer conjugates can eventually find their way to clinical cancer trials.
Acknowledgments
The authors gratefully acknowledge the assistance of Dr. Shannon Weigum with image analysis using ImageJ. This study was supported by internal funding from Operational Technologies Corp. and a National Science Foundation MRI award (NSF DBI 0821252) to JK and DG.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- BARRETT A. ROQUES T. SMALL M. SMITH R.D. How much will Herceptin really cost? Br. Med. J. 2006;333:1118–1120. doi: 10.1136/bmj.39008.624051.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BITTAR N. KOKE J.R. BERKOFF H.A. KAHN D.R. Histochemical and ultrastructural changes in human myocardial cells after cardiopulmonary bypass. Circulation. 1975;52:16–25. [PubMed] [Google Scholar]
- BOUCHARD P.R. HUTABARAT R.M. THOMPSON K.M. Discovery and development of therapeutic aptamers. Ann. Rev. Pharmacol. Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- BRUNO J.G. Aptamer-biotin-streptavidin-C1q complexes can trigger the classical complement pathway to kill cancer cells. In vitro Cell. Dev. Biol. Animal. 2010;46:107–113. doi: 10.1007/s11626-009-9257-7. [DOI] [PubMed] [Google Scholar]
- BRUNO J.G. CARRILLO M.P. PHILLIPS T. In vitro antibacterial effects of antilipopolysaccharide DNA aptamer-C1qrs complexes. Folia Microbiol. (Praha) 2008;53:295–302. doi: 10.1007/s12223-008-0046-6. [DOI] [PubMed] [Google Scholar]
- BRUNO J.G. CROWELL R. Selective glutaraldehyde-mediated coupling of proteins to the 3'-adenine terminus of polymerase chain reaction products. J. Biomol. Tech. 2008;19:177–183. [PMC free article] [PubMed] [Google Scholar]
- BRUNO J.G. MINER J.C. Therapeutic Nucleic Acid-3'–Conjugates. 2011. U.S. Patent No. 7,910,297.
- CHAN A.K. LOCKHART D.C. VON BERNSTORFF W. SPANJAARD R.A. JOO H.G. EBERLEIN T.J. GOEDEGEBUURE P.S. Soluble MUC1 secreted by human epithelial cancer cells mediates immune suppression by blocking T-cell activation. Int. J. Cancer. 1999;82:721–726. doi: 10.1002/(sici)1097-0215(19990827)82:5<721::aid-ijc16>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- CHU T.C. MARKS J.W. LAVERY L.A. FAULKNER S. ROSENBLUM M.G. ELLINGTON A.D. LEVY M. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- CROMEY D.W. Avoiding twisted pixels: ethical guidelines for the appropriate use and manipulation of scientific digital images. Sci. Eng. Ethics. 2010;16:639–667. doi: 10.1007/s11948-010-9201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONIN N. JURIANZ K. ZIPOREN L. SCHULTZ S. KIRSCHFINK M. FISHELSON Z. Complement resistance of human carcinoma cells depends on membrane regulatory proteins, protein kinases and sialic acid. Clin. Exp. Immunol. 2003;131:254–263. doi: 10.1046/j.1365-2249.2003.02066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOUGAN H. LYSTER D.M. VO C.V. STAFFORD A. WEITZ J.I. HOBBS J.B. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl. Med. Biol. 2000;27:289–297. doi: 10.1016/s0969-8051(99)00103-1. [DOI] [PubMed] [Google Scholar]
- FARKAS I. BARANYI L. ISHIKAWA Y. OKADA N. BOHATA C. BUDAI D. FUKUDA A. IMAI M. OKADA H. CD59 blocks not only the insertion of C9 into MAC but inhibits ion channel formation by homologous C5b-8 as well as C5b-9. J. Physiol. 2002;539:537–545. doi: 10.1113/jphysiol.2001.013381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERREIRA C.S. MATTHEWS C.S. MISSAILIDIS S. DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumour Biol. 2006;27:289–301. doi: 10.1159/000096085. [DOI] [PubMed] [Google Scholar]
