SUMMARY
The mammalian Protocadherin (Pcdh) alpha, beta, and gamma gene clusters encode a large family of cadherin-like transmembrane proteins that are differentially expressed in individual neurons. The 22 isoforms of the Pcdhg gene cluster are diversified into A-, B- and C-types, and the C-type isoforms differ from all other clustered Pcdhs in sequence and expression. Here we show that mice lacking the 3 C-type isoforms are phenotypically indistinguishable from the Pcdhg null mutants, displaying virtually identical cellular and synaptic alterations resulting from neuronal apoptosis. By contrast, mice lacking 3 A-type isoforms exhibit no detectable phenotypes. Remarkably, however, genetically blocking apoptosis rescues the neonatal lethality of the C-type isoform knockouts, but not that of the Pcdhg null mutants. We conclude that the role of the Pcdhg gene cluster in neuronal survival is primarily, if not specifically mediated by its C-type isoforms, whereas a separate role essential for postnatal development, likely in neuronal wiring, requires isoform diversity.
INTRODUCTION
Protocadherins (Pcdhs) are the largest subgroup of the cadherin superfamily of cell adhesion proteins. Of the ~70 Pcdh genes identified in mammalian genomes, over 50 are located in three tightly linked gene clusters (Pcdha, Pcdhb, and Pcdhg) on a single chromosome (Wu and Maniatis, 1999). These clustered Pcdh genes are found exclusively in vertebrates, and are predominantly expressed in the nervous system. Distinct subsets of Pcdh genes are differentially expressed in individual neurons, and enormous cell surface diversity may result from combinatorial expression (Esumi et al., 2005; Kaneko et al., 2006; Kohmura et al., 1998; Wang et al., 2002a). A subset of Pcdhg isoforms have been shown to engage in intercellular interactions that are strictly homophilic (Schreiner and Weiner, 2010). The molecular diversity as well as the binding specificity of clustered Pcdhs has led to the proposal that they provide a synaptic address code for neuronal connectivity, or a single cell barcode for self-recognition and self-avoidance similar to that ascribed to Dscam1 proteins of invertebrates (Junghans et al., 2005; Serafini, 1999; Shapiro and Colman, 1999; Zipursky and Sanes, 2010).
Genetic manipulations of individual Pcdh gene clusters in mice have provided functional evidence that the clustered Pcdhs are required for normal development of the nervous system. Mutations in the Pcdha gene cluster have been reported to result in defects in olfactory sensory neuron axon coalescence and serotonergic axonal arborization as well as behavioral perturbations (Fukuda et al., 2008; Hasegawa et al., 2008; Katori et al., 2009). By contrast, abolishing Pcdhg function leads to neuronal apoptosis and synaptic loss in the spinal cord and retina (Lefebvre et al., 2008; Prasad et al., 2008; Wang et al., 2002b; Weiner et al., 2005). Although these genetic studies have provided interesting insights into the roles of clustered Pcdhs in the nervous system, the functional significance of the diverse isoforms encoded by the three gene clusters is not understood. For example, it is unclear whether individual Pcdh isoforms within each cluster are functionally equivalent, or whether certain isoforms may play distinct roles. The unique and highly conserved genomic organization of Pcdh gene clusters suggests that the isoform diversity and evolutionary diversification of Pcdh genes are central to understanding their function.
