Abstract
Transplantation of acute myeloid leukemia (AML) patients with grafts from related haploidentical donors has been shown to result in a potent graft-versus-leukemia effect. This effect is mediated by NK cells because of the lack of activation of inhibitory killer cell immunoglobulin-like receptors (KIRs) which recognize HLA-Bw4 and HLA-C alleles. However, conflicting results have been reported about the impact of KIR ligand mismatching on the outcome of unrelated HLA-mismatched hematopoietic stem cells transplants (HSCT) to leukemic patients. The interpretation of these conflicting results is hampered by the scant information about the level of expression of HLA class I alleles on leukemic cells, although this variable may affect the activation of inhibitory KIRs. Therefore in the present study, utilizing a large panel of human monoclonal antibodies we have measured the level of expression of HLA-A, -B and -C alleles on 20 B-chronic lymphoid leukemic (B-CLL) cell preparations, on 16 B-acute lymphoid leukemic (B-ALL) cell preparations and on 19 AML cell preparations. Comparison of the level of HLA class I antigen expression on leukemic cells and autologous normal T cells identified selective downregulation of HLA-A and HLA-B alleles on 15 and 14 of the 20 B-CLL, on 2 and 5 of the 16 B-ALL and on 7 and 11 of the 19 AML patients tested, respectively. Most interestingly HLA-C alleles were markedly downregulated on all three types of leukemic cells; the downregulation was most pronounced on AML cells. The potential functional relevance of these abnormalities is suggested by the dose-dependent enhancement of NK cell activation caused by coating the HLA-HLA-Bw4 epitope with monoclonal antibodies on leukemic cells which express NK cell activating ligands. Our results suggest that besides the HLA and KIR genotype, expression levels of KIR ligands on leukemic cells should be included among the criteria used to select the donor-recipient combinations for HSCT.
Keywords: NK cells, HLA class I molecules, Killer cell immunoglobulin-like receptors, Leukemia, Flow cytometry, Hematopoietic stem cell transplantation
Introduction
Transplantation of acute myeloid leukemia (AML) patients with grafts from related haploidentical donors has been shown to result in a potent graft-versus-leukemia effect [1–3]. This effect is mediated by NK cells which are activated because of the lack of interaction of inhibitory killer cell immunoglobulin-like receptors (KIRs) with their ligands HLA-Bw4 and HLA-C alleles. HLA-B molecules containing the serologically defined HLA-Bw4 epitope determined by the amino acids at positions 77–83 interact with the inhibitory KIR3DL1 [4]. HLA-C antigens are the main ligands for the inhibitory KIRs, the specificity of the interaction being determined by a genetic polymorphism at positions 77 and 80. Group 2 HLA-C molecules (HLA-C2) which have Asp at position 77 and Lys at position 80 are bound by KIR2DL1, while group 1 HLA-C molecules (HLA-C1), which have Ser at position 77 and Asp at position 80, are bound by KIR2DL2 and KIR2DL3 [5–7]. However, conflicting results have been reported about the impact of the interactions of KIRs with their ligands HLA-Bw4 and HLA-C alleles on the outcome of unrelated HLA-mismatched hematopoietic stem cell transplants (HSCT) to leukemic patients [8–12]. Heterogeneity in the treatment procedures (conditioning regimen, graft composition and post-transplant immune suppression) and in the patient population (disease, risk category) may account for this discrepancy. However, the interpretation of these conflicting results is hampered by the scant and conflicting information about the level of HLA class I allele expression on leukemic cells, although this variable may affect the activation of inhibitory KIRs.
To the best of our knowledge, HLA class I antigen expression has been investigated only on acute leukemic cells [13–17] and on B lineage chronic lymphoid leukemic (B-CLL) cells [18]. Staining of 49 newly diagnosed and 5 relapsed AML and 13 B-CLL samples with mAbs recognizing monomorphic determinants of HLA class I antigens did not detect defects in their expression in most of the samples [13, 14, 18]. However, we cannot exclude HLA class I allele downregulation or loss in these samples, since these defects are not detected by the mAb used. In addition, a low frequency of HLA-A allele downregulation has been recently described in leukemic samples analyzed at initial diagnosis with a few mAb which recognize HLA class I allospecificities and mAb which recognize monomorphic determinants [17]. In contrast, testing of nine AML and six B-ALL samples with allo-antisera detected downregulation of allospecificities in about 60% of the samples analyzed [14]. Furthermore, we have also found that 35% of HLA-A alleles and 38% of HLA-B alleles were downregulated in 32 leukemic patients in comparison to their autologous T cells [16].
The paucity of information about HLA class I antigen expression on leukemic cells has a negative impact on our ability to optimize the selection of donor-recipient combinations for HSCT. Therefore in the present study taking advantage of a large panel of human mAb which recognize HLA class I allospecificities [19, 20], we have analyzed the HLA-A, -B and -C allele expression level on leukemic cells. Furthermore, we have compared the HLA class I allele expression level on myeloid and lymphoid leukemic cells, since NK cells have been shown to inhibit the growth of AML cells, but to have no detectable effect on that of acute lymphoid leukemic (ALL) cells [1, 2]. Last, to assess the functional significance of HLA class I allele downregulation on leukemic cells, we have investigated the effect of the HLA class I allele expression level on leukemic cells on their susceptibility to lysis by allogeneic NK cells.
Materials and methods
Patients and healthy volunteers
After informed consent, peripheral blood was collected with EDTA and citrate from 55 consecutive leukemic patients at the Department of Hemato-Oncology of the UZ Brussel and UZ Gent (Brussel and Gent, Belgium), respectively, between March 2003 and November 2006. The study protocol was approved by the Ethical Committee of the UZ Brussel.
The leukemic patients included 19 with AML (10 males and 9 females, with a median age of 61.5 years and range of 28–82 years), 16 with B-ALL (7 males and 9 females with a median age of 23.5 years and range of 1–72 years) and 20 with B-CLL (11 males and 9 females with a median age of 71.0 years and range of 42–93 years). Fourteen of the 19 AML patients were newly diagnosed patients; four patients represented a progression of refractory anemia with excess of blasts (RAEB) to AML with more than 20% blasts and one was a relapse. The latter five patients received neither chemo- nor radio-therapy at the time of blood collection for this study. However, two patients with a progression of RAEB to AML had received chemotherapy previously. All patients with B-ALL were newly diagnosed patients who had received no therapy. Blast counts in peripheral blood ranged from 9 to 96% (median 45%) in the AML patients and from 14 to 89% (median 53%) in the B-ALL patients. Finally, follow-up samples were taken from all B-CLL patients, except one; malignant B cells in peripheral blood ranged from 36 to 99% (median 81%).