- FERREIRA C.S. PAPAMICHAEL K. GUILBAULT G. SCHWARZACHER T. GARIEPY J. MISSAILIDIS S. DNA aptamers against the MUC1 tumour marker: design of aptamer-antibody sandwich ELISA for the early diagnosis of epithelial tumours. Anal. Bioanal. Chem. 2008;390:1039–1050. doi: 10.1007/s00216-007-1470-1. [DOI] [PubMed] [Google Scholar]
- FERREIRA C.S.M. CHEUNG M.C. MISSAILIDIS S. BISLAND S. GARIEPY J. Phototoxic aptamers selectively enter and kill epithelial cancer cells. Nucl. Acids Res. 2009;37:866–876. doi: 10.1093/nar/gkn967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOLTS J.D. SHUG A.L. KOKE J.R. BITTAR N. Protection of the ischemic myocardium with L-carnitine. Amer. J. Cardiol. 1978;41:1209–1214. doi: 10.1016/0002-9149(78)90877-9. [DOI] [PubMed] [Google Scholar]
- HALULINEN J. MERI S. Complement-mediated killing of microtumors in vitro. Am. J. Pathol. 1998;153:845–855. doi: 10.1016/S0002-9440(10)65626-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARDING F.A. STICKLER M.M. RAZO J. DUBRIDGE R.B. The immunogenicity of humanized and fully human antibodies: Residual immunogenicity resides in the CDR regions. MAbs. 2010;2:256–265. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAYASENA S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- JURIANZ K. MASLAK S. GARCIA-SCHÜLER H. FISHELSON Z. KIRSCHFINK M. Neutralization of complement regulatory proteins augments lysis of breast carcinoma cells targeted with rhumAb anti-HER2. Immunopharmacol. 1999;42:209–218. doi: 10.1016/s0162-3109(99)00006-5. [DOI] [PubMed] [Google Scholar]
- KOKE J.R. SHUG A.L. FOLTS J.D. BITTAR N. Ultrastructural and physiological changes induced in the canine myocardium by atractyloside. Cytobios. 1976;17:211–229. [PubMed] [Google Scholar]
- LI S. XU H. DING H. HUANG Y. CAO X. YANG G. LI J. XIE Z. MENG Y. LI X. ZHAO Q. SHEN B. SHAO N. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J. Pathol. 2009;218:327–336. doi: 10.1002/path.2543. [DOI] [PubMed] [Google Scholar]
- MENG L. YANG L. ZHAO X. ZHANG L. ZHU H. LIU C. TAN W. Targeted delivery of chemotherapy agents using a liver cancer-specific aptamer. PLoS One. 2012;7:e33434. doi: 10.1371/journal.pone.0033434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PENDERGAST P.S. MARSH H.N. GRATE D. HEALY J.M. STANTON M. Nucleic acid aptamers for target validation and therapeutic applications. J. Biomolec. Techniques. 2005;16:224–234. [PMC free article] [PubMed] [Google Scholar]
- WANG S.Y. WEINER G. Complement and cellular cytotoxicity in antibody therapy of cancer. Expert Opin. Biol. Ther. 2008;8:759–768. doi: 10.1517/14712598.8.6.759. [DOI] [PubMed] [Google Scholar]
- WEIGUM S.E. FLORIANO P.N. REDDING S.W. YEH C.K. WESTBROOK S.D. MCGUFF H.S. LIN A. MILLER F.R. VILLARREAL F. ROWAN S.D. VIGNESWARAN N. WILLIAMS M.D. MCDEVITT J.T. Nano-bio-chip sensor platform for examination of oral exfoliative cytology. Cancer Prev. Res. (Phila) 2010;3:518–528. doi: 10.1158/1940-6207.CAPR-09-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN J. ALLENDORF D.J. LI B. YAN R. HANSEN R. DONEV R. The role of membrane complement regulatory proteins in cancer immunotherapy. Adv. Exp. Med. Biol. 2008;632:159–174. [PubMed] [Google Scholar]