In mice, the three Pcdh gene clusters each contain 14–22 homologous “variable” exons arrayed in tandem. Each variable exon is transcribed from its own promoter, and encodes the entire extracellular domain, a transmembrane domain, and a short intracellular domain of the corresponding Pcdh protein. In Pcdha and Pcdhg clusters (but not Pcdhb cluster), these variable exons are followed by a set of three “constant” exons, which are joined to each variable exon via cis-splicing to encode a common distal intracellular domain (Tasic et al., 2002; Wang et al., 2002a). An interesting feature of the genomic organization of the Pcdh gene clusters is that the last two variable exons in the Pcdha cluster, as well as the last three variable exons in the Pcdhg cluster, are more similar to each other than to other variable exons within their respective cluster (Wu et al., 2001). These 5 Pcdh genes (Pcdhac1, Pcdhac2, Pcdhgc3, Pcdhgc4, and Pcdhgc5) are designated C-type genes, to be distinguished from A-type and B-type genes of the Pcdhg cluster. The C-type isoforms bear several unique features among all Pcdhs: 1) While all other Pcdhs are more closely related to members within their own cluster, C-type isoforms are evolutionarily divergent, forming a separate branch in the phylogenetic tree (Wu and Maniatis, 1999; Wu et al., 2001); 2) Three out of the five C-type genes (Pcdhac2, Pcdhgc4 and Pcdhgc5) lack the conserved sequence element (CSE) found in the promoters of all other Pcdh genes (except Pcdhb1), suggesting that these genes are regulated differently (Wu et al., 2001). 3) Single cell RT-PCR experiments indicated that, while other Pcdh genes are stochastically and monoallelically expressed in Purkinje neurons, every neuron expresses all five C-type genes from both chromosomes (Esumi et al., 2005; Kaneko et al., 2006). Taken together, these observations suggest that the C-type isoforms play unique and essential roles among all clustered Pcdhs. To investigate this possibility, we generated mutant mice lacking the 3 C-type genes (Pcdhgc3, Pcdhgc4, Pcdhgc5) in the Pcdhg cluster.
RESULTS
Mice lacking the C-type Pcdhg genes are phenotypically indistinguishable from mice lacking the entire Pcdhg cluster
The triple C-type isoform knockout (TCKO) allele was generated by deleting the three variable exons (Figures 1A–B and S1A), which specifically removes the C-type genes without affecting the splicing of the remaining 19 A-type and B-type Pcdhg variable exons (see below). Pcdhgtcko/tcko mutants are born alive at the normal Mendelian ratio, but invariably die during the first day after birth. The mutant mice are readily distinguishable from wild type and heterozygous littermates by a characteristic hunched posture and limb tremors, as well as by severely compromised voluntary movements and reflexes (Figure 1C and Movie S1). Remarkably, these phenotypes are identical to those described for the Pcdhg full cluster deletion mice (Figure 1D and Movie S1), in which all Pcdhg isoforms are abolished (Wang et al., 2002b). In addition to the common phenotypes described above, we found that both lines of mutants also exhibit intense muscle stiffness (Figures 1F–F”) and umbilical hernia (Figures 1G–G”). Interestingly, these phenotypes closely resemble those of mutant mice deficient in VGAT (Wojcik et al., 2006), GAD67 (Asada et al., 1997), and Gephyrin (Feng et al., 1998), which are essential components for GABA and glycine production and transmission.
Figure 1. Pcdhg mutant alleles and phenotypes.
(A) Schematic representations of Pcdhg wild type and mutant alleles. (B) Validation of deletions in homozygous animals by genomic PCR using exon-specific primers. (C–E) Gross phenotypes of Pcdhgtcko/tcko (P0), Pcdhgdel/del (P0) and Pcdhgtako/tako (P60) mutants as compared to littermate controls. Homozygous mutants are indicated with a red arrow. (F–G) Striking phenotype similarities between Pcdhgtcko/tcko and Pcdhgdel/del mutants at P0. Both have hunched posture and lack of voluntary movements and reflexes, exhibiting limb tremor, intense muscle stiffness and umbilical hernia (arrow), and die within hours after birth.
While the virtually identical phenotypes of the Pcdhgtcko/tcko and Pcdhgdel/del mutants demonstrate that C-type isoforms are essential, it is also possible that the entire repertoire of Pcdhg genes are required; that is, each isoform is indispensable. Indeed, essentially every Pcdhg gene in humans has an ortholog in the mouse, in contrast to the Pcdha and Pcdhb genes (Wu et al., 2001). To address this possibility, we deleted Pcdhga1, Pcdhga2 and Pcdhga3 genes in the mouse (Figures 1A–B and S1B). The triple A-type isoform knockout (TAKO) mutants are viable and fertile, and survive to adulthood with no discernible abnormalities (Figure 1E and Movie S1).