Peripheral blood was also collected from 29 unrelated, randomly selected Belgian Caucasoids apparently healthy volunteers (HV) (9 males and 20 females, with a median age of 37.0 years and range of 22–54 years) after informed consent.
Peripheral blood mononuclear cells (PBMC) were isolated from citrate and EDTA peripheral blood by Ficoll-Hypaque density gradient centrifugation.
Cell lines
The AML ML2 cell line, the erythroleukemia K562 cell line, the monoblastic leukemia MONOMAC-6 cell line, and the T-ALL RPMI 8402 cell line were cultured in RPMI 1640 medium (Invitrogen Life technologies, Merelbeke, Belgium) supplemented with 1% l-glutamine (Invitrogen Life technologies), 1% penicillin–streptomycin (Invitrogen Life technologies) and 10% fetal calf serum (A&E Scientific, Marcq, Belgium).
Antibodies
The human mAb included the HLA-Cw1-specific mAb VP6G3 (an IgM) [20]; the mAb WK4C11 (an IgM), which recognizes a determinant shared by HLA-Cw1, -Cw3, -Cw4, -Cw*0801, -Cw*1202 and -Cw*1402 alleles; the mAb TRA2G9 (an IgM), which recognizes a determinant shared by HLA-Cw1, -Cw3, -Cw4, and -Cw*1402 alleles [20]; the mAb MUS4H4 (an IgG1), which recognizes a determinant shared by HLA-A24, -A25, -A32 and -HLA-Bw4 alleles; the mAb VDK8F7 (an IgM), which recognizes a determinant shared by HLA-A23, A24 and HLA-Bw4 alleles, but not by B51, B53, B13, B63, and B49; the HLA-Bw6-specific mAb OUW4F11 (an IgG1), and 30 allele-specific HLA mAbs. All these mAbs were developed and characterized by Dr. A. Mulder and most of them have been described elsewhere [16, 19, 20]. FITC-conjugated F(ab′)2 fragments of rabbit anti-human IgM and IgG antibodies were purchased from DakoCytomation (Heverlee, Belgium).
The mouse mAbs included the PE-anti-CD3 mAb SK7 (an IgG1), the PE-anti-CD19 mAb 4G7 (an IgG1), the PE-anti-CD34 mAb 8G12 (an IgG1), the Cy5-anti-CD107a mAb H4A3 (an IgG1), and the PE-anti-CD56 mAb B159 (an IgG1) which were all purchased from BD Pharmingen, (Erembodegem, Belgium); the RPE-Cy5-anti-CD3 mAb S4.1 (an IgG2a), the PE-anti-CD45 mAb F10-89-4 (an IgG2a), and the anti-CD112 mAb R2.525 (an IgG1) which were all purchased from Serotec (Oxford, United Kingdom); the PE-anti-CD19 mAb B19 (an IgG2a), which was purchased from Chemicon (Heule, Belgium); the FITC-anti-CD54 mAb 84H10 (an IgG1) and the FITC-anti-CD48 mAb J4-57 (an IgG1), which were both purchased from Beckman Coulter (Suarlée, Belgium); and the anti-CD155 mAb 300907 (an IgG1), the anti-MICA/B mAb 159207(an IgG2a), the anti-ULBP1 mAb 170818 (an IgG2a), the anti-ULBP2 mAb JQE01 (an IgG2a) and the anti-ULBP3 mAb JFY02 (an IgG2a), which were all purchased from R&D Systems (Abingdon, United Kingdom). FITC-conjugated F(ab′)2 fragments of goat anti-mouse IgG1 and IgG2a antibodies were purchased from ImTech Diagnostics (Antwerpen, Belgium).
Serologic HLA typing
The HLA-A, -B and -C phenotype was determined by testing PBMC with HLA typing trays (Biotest Seralc, Ternat, Belgium) in the complement dependent lymphocytotoxicity assay. This assay was performed by an experienced technician ensuring reproducibility and reliability of the results. In case of serological results compatible with HLA-A, -B or -C homozygosity of leukemic patients and HV, additional molecular typing was done utilizing a reverse hybridization technique (InnoLiPA, Innogenetics, Belgium) following the manufacturer’s instructions.
Flow cytometry quantification of HLA class I molecules
Peripheral blood mononuclear cells were prestained for 20 min at room temperature with PE- or Cy5-conjugated anti-CD mAbs to distinguish normal T cells from leukemic cells in patients and from normal B cells in HV. Cell surface staining of cells was performed as described. Briefly, unfixed cells (5 × 105) were incubated for 25 min at room temperature with 25 μl of a human mAb preparation (10 μg/ml). Following two washings with phosphate-buffered saline (PBS) supplemented with an optimal amount of bovine serum albumin (BSA) (A&E Scientific, Marcq, Belgium), an optimal amount of FITC-anti-human IgM or IgG antibodies was added and incubation was continued for an additional 25 min at room temperature. Negative controls included samples incubated with an isotype-matched mAb recognizing a HLA class I allele not expressed on the sample being tested and cells incubated only with the secondary labeled mAb. Samples were analyzed with a Coulter Epics XL-MCL cytometer using the Coulter System II software v. 3.0. Cells were gated first by forward versus sideward scatter. Then, leukemic B cells in patients with B-CLL were gated on high CD19 expression versus sideward scatter whereas the blasts in the AML patients were gated on high CD34 or on low CD45 expression versus side scatter. Leukemic blasts in B-ALL patients were gated on CD34 or on CD19 expression (CD34 negative) versus side scatter. The expression of HLA class I molecules by these leukemic cell populations was compared to that by autologous normal CD3+ T cells. In addition, PBMCs from 29 HV were also used to determine the range of expression of HLA class I molecules by normal CD19+ B and CD3+ T cells. The expression of the analyzed molecules was defined by the mean fluorescence intensity (MFI) and the cell surface antigen density was calculated by determining the MFI/mean forward scatter (MFSC) ratio. All values were corrected for cell size. In addition, the results were corrected with the background control values.
NK cell isolation
NK cells were isolated by immunomagnetic depletion of T cells, B cells, dendritic cells, monocytes, granulocytes, platelets, and basophils from HV PBMCs using a cocktail of biotin-conjugated CD3, CD19, CD14, CD36 and anti-IgE mAbs, and the NK cell Micro Bead Cocktail (Myltenyi Biotec, Utrecht, The Netherlands). Purity of NK cells ranged between 85 and 90%.