Pcdhgtcko/tcko and Pcdhgdel/del mutants display similar levels and patterns of neuronal cell loss in the spinal cord
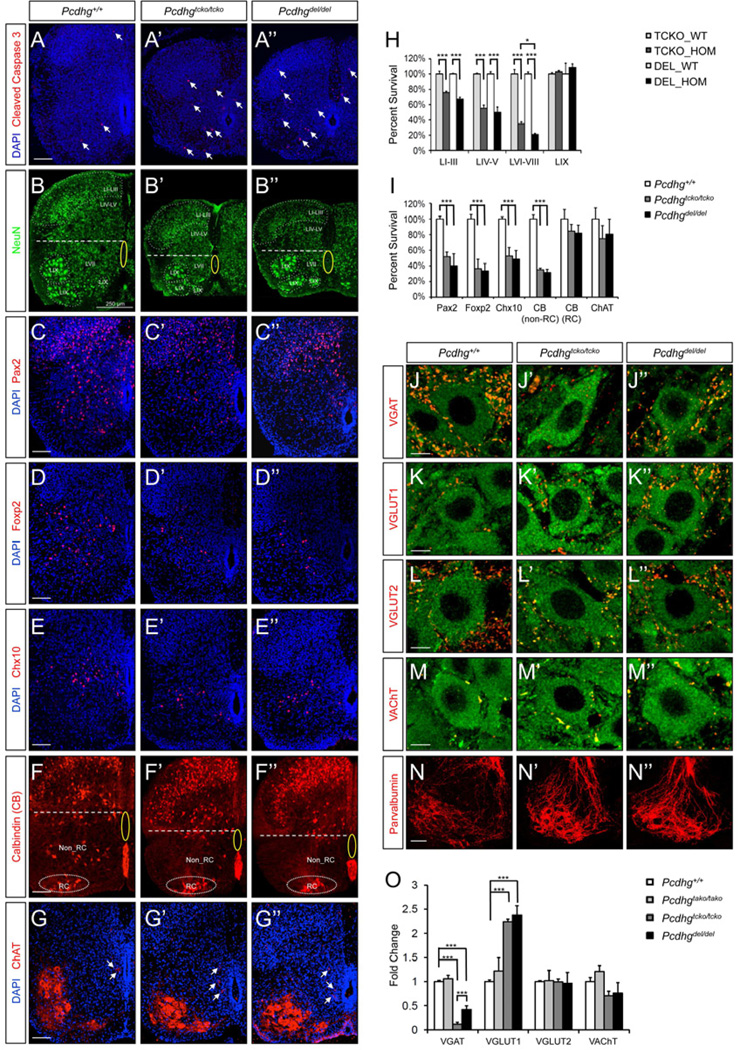
Previous studies showed that deletion of the Pcdhg cluster leads to extensive apoptosis and eventual loss of specific subpopulations of spinal interneurons (Prasad et al., 2008; Wang et al., 2002b; Weiner et al., 2005). To determine whether these changes also occur in TCKO mutants, we labeled cells undergoing apoptosis with anti-cleaved caspase-3 in P0 spinal cords. As expected, the number of apoptotic profiles is markedly increased in the spinal cord of both Pcdhgtcko/tcko and Pcdhgdel/del mutants (Figures 2A–A”). Concurrently, the spinal cords of both mutants exhibit similar levels of astrogliosis and microglia activation (Figures S2A), which typically accompany neuronal cell death. To compare the extent of neuronal cell loss in different Pcdhg mutant lines, we quantified the surviving NeuN+ neurons in different spinal regions at P0. The spinal cords of Pcdhgtcko/tcko and Pcdhgdel/del mutants have a similarly reduced cross sectional area compared to those of the wild type littermates, particularly in the ventral horn (LVI-VIII) and in the deep dorsal horn (LIV-V). Superficial dorsal horn (LI-III) and motor pools (LIX), however, appear relatively normal (Figures 2B–B” and S2B). Consistently, the most severe neuronal loss was detected in the ventral horn and to a lesser extent in the deep dorsal horn (~70% and ~50%, respectively). We also observed ~30% interneuron cell loss in the superficial dorsal horn, which was not reported previously. By contrast, motor neuron (LIX) counts in both mutants are the same as those in wild type controls (Figures 2H and S2B). As expected, Pcdhgtako/tako spinal cords are indistinguishable from the wild type controls, and neuronal cell counts in each of the 4 specified regions are normal (Figure S2B).
Figure 2. Similar levels and patterns of neuronal loss and synaptic changes in the spinal cord of Pcdhgtcko/tcko and Pcdhgdel/del mutants.
(A–A”) Both mutants exhibit increased levels of apoptosis as indicated by the increased numbers of cleaved caspase 3+ profiles (arrows). Scale bar: 100 µm. (B–B”) Quantitative analyses of surviving neuronal populations in different lamina. NeuN+ neurons of distinct laminar regions are indicated. The central canal area is highlighted with a yellow oval, and the spinal cord is horizontally bisected with a dashed line to indicate the dorsal horn and the ventral horn areas. Scale bar: 250 µm. (C–G) Interneuron subpopulations are differentially affected to similar extents in both mutants. Shown are representative images of Pax2 (C-C”), Foxp2 (D-D”), Chx10 (E-E”), CB (F-F”) and ChAT (G-G”) immunolabeled P0 spinal cords of Pcdhg+/+, Pcdhgtcko/tcko, Pcdhgdel/del mice. Clusters of Renshaw cells (RC, F–F”) are encircled, and partition cells (G-G”) are indicated with arrows. Note the motor neurons are prominently labeled with anti-ChAT (G-G”). Scale bars: 100 µm. (H) Percent survival of NeuN+ neuronal populations according to their laminar locations (LI-LIX) in homozygous mutants (TCKO_HOM or DEL_HOM), calculated by normalizing neuronal counts to wild type littermate controls (TCKO_WT and DEL_WT). Details of the quantitative analyses can be found in Figure S2B. *p<0.05, ***p<0.001. (I) Percent survival of interneuron subpopulations in the ventral horn. ***p<0.001. (J–M) Specific types of synaptic inputs onto motor neurons of wild type and mutant spinal cords at P0. Motor neurons are labeled with anti-ChAT in green, and synaptic inputs with antibodies against different vesicular transporters in red. Scale bars: 5 µm. (N-N”) Terminal arborization of Parvalbumin+ Ia primary afferents surrounding motor pools is altered in both lines of mutants. Scale bar: 50 µm. (O) Fold changes in synaptic density for each type of synapse in the mutants as calculated by normalizing to wild type controls. Details of the quantitative analyses can be found in Figure S2C. ***p<0.001.
To investigate whether neuronal subpopulations are similarly affected in Pcdhgtcko/tcko and Pcdhgdel/del mutants, we examined several classes of interneurons in the ventral spinal cord at P0. Interestingly, while Pax2+ and Foxp2+ inhibitory interneurons, as well as Chx10+ excitatory interneurons are similarly reduced in number in both mutants, V1-derived Calbindin (CB)+ Renshaw cells and V0-derived cholinergic ChAT+ partition cells are spared (Figures 2C–G, 2I). In conclusion, the Pcdhgtcko/tcko and Pcdhgdel/del mutants display similar levels and patterns of neuronal cell loss in the spinal cord, and interneuron subpopulations are differentially affected in both mutants.
Pcdhgtcko/tcko and Pcdhgdel/del mutants display similar alterations in specific synaptic inputs onto motor neurons
In addition to neuronal cell loss, a general reduction in the numbers of both excitatory and inhibitory synapses was observed in the neuropil of Pcdhgdel/del spinal cords using generic synaptic markers (Wang et al., 2002b; Weiner et al., 2005). It was unclear, however, whether all synapses are similarly affected, or if certain types are spared or even increased in number. To distinguish between these possibilities, we examined major classes of synaptic inputs onto motor neurons, a cell type that receives defined synaptic inputs and survives in both Pcdhgtcko/tcko and Pcdhgdel/del mutants. Four type-specific pre-synaptic markers were used, which respectively label synaptic vesicular transporters for the neurotransmitters GABA and glycine (VGAT), glutamate (VGLUT1 and VGLUT2), and acetylcholine (VAChT). We found that the average linear density of VGAT+ contacts was markedly decreased in both Pcdhgtcko/tcko and Pcdhgdel/del mutants (Figures 2J–J”, 2O), whereas the number of VGLUT1+ proprioceptive primary afferent inputs was surprisingly increased, more than double the number in wild type controls (Figures 2K-K”, 2O). By contrast, the densities of VGLUT2+ and VAChT+ contacts on motor neurons remain constant (Figures 2L-M, 2O). As expected, all four types of synapses are unaltered in Pcdhgtako/tako mutants (Figures 2O and S2C).