NK cell mediated cytotoxicity assay
This assay was performed as previously described [21]. Briefly, cell lines and leukemic cells isolated from patients who had at least 85% leukemic cells in their peripheral blood were used as targets. The viability of target cells was assessed by trypan blue exclusion. HLA-Bw4 was masked by incubating target cells with the HLA-Bw4-specific mAb MUS4H4 (5 μg/ml) (IgG1) for 20 min at 4°C. Cells incubated with an isotype-matched unrelated mAb under the same experimental conditions were used as specificity controls. To minimize the interference of antibody dependent cell mediated cytotoxicity (ADCC), NK cells were incubated with 50 μl heat-inactivated serum from healthy AB+ blood donors for 10 min at 4°C before the addition of target cells.
Target cells (1 × 106 per ml) were prestained with 10 μl lipophilic tracer 3,3′-dioctadecyloxacarbocyanine perchlorate (DIOC18 at 3 mM, Invitrogen Life technologies) for 20 min at 37°C, washed and diluted to 1 × 105 per ml. Subsequently, NK cells were added to 1 × 104 target cells to yield an effector (E):target (T) ratio of 10:1 in a final volume of 200 μl. After a 4-h incubation at 37°C in a 5% CO2 atmosphere, cells were stained with propidium iodide (PI) (10 μg/ml, Invitrogen Life technologies). A control sample with only target cells was included to monitor spontaneous cell death.
CD107a assay
The antitumor activity of NK cells was assessed by measuring the expression of the lysosome-associated membrane protein CD107a as a marker of degranulation. Purified NK cells were mixed at the effector:target ratio of 1:2 with target cells (5 × 104) in a 96-well microtiter plate in a total volume of 200 μl of RPMI1640 medium. Following the addition of 5 μl of a CD107a-specific cy5-mAb H4A3 preparation, the plate was incubated for 4 h at 37°C in a 5% CO2 incubator. At the end of the incubation, cells were washed twice with PBS supplemented with 1% BSA and stained with PE-anti-CD56 mAb B159 and FITC-anti-CD3 mAb SK7. A control sample with only effector cells was included in each experiment to detect spontaneous degranulation. Cells were analyzed with a flow cytometer. The percentage of CD107a positive cells of the gated NK cells (CD3− and CD56+) was determined.
Statistics
Differences in the MFI values for HLA antigens between leukemic and normal cells were analyzed utilizing the nonparametric Mann–Whitney U test.
Results
Differential HLA-A, -B, -C allele cell surface expression on AML, B-ALL and B-CLL cells
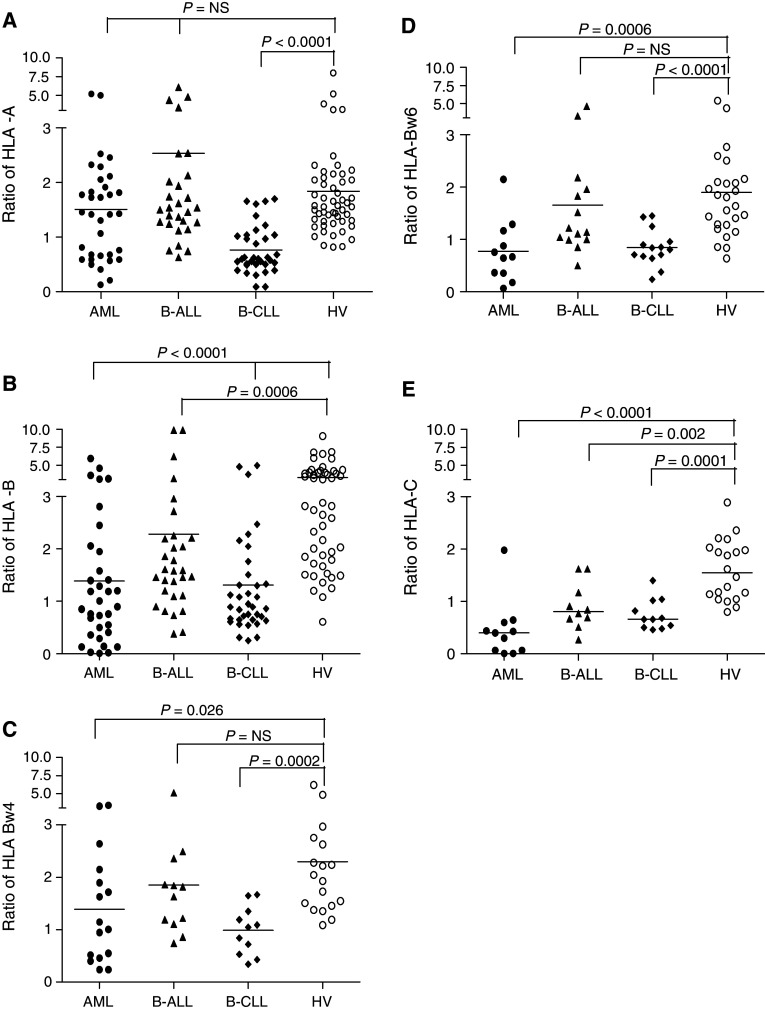
Peripheral blood mononuclear cells from leukemic patients and from HV were stained with human mAb which recognize HLA class I allospecificities and analyzed by flow cytometry. Because of the individual genetic variability of HLA class I allele expression, comparison of the level of HLA class I allele expression on leukemic cells and on PBMC from HV did not utilize the raw MFI values, but the MFI ratios. The latter were calculated by dividing the MFI values derived from the analysis of HLA class I alleles on leukemic or B cells by the MFI values derived from the analysis of autologous normal T cells. The number of HLA class I alleles tested in each group depended on the availability of HLA class I allele-specific mAb with the appropriate specificity, the number of patients included, and the hetero- or homozygosity for the HLA class I locus tested (Fig. 1). No significant differences were found in the expression of the gene products of HLA-A, -B and -C loci between T cells from HV, and those from leukemic patients (data not shown). In addition in HV, the MFI values related to HLA class I alleles on B cells were generally higher than those on autologous T cells. As a result, the MFI ratios were greater than 1 (Fig. 1).
Fig. 1.
Comparison of HLA class I allele expression between leukemic patients and HV. To study the expression of HLA-A, -B and -C alleles, a mAb was selected depending on the recognition of a particular allele without cross-reactivity with other alleles present in that patient. Leukemic blasts and lymphocytes were stained with a HLA-A allele-specific human mAbs; 34, 29, 35 and 54 HLA-A alleles were tested for 18 AML patients, 16 B-ALL patients, 19 B-CLL patients and 29 HV, respectively; b HLA-B allele-specific human mAbs; 34, 30, 36 and 51 HLA-B alleles tested for 18 AML patients, 15 B-ALL patients, 20 B-CLL patients and 29 HV, respectively; c mAb MUS4H4 specific for Bw4 epitope or mAb VDK8F7 specific for HLA-Bw4 but not for HLA-B51, -B53, -B13, -B63 and -B49; 16, 12, 11 and 18 of HLA-Bw4 molecules tested for 16 AML patients, 12 B-ALL patients, 11 B-CLL patients and 18 HV, respectively; d HLA-HLA-Bw6-specific human mAb OUW4F11; 11, 14, 14 and 26-HLA-Bw6 molecules tested for 11 AML patients, 14 B-ALL patients, 14 B-CLL patients and 26 HV, respectively; e VP6G3 specific for Cw1; mAb WK4C11 specific for Cw1, Cw3, Cw4, Cw0801, Cw1202, Cw1402; or mAb TRA2G9 specific for Cw1, Cw3, Cw4, Cw1402; 11, 10, 11 and 20 HLA-C alleles tested for 10 AML patients, 9 B-ALL patients, 10 B-CLL patients and 19 HV, respectively. Every dot represents the individual ratio for each mAb; the ratio is determined by MFI/MFSC expressed on leukemic cells or normal B lymphocytes in HV divided by the MFI/MFSC expressed on autologous T lymphocytes. The horizontal lines indicate means. The results were compared with the Mann–Whitney U test
The MFI ratios related to HLA-A, -B, -C alleles in patients with AML, B-ALL and B-CLL were compared to those in HV. The MFI values related to HLA-A alleles on the leukemic blasts in patients with ALL and AML were mostly higher than those on the corresponding autologous T cells (Ratio > 1); the mean MFI ratio values were not different from those in HV. It is noteworthy that in seven AML patients HLA-A alleles had a lower expression on blasts than on autologous T cells (Fig. 1a). HLA-A antigen downregulation may provide myeloid blasts with a mechanism to escape from cytotoxic T lymphocyte (CTL) destruction; as a result, leukemic blast count may be increased. We therefore compared the percentage of leukemic blasts in patients with and without HLA-A antigen downregulation. The median blast count was 64.0% in the subgroup of seven AML patients with a MFI ratio lower than one and 41.5% in the remaining patients. However, this difference did not reach the level of statistical significance (P = 0.181). In contrast, the expression of HLA-A alleles on B-CLL cells was downregulated in 15 patients.
HLA-B alleles had a significantly lower expression on the three types of leukemic cells than on B cells from HV (Fig. 1b). In addition analysis of the expression of the HLA-Bw4 and HLA-Bw6 group alleles showed that the MFI ratios related to HLA-Bw4 and HLA-Bw6 alleles on B-ALL cells are similar to those on B cells in HV (Fig. 1c, d). In contrast, the expression of both HLA-Bw4 and HLA-Bw6 group alleles is significantly lower on both B-CLL and AML cells than on B cells from HV (Fig. 1c, d). The downregulation was especially marked for HLA-Bw6 group alleles on AML cells. It is noteworthy that the mean MFI ratio for HLA-Bw4 group alleles is greater than 1 on both B-CLL and AML cells. This result indicates that HLA-Bw4 and HLA-Bw6 group alleles are differentially expressed on B-CLL and AML cells and that HLA-HLA-Bw4 group alleles are not downregulated on 5 of the 11 B-CLL and 9 of the 16 AML cells.
To determine whether HLA class I alleles are differentially downregulated on leukemic cells, we compared the frequency of downregulation of some of the HLA-A and -B alleles tested in each leukemic patient group (Table 1). This analysis could be performed only for the most frequent HLA-A and -B alleles since the number of samples available for the less frequent HLA-A and -B alleles and for HLA-C alleles was too low for such an analysis. The HLA-B44 allele had a significantly (P < 0.01) higher frequency of downregulation than the other HLA class I alleles analyzed in AML patients. The HLA-B7 allele had also a higher frequency of downregulation than the HLA class I analyzed in AML patients. However, this difference did not reach the level of statistical significance. In B-ALL the HLA-B5, -B8 and -B44 alleles had a higher frequency of downregulation than the other HLA class I alleles analyzed; however, none of the differences reached the level of statistical significance. Last, all the HLA-A and -B alleles analyzed in B-CLL, with the exception of HLA-B7, were downregulated in at least 60% of the patients.
Table 1.
Comparison of the frequency of downregulation of the HLA-A and -B alleles most frequently expressed on leukemic cells
| Patients | Frequency of HLA class I allele downregulation (MFI ratio < 1) (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | A24 | A28 | B5 (B51, 52) | B7 | B8 | B44 | |
| AML | 0/3 (0.0) | 2/9 (22.2) | 4/9 (44.4) | 2/4 (50.0) | 1/3 (33.3) | 1/6 (16.7) | 4/5 (80.0) | 0/3 (0.0) | 6/6* (100.0) |
| B-ALL | 1/4 (25.0) | 1/6 (16.7) | 0/4 (0.0) | 0/1 (0.0) | 0/4 (0.0) | 2/4 (50.0) | 0/3 (0.0) | 1/2 (50.0) | 1/2 (50.0) |
| B-CLL | 5/6 (83.6) | 9/11 (81.8) | 5/6 (83.3) | 2/2 (100.0) | 1/1 (100.0) | 3/3 (100.0) | 0/2 (0.0) | 4/5 (80.0) | 4/6 (66.7) |
Differences in the frequency of downregulation between common HLA alleles in each leukemic patient group were tested by Fisher exact test. Significance is indicated using *
The analysis of HLA-C allele expression was restricted to HLA-Cw1, Cw3, Cw4, -Cw0801, -Cw1202, and -Cw1402 allospecificities; the expression of the other HLA-C allospecificities could not be analyzed because of the lack of mAb with the appropriate specificity. As a result, the expression of only 33.0% of the HLA-C alleles on leukemic cells could be analyzed. All of the tested HLA-C alleles were markedly downregulated on all three types of leukemic cells analyzed. Moreover, the mean values of the MFI ratios were all lower than 1, pointing out that HLA-C allele expression on nearly all leukemic cells was markedly lower than on their autologous normal T cells. Last, HLA-C alleles were barely detectable on leukemic blasts in four AML patients (Fig. 1e).
In conclusion, the expression of one or more HLA class I alleles on leukemic cells, as determined by comparison with autologous T cells, was downregulated in 85.0, 62.5, and 68.4% of the B-CLL, B-ALL, and AML samples analyzed, respectively. In contrast, HLA class I allele loss on leukemic cells was not detected in this study.