The significant decrease in VGAT+ synapses on motor neurons in both Pcdhgtcko/tcko and Pcdhgdel/del mutants is consistent with our observation that the two mutants display identical motor defects, which closely resemble those found in the VGAT (Wojcik et al., 2006), GAD67 (Asada et al., 1997), and Gephyrin (Feng et al., 1998) knockouts. Key features of the common phenotypes are muscle stiffness and immobility, which can be explained by tetanic motor neuron activation due to compromised inhibitory neurotransmission. The reduced density of VGAT+ contacts, as well as the normal numbers of VAChT+ synapses in Pcdhgtcko/tcko and Pcdhgdel/del mutants correlate well with the significant reduction of inhibitory interneurons and unaltered numbers of cholinergic partition cells in both mutants. By contrast, VGLUT2+ synaptic density is normal despite the reduction of certain premotor glutamatergic interneurons (e.g. Chx10+ V2a interneurons), which suggests that alternative neuronal sources or compensatory mechanisms might be involved in the development of these synapses.
The increased densities of VGLUT1+ contacts in both Pcdhgtcko/tcko and Pcdhgdel/del mutants indicate alterations in the stretch reflex circuit, where proprioceptive sensory afferents (Ia primary afferents, IaPA) establish monosynaptic contacts with spinal motor neurons innervating the same muscle (Chen et al., 2003). Centrally projecting IaPA axons (Parvalbumin+) in wild type spinal cords are distributed in an orderly fashion around motor pools, but in both mutants they appear clumped and more densely surround motor neurons, consistent with the observed increase in the density of VGLUT1+ contacts (Figures 2N-N”). The percentage of Parvalbumin+ neurons in mutant dorsal root ganglia (DRG) is similar to those of wild type animals (L2 DRG, 23.5±1.3% in Pcdhgdel/del and 21.8±1.7% in Pcdhg+/+, p>0.05), indicating that the increased density of the central projections of IaPAs is not due to increased numbers of proprioceptive neurons, but rather to a defect in their terminal arborization resulting from the loss of interneurons they interact with (Jankowska, 1992). Taken together, these observations indicate that similar alterations in specific types of synapses on motor neurons occur in both Pcdhgtcko/tcko and Pcdhgdel/del mutants.
Conditionally inactivating the C-type Pcdhg genes in the retina recapitulates the Pcdhg null phenotype
We next extended the genetic analysis to the retina. We have shown previously that Pcdhg genes are required for the survival of many neuronal subpopulations in the developing inner retina, but appear to be dispensable for synapse formation and function (Lefebvre et al., 2008). To generate retina-specific knockouts of TCKO and TAKO mutants, trans-heterozygous animals containing one conditional Pcdhgfcon3 allele (Lefebvre et al., 2008; Prasad et al., 2008) and one Pcdhgtcko or Pcdhgtako allele were produced. Upon retina-specific recombination of Pcdhgfcon3 that leads to a functionally null allele, the remaining Pcdhg isoforms are only expressed from the Pcdhgtcko or Pcdhgtako allele, providing a convenient way to conditionally inactivate the three C-type or A-type isoforms in postnatal animals.