The results of HLA class I typing obtained by flow cytometric analysis of cells stained with HLA class I allele-specific human mAb were corroborated by those obtained by molecular typing and by testing with HLA typing trays in the CDC assay. It is of interest that in one B-CLL, one B-ALL, and six AML samples HLA class I alleles which were barely detectable on leukemic blasts by flow cytometry were readily detectable by CDC.
Association of chromosomal abnormalities with HLA class I antigen downregulation
We tested whether HLA-A, -B, or -C allele downregulation was associated with molecular abnormalities (translocation, deletion, inversion and abnormal karyotype) in 15 of the 19 AML and 15 of the 16 B-ALL patients (Table 2). This analysis could not be performed in the group of B-CLL patients because most of them were not screened routinely for genetic aberrations.
Table 2.
Chromosomal abnormalities in B-ALL and AML patients
| Chromosomal aberration | Abnormal karyotype | Normal karyotype | |
|---|---|---|---|
| AML (n = 15) | |||
| Translocation | t(15;17) (q2;q21) | 0 | 1 |
| Deletion |
−5/del(5q) −7/del(7) + t(3,3) |
1 1 |
0 0 |
| Inversion | Inv(16) | 0 | 1 |
| Other | 2 | 0 | |
| None detected | 0 | 9 | |
| B-ALL (n = 15) | |||
| Translocation | t(9;22) (q34;q11) |
0 2 |
1 0 |
|
t(9;22) and t(5;10) t(12;21) |
1 1 |
0 1 |
|
| Trisomy 21 | +21 | 1 | 0 |
| Other | 5 | 0 | |
| None detected | 0 | 3 | |
The number of patients with molecular aberrations in the group of AML patients with a MFI ratio of HLA-A, -B, or -C alleles lower than one was not significantly different from that in the group of patients with a MFI ratio of at least one. It is of interest that the AML patient with chromosome 16 inversion showed the lowest HLA-A, -B, and -C allele expression level.
All B-ALL patients (n = 5) with a MFI ratio lower than one for HLA-B alleles had an abnormal karyotype compared to 40% of the remaining patients (4/10). We did not measure the HLA-B expression of one B-ALL patient with an abnormal karyotype. In addition, three of these 5 B-ALL patients were positive for a translocation [t(9;22), t(5;10), or t(12;21)]. In contrast, no association between molecular abnormalities and HLA-A or -C allele downregulation was found in B-ALL patients.
Impact of the HLA-Bw4 allele level on the susceptibility of leukemic cells to lysis mediated by allogeneic NK cell-mediated lysis
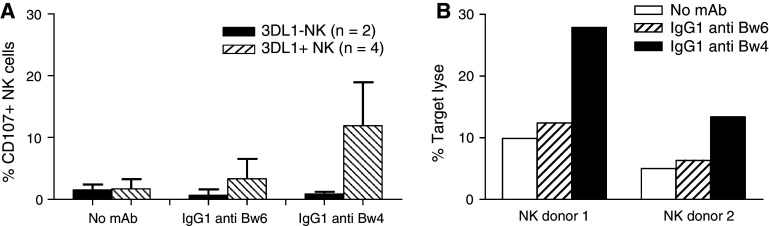
To assess the functional relevance of the HLA class I allele expression level by leukemic cells, the insusceptibility to allogeneic NK cell-mediated lysis was investigated. B-CLL cells with a high HLA-Bw4 allele expression were used as targets in the CD107a and in the cytotoxicity assay (Fig. 2a, b). Polyclonal NK cells from four donors who express KIR3DL1 and from two donors who do not express this inhibitory receptor were used as effectors (Fig. 2a). HLA typing of both NK cell donors and of the B-CLL patient ruled out NK cell alloreactivity triggered by a mismatch between inhibitory KIRs on donor NK cells and the HLA class I molecules on leukemic targets. Neither KIR3DL1+ nor KIR3DL1− NK cells showed CD107 surface expression when they were incubated with B-CLL cells. Incubation of B-CLL cells with the HLA-Bw4-specific mAb MUS4H4 enhanced the percentage of CD107a positive NK cells in KIR3DL1+ donors, but had no marked effect on the activation of NK cells in KIR3DL1− donors (Fig. 2a). Furthermore, incubation of B-CLL cells with the HLA-Bw6-specific mAb OUW4F11 had no detectable effect on the CD107a expression on KIR3DL1+ and KIR3DL1− NK cells (Fig. 2a). These findings were corroborated by the results of cytotoxicity assays performed in the presence of HLA-Bw4-specific mAb MUS4H4 to inhibit KIR3DL1-HLA-Bw4 allele interactions. NK cells from two KIR3DL1+ donors were used as effectors. As shown in Fig. 2b, the HLA-Bw4-specific mAb MUS4H4 markedly enhanced the extent of lysis, while the HLA-Bw6-specific mAb OUW4F11 had no marked effect. Moreover, the percentage of target cell lysis in the presence of HLA-Bw4-specific mAb correlated with the percentage of NK cells expressing KIR3DL1 (20.0% 3DL1+ NK clones in donor 1 vs. 11.8% 3DL1+ NK clones in donor 2).
Fig. 2.
Increase by HLA-Bw4-specific mAb MUS4H4 of the susceptibility of high HLA-Bw4-expressing B-CLL cells to NK cell-mediated lysis. a Percentage of CD107+ NK cells from KIR3DL1-positive (n = 4) (stripped bars) and KIR3DL1 negative (n = 2) (black bars) donors following incubation with B-CLL cells in the absence or in the presence of saturating amounts of HLA-Bw4-specific mAb MUS4H4 (IgG) or HLA-Bw6-specific mAb OUW4F11 (IgG). Polyclonal NK cells were isolated and incubated with B-CLL cells at the E:T ratio of 1:2. Error bars indicate standard deviation (SD) b lysis of B-CLL cells by NK cells from two 3DL1-positive donors in the absence (white bars) or in the presence of HLA-Bw4-specific mAb MUS4H4 (IgG) (black bars) or HLA-Bw6-specific mAb OUW4F11 (IgG) (stripped bars). Polyclonal NK cells were isolated and incubated with B-CLL cells at the E:T ratio of 1:10. NK cells were pre-incubated with 50 μl heat-inactivated human AB serum to minimize ADCC
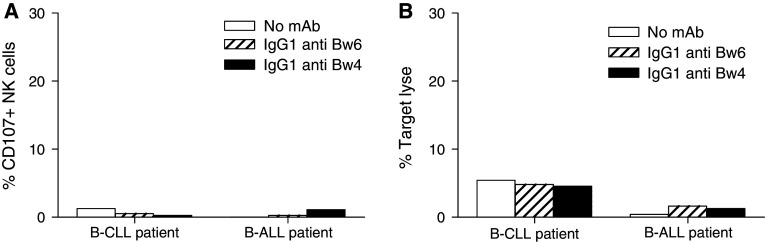
Unexpectedly, when B-CLL cells with HLA-HLA-Bw4 allele downregulation and B-ALL cells with a HLA-HLA-Bw4 allele expression level within normal range were used as targets, the extent of lysis (Fig. 3b) and the percentage of activated NK cells (Fig. 3a) from a 3DL1+ donor were very low. Whether the latter findings reflect the role of other resistance mechanisms to NK cell recognition could not be investigated, because leukemic cells from these patients were not available to perform these studies.
Fig. 3.
Lack of detectable effect of HLA-Bw4-specific mAb MUS4H4 on the susceptibility of B-ALL cells and low HLA-Bw4-expressing B-CLL cells to NK cell-mediated lysis. Percentage of CD107+ NK cells from a KIR3DL1-positive donor (a) and NK cell mediated lysis (b) of B-CLL cells with HLA-Bw4 downregulation or against blasts from a B-ALL patient with normal HLA-Bw4 expression in the absence (white bars) or in the presence of HLA-Bw4-specific mAb MUS4H4 (IgG) (black bars) or HLA-Bw6-specific mAb OUW4F11 (IgG) (stripped bars). Polyclonal NK cells were isolated and incubated with B-CLL cells and B-ALL cells at the E:T ratio of 1:2 and 1/10, respectively. NK cells were obtained from the same patient used in the experiments shown in Fig. 2. NK cells were pre-incubated with 50 μl heat-inactivated human AB serum to minimize ADCC
Therefore, the studies were continued with the cell lines K562, ML2, MONOMAC-6, and RPMI 8402. Polyclonal NK cells from two healthy donors were used as effectors. Phenotyping of the two donors showed that both of them are heterozygous for HLA-Bw4 and HLA-Bw6 alleles and for group 1 and 2 of HLA-C ligands. HLA-Bw4 alleles are the ligands for the inhibitory 3DL1 receptor, while HLA-C alleles are the ligands for the inhibitory 2DL receptors. Moreover, both donors possessed all inhibitory KIR receptor genes, i.e., 2DL1, 2DL2, 2DL3, 3DL1, and 3DL2. To exclude the presence of the 3DL1*004 deletion variant, the expression of KIR3DL1 was confirmed by flow cytometry (data not shown). HLA class I phenotyping of the four cell lines by flow cytometry showed that all of them with the exception of the K562 cells line, express the three major KIR ligands, i.e., HLA-Bw4, HLA-C1, and HLA-C2 alleles. In this way, we excluded donor NK cell alloreactivity caused by a mismatch between the inhibitory KIRs on donor NK cells and the HLA class I molecules on leukemic cell lines. Incubation of the four cell lines with increasing concentrations (0.5, 1, 5, 20 and 100 μg/ml) of the HLA-Bw4-specific mAb MUS4H4 caused a dose-dependent increase of the percentage of activated NK cells when they were incubated with the cell lines ML2 and MONOMAC (Fig. 4).
Fig. 4.
Role of HLA-Bw4 expression on target cells in their susceptibility to NK cell mediated lysis. NK cells from two KIR3DL1-positive healthy donors were incubated with the leukemia cell lines K562, ML2, MONOMAC-6 and RPMI 8402 in the presence of the HLA-Bw4-specific mAb MUS4H4 (IgG) (0.5, 1, 5, 20 or 100 μg/ml). Polyclonal NK cells were isolated and incubated with target cells at the E:T ratio of 1:2. NK cells were pre-incubated with 50 μl heat-inactivated human AB serum for 10 min on 4°C to minimize ADCC
The percentage of activated NK cells appears uniformly high when they were incubated with the K562 cell line and was not affected by the incubation with the mAb MUS4H4 at all the concentrations tested. In contrast, no activation was detected when NK cells were incubated with the RPMI 8042 cell line (Fig. 4). Additional experiments investigated whether the differential sensitivity of the four cell lines analyzed for NK cell recognition was caused by the differential expression of the ligands for the activating NK cell receptors. Cytofluorographic analysis of the cell lines stained with mAbs showed that PVR, Nectin-2 and the adhesion molecule ICAM-1 were expressed on K562, ML2, and MONOMAC-6 cells, but were not detectable on RPMI 8042 cells (data not shown). Furthermore the NKG2D ligands, MICA/B was weakly expressed only on K562 cells and ULBP-2 and -3 were expressed only on MONOMAC-6 cells (data not shown). Finally, CD48, the ligand for the 2B4 receptor was not detected on any of the cell lines. All together, these data show that all known ligands for the activating NK cell receptors were not detectable on the RPMI 8042 cell line. This phenotype may provide a mechanism for its lack of susceptibility to NK cell recognition.
Discussion
The scant information in the literature pertaining to the level of HLA class I alleles on leukemic cells and its potential role in the outcome of allogeneic HLA-mismatched HSCT to leukemic patients have prompted us to investigate this topic in B-ALL, AML, and B-CLL patients. Our study has shown that total HLA class I loss is rare, while selective HLA class I allele downregulation on leukemic cells occurs frequently and is most pronounced in B-CLL patients. From a methodological view point, two points are noteworthy. Defects in HLA class I allele expression could be identified in our study, since we utilized HLA class I allele-specific mAbs. This conclusion is in line with the evidence derived from studies of solid tumors that has shown that selective HLA class I allele downregulation cannot be detected by mAbs recognizing monomorphic determinats expressed on the gene products of HLA-A, -B, and -C loci [22, 23]. Furthermore, the complement dependent cytotoxicity assay appears to be much more sensitive than flow cytometric analysis in the analysis of HLA class I antigen expression by leukemic cells. This difference is likely to reflect the high sensitivity of leukemic cells to complement dependent lysis and/or the polyclonal nature of the HLA typing sera utilized in the complement dependent cytotoxicity assay.
Our findings are in agreement with our previous data [16], but are in apparent conflict with most of the results reported by Brouwer et al. [14] and Masuda et al. [17], since both groups of investigators described a low frequency of HLA class I allele downregulation in the leukemic patient population analyzed. The apparent discrepancy between the results obtained by the latter two investigators and by ourselves is likely to reflect the different criteria used to select the patient population to be included in the study and the HLA class I alleles included in the analysis. Brouwer et al. [14] who like us described a low frequency of total HLA class I loss in leukemic cells, utilized flow cytometry to analyze only leukemic patients in whom single HLA class I allele loss had been detected by CDC assay; as a result, they could not detect HLA class I allele downregulation. Moreover, Masuda et al. [17] investigated only HLA-A allele expression mainly in AML patients and only in one B-CLL patient. In our study HLA-A allele expression was only significantly downregulated in B-CLL patients.
The frequency of HLA-B allele downregulation which was high in patients with B-CLL and AML, was mostly restricted to those HLA-B allele with the HLA-Bw6 epitope. The data we have obtained about the expression of HLA-B alleles with the HLA-Bw4 epitope parallel, but are not superimposable onto those we previously described [16] Specifically the frequency of HLA-Bw4 downregulation in the present study is higher than that in the previous one [16]. However, in both studies the level of HLA-Bw4 alleles is higher than that of HLA-Bw6 alleles; this difference may reflect the outgrowth of leukemic cells with high HLA-Bw4 allele expression because of the selective pressure imposed by NK cells on the leukemic cell population. In addition, we observed that the HLA-B7 and -B44 alleles were downregulated more frequently than others in AML patients. Although the number of AML patients analyzed for HLA-B7 and -B44 alleles is too low to draw definitive conclusions, it is intriguing that HLA-B44 has been reported to be lost more frequently than other HLA class I alles in solid tumors [24]; whether this finding reflects the frequent use by malignant cells of HLA-B44 as a restricting element in their interactions with tumor antigen-specific CTL remains to be determined.
The analysis of HLA-C allele expression was limited to 33.0% of the known alleles because of the lack of mAb with the appropriate specificity. Nevertheless, our study has shown that HLA-C allele downregulation is frequent in all the three types of leukemic cells investigated and was most pronounced in the AML samples analyzed. Most AML cells showed a strong downregulation and in some cases even a complete loss of HLA-C alleles in comparison with their autologous T cells. This result might explain why donor NK cell alloreactivity based on HLA-C mismatch in AML patients does not always result in a markedly reduced incidence of relapse [9–11], especially since Almeida and Davis [25] have recently showed that a high density of HLA-C proteins on target cells is required to inhibit NK cell cytotoxicity. In agreement with these results, our studies in progress suggest that HLA-C downregulation can indeed induce leukemic target lysis by allogeneic donor NK cells that are matched with a patient’s KIR ligands (HLA-C and -Bw4). However, the functional significance of this finding has to be interpreted with caution, because only a few NK cell cytotoxicity assays have been performed. Furthermore, no mAb that specifically blocks the HLA-C1 or -C2 epitopes was available to investigate the impact of HLA-C downregulation on NK cell activity.
What is the functional relevance of the defects in HLA class I allele expression we have found in leukemic cells? The “missing self” hypothesis would predict that leukemic cells with lost or downregulated HLA class I allele become susceptible to NK cell-mediated lysis [26] Our data show that HLA class I allele downregulation or loss on leukemic cell lines and B-CLL cells enhance NK cell activation and target cell lysis, provided that NK cell activating ligands are expressed by leukemic cells. These results parallel the susceptibility of melanoma cells with total HLA class I antigen loss to NK cell-mediated lysis only if NK cell activating ligands were expressed on melanoma cells [27]. Furthermore, NK cell-mediated lysis of B-ALL cells was not detected when HLA-Bw4 antigens were coated with mAb. This result may reflect the resistance of this leukemic cell type to NK cells because of the lack of expression of the LFA-1 adhesion molecule or other NK cell activating ligands [1, 28, 29]. A study reported by Pende et al. [30] and unpublished data from our laboratory found that the expression of different NK cell activating ligands is variable among different leukemic patients and therefore should be determined in order to define the susceptibility of leukemic cells to NK cell-mediated lysis. In addition, we cannot exclude that the resistance of leukemic cells to NK cell-mediated lysis is caused by the expression of the non classical HLA class I antigen, HLA-G, although, to the best of our knowledge, this antigen has not been detected on leukemic cells [31, 32].
Our study has clearly shown that besides the HLA and KIR genotype, expression levels of KIR ligands and of activating ligands on leukemic cells affect their interactions with NK cells. If these in vitro data are paralleled by in vivo data, these variables should be taken into account in the selection of donor-recipient combinations for HSCT to leukemic patients. In addition, interpretation of the conflicting results pertaining to NK cell-mediated GvL effects in related and unrelated allogeneic HSCT, which have been described in the literature, may benefit from the inclusion of HLA class I allele expression and expression level of NK cell activating ligands on leukemic cells in the analysis of the data.
Acknowledgments
This work was supported by grants awarded by the Scientific Fund Willy Gepts-UZ Brussel (S. V.); Fonds voor Wetenschappelijk Onderzoek Vlaanderen (G.0481.06) (C. D.); “Stichting tegen Kanker (SCIE-2005-33) (C. D.) and by the National Cancer Institute, DHHS PHS grants RO1CA110249 and RO1CA113861 (S. F.).
References
- 1.Ruggeri L, Capanni M, Casucci M, et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94:333–339. [PubMed] [Google Scholar]
- 2.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2101. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 3.Ruggeri L, Mancusi A, Capanni M, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110:433–440. doi: 10.1182/blood-2006-07-038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The HLA-Bw4 public epitope of HLA-B molecules confers reactivity with NK cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moretta A, Vitale M, Bottino C, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells: anti-P58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colonna M, Brooks EG, Falco M, Ferrara GB, Strominger JL. Generation of allospecific natural killer cells by stimulation across a polymorphism of HLA-C. Science. 1993;260:1121–1124. doi: 10.1126/science.8493555. [DOI] [PubMed] [Google Scholar]
- 7.Biassoni R, Falco M, Cambiaggi A, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaffer M, Malmberg KJ, Ringdén O, Ljunggren HG, Remberger M. Increased infection-related mortality in KIR-ligand-mismatched unrelated allogeneic hematopoietic stem-cell transplantation. Transplantation. 2004;78:1081–1085. doi: 10.1097/01.TP.0000137103.19717.86. [DOI] [PubMed] [Google Scholar]
- 9.Bishara A, De Santis D, Witt CC, et al. The beneficial role of inhibitory KIR genes of HLA class I NK epitopes in haploidentically mismatched stem cell allografts may be masked by residual donor-alloreactive T cells causing GVHD. Tissue Antigens. 2004;63:204–211. doi: 10.1111/j.0001-2815.2004.00182.x. [DOI] [PubMed] [Google Scholar]
- 10.Bornhauser M, Schwerdtfeger R, Martin H, Frank KH, Theuser C, Ehninger G. Role of KIR ligand incompatibility in hematopoietic stem cell transplantation using unrelated donors. Blood. 2004;103:2860–2862. doi: 10.1182/blood-2003-11-3893. [DOI] [PubMed] [Google Scholar]
- 11.Davies SM, Ruggieri L, DeFor T, et al. Evaluation of KIR ligand incompatibility in mismatched unrelated donor hematopoietic transplants. Blood. 2002;100:3825–3827. doi: 10.1182/blood-2002-04-1197. [DOI] [PubMed] [Google Scholar]
- 12.Bignon JD, Gagne K. KIR matching in hematopoietic stem cell transplantation. Curr Opin Immunol. 2005;17:553–559. doi: 10.1016/j.coi.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Wetzler M, Baer MR, Stewart SJ, et al. HLA class I antigen cell surface expression is preserved on acute myeloid leukemia blasts at diagnosis and at relapse. Leukemia. 2001;15:128–133. doi: 10.1038/sj.leu.2401982. [DOI] [PubMed] [Google Scholar]
- 14.Brouwer RE, van der Heiden P, Schreuder GM, et al. Loss or downregulation of HLA class I expression at the allelic level in acute leukemia is infrequent but functionally relevant, and can be restored by interferon. Hum Immunol. 2002;3:200–210. doi: 10.1016/S0198-8859(01)00381-0. [DOI] [PubMed] [Google Scholar]
- 15.Vollmer M, Li L, Schmitt A, et al. Expression of human leucocyte antigens and co-stimulatory molecules on blasts of patients with acute myeloid leukaemia. Br J Haematol. 2003;120:1000–1008. doi: 10.1046/j.1365-2141.2003.04212.x. [DOI] [PubMed] [Google Scholar]
- 16.Demanet C, Mulder A, Deneys V, Worsham MJ, Claas FH, Ferrone S. Down-regulation of HLA-A and HLA-Bw6, but not HLA-Bw4, allospecificities in leukemic cells: an escape mechanism from CTL and NK attack? Blood. 2004;103:3122–3130. doi: 10.1182/blood-2003-07-2500. [DOI] [PubMed] [Google Scholar]
- 17.Masuda K, Hiraki A, Fujii N, et al. Loss or down-regulation of HLA class I expression at the allelic level in freshly isolated leukemic blasts. Cancer Sci. 2007;98:102–108. doi: 10.1111/j.1349-7006.2006.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nouri AM, Smith S, Oliver TR, Newland AC, Macey MG. Comparative expression of major histocompatibility complex (MHC) antigens on CD5+ and CD5-B cells in patients with chronic lymphocytic leukaemia (CLL) Eur J Cancer. 1998;34:1618–1622. doi: 10.1016/S0959-8049(98)00158-0. [DOI] [PubMed] [Google Scholar]
- 19.Mulder A, Kardol M, Regan J, Buelow R, Claas FH. Reactivity of twenty-two cytotoxic human monoclonal HLA antibodies towards soluble HLA class I in an enzyme-linked immunosorbent assay (PRA-STAT) Hum Immunol. 1997;56:106–113. doi: 10.1016/S0198-8859(97)00146-8. [DOI] [PubMed] [Google Scholar]
- 20.Mulder A, Kardol M, Uit het Broek CM, Tanke-Visser J, Young NT, Claas FH. A human monoclonal antibody against HLA-Cw1 and a human monoclonal antibody against an HLA-A locus determinant derived from a single uniparous female. Tissue Antigens. 1998;52:393–396. doi: 10.1111/j.1399-0039.1998.tb03062.x. [DOI] [PubMed] [Google Scholar]
- 21.Chang L, Gusewitch GA, Chritton DB, Folz JC, Lebeck LK, Nehlsen-Cannarella SL. Rapid flow cytometric assay for the assessment of natural killer cell activity. J Immunol Methods. 1993;166:45–54. doi: 10.1016/0022-1759(93)90327-4. [DOI] [PubMed] [Google Scholar]
- 22.Kageshita T, Wang Z, Calorini L, et al. Selective loss of human leukocyte Class I allospecificities and staining of melanoma cells by monoclonal antibodies recognizing monomorphic determinants of Class I human leukocyte antigens. Cancer Res. 1993;53:3349–3354. [PubMed] [Google Scholar]
- 23.Wang Z, Marincola FM, Rivoltini L, Parmiani G, Ferrone S. Selective histocompatibility leukocyte antigen (HLA)-A2 loss caused by aberrant pre-mRNA splicing in 624MEL28 melanoma cells. J Exp Med. 1999;190:205–215. doi: 10.1084/jem.190.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabrera T, Maleno I, Lopez-Nevot MA, Redondo M, Fernandez MA, Collado A, Garrido F. High frequency of HLA-B44 allelic losses in human solid tumors. Hum Immunol. 2003;60:941–950. doi: 10.1016/S0198-8859(03)00164-2. [DOI] [PubMed] [Google Scholar]
- 25.Almeida CR, Davis DM. Segregation of HLA-C from ICAM-1 at NK cell immune synapses is controlled by its cell surface density. J Immunol. 2006;177:6904–6910. doi: 10.4049/jimmunol.177.10.6904. [DOI] [PubMed] [Google Scholar]
- 26.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-S. [DOI] [PubMed] [Google Scholar]
- 27.Pende D, Rivera P, Marcenaro S, et al. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62:6178–6186. [PubMed] [Google Scholar]
- 28.Romanski A, Bug G, Becker S, et al. Mechanisms of resistance to natural killer cell-mediated cytotoxicity in acute lymphoblastic leukemia. Exp Hematol. 2005;33:344–352. doi: 10.1016/j.exphem.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Verheyden S, Demanet C. NK cell receptors and their ligands in leukemia. Leukemia. 2008;22:249–257. doi: 10.1038/sj.leu.2405040. [DOI] [PubMed] [Google Scholar]
- 30.Pende D, Spaggiari GM, Marcenaro S, et al. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112) Blood. 2005;105:2066–2073. doi: 10.1182/blood-2004-09-3548. [DOI] [PubMed] [Google Scholar]
- 31.Amiot L, Onno M, Drenou B, Monvoisin C, Fauchet R. HLA-G class I gene expression in normal and malignant hematopoietic cells. Hum Immunol. 1998;59:524–528. doi: 10.1016/S0198-8859(98)00041-X. [DOI] [PubMed] [Google Scholar]
- 32.Poláková K, Krcova M, Kuba D, Russ G. Analysis of HLA-G expression in malignant hematopoetic cells from leukemia patients. Leuk Res. 2003;27:643–648. doi: 10.1016/S0145-2126(02)00228-X. [DOI] [PubMed] [Google Scholar]