We first examined the retinal architecture in conditional trans-heterozygous and Pcdhg null mutant animals. Retina lamination was normal and the ONL and OPL layers were unaffected in all three types of mutants. However, while the INL and IPL layers in Pcdhgfcon3/tako mutant retinas were indistinguishable from those in control, they were significantly thinner in both Pcdhgfcon3//tcko and Pcdhgfcon3/fcon3 mutant retinas (Figures 3A-A”’, 3D). Quantification of retina interneurons using cell-type specific markers revealed that INL thinning in Pcdhgfcon3//tcko mutants is due to reduced numbers of bipolar (Chx10+) and amacrine (Pax6+) interneurons (Figures 3B-B”’, 3E) as in the null (Lefebvre et al., 2008). Likewise, the loss of RGC projection neurons (Brn3a+) in Pcdhgfcon3/tcko trans-heterozygous retinas is equal in severity to that of Pcdhgfcon3/fcon3 mutants (Figures 3C-C”’, 3F). By contrast, we found no change in retina cell number in conditional Pcdhgfcon3/tako trans-heterozygous retinas (Figures 3D–F). Like the 3 C-type genes, the 3 A-type genes are also expressed in these cell types in the developing retina (Figures S3A-B), indicating that the full spectrum of Pcdhg isoforms are not required for neuronal survival. Increased levels of apoptosis observed in Pcdhgfcon3/tcko trans-heterozygous retinas (Figures 3G-G’) are consistent with the previous finding in Pcdhgfcon3/fcon3 mutants that cell loss and the consequent thinning of IPL is due to elevated programmed cell death (Lefebvre et al., 2008). As with Pcdhgfcon3/fcon3 mutants, targeting of interneurons and RGC dendrites to appropriate IPL laminae was intact in Pcdhgfcon3/tcko trans-heterozygous mutants (Figures 3H-H’). Therefore, in the retina as in the spinal cord, removal of the 3 C-type isoforms results in elevated neuronal apoptosis and loss of specific neuronal subtypes at similar levels and patterns as deletion of all 22 Pcdhg isoforms.
Figure 3. Similar levels and patterns of neuronal loss in the retina of Pcdhgtcko/tcko and Pcdhgdel/del mutants.
(A-A”’) Nuclear and synaptic layers of retina sections from P18 control and mutants are labeled with Po-Pro1 or anti-Bassoon, respectively. Retinal lamination is normal, and the ONL and OPL thickness are unaffected. INL and IPL thickness are similarly reduced in Pcdhgfcon3/tcko and Pcdhgfcon3/fcon3 mutants but are unaffected in Pcdhgfcon3/tako mutants. (B–C) Numbers of Chx10+ bipolar (red), Pax6+ GABAergic amacrine interneurons (green), as well as Brn3a+ projection retinal ganglion cells (magenta) are similarly reduced in Pcdhgfcon3/tcko and Pcdhgfcon3/fcon3 mutants. (D) Quantification of INL and IPL thickness in control and mutant retinas. (E, F) Quantification of bipolar interneurons (Chx10+ BP) and projection retinal ganglion cells (Brn3a+ RGC). (F-F”) Increased number of apoptotic cells (cleaved caspase 3+) is detected in the GCL and INL of the mutant retina (P4), as seen in Pcdhg null retinas. (G-G”) Syt2+ OFF bipolar processes (red) and Gγ13+ ON bipolar processes (green) ramify in the upper and lower regions of the IPL, respectively (P18). Laminar specificity is retained in mutants deficient in C-type Pcdhg isoforms, but marker-laminae are reduced and disrupted, identical as shown for Pcdhg null mutants. ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. *p < 0.05, *** p < 0.001. Scale bars: 50 µm.
Expression and function of A-type and B-type Pcdhg genes are not appreciably affected in the absence of C-type isoforms
We considered the possibility that the indistinguishable phenotypes of Pcdhgtcko/tcko and Pcdhgdel/del mutants arise because the deletion of C-type exons, which are located immediately upstream of the constant exons, interferes with transcription and splicing of other Pcdhg genes, leading to a severely hypomorphic Pcdhg allele. To address this possibility, we first examined the expression of the remaining 19 A- and B-type Pcdhg genes in Pcdhgtcko/tcko brains. RT-PCR using exon-specific primers revealed that all 19 variable exons are expressed and correctly spliced to constant exons (Figures 4A-B). Western blotting indicated that Pcdhg total protein levels in Pcdhgtcko/tcko brains are similar to the wild type and even higher than those in Pcdhg full cluster deletion heterozygotes, which are phenotypically normal (Figure 4C). We next asked whether the remaining A- and B-type proteins in Pcdhgtcko/tcko mutants are functional. Several studies indicate that Pcdhg and Pcdha proteins interact, and may form multimeric complexes with Pcdhb proteins (Han et al., 2010; Murata et al., 2004; Schalm et al., 2010). Moreover, both Pcdha and Pcdhg proteins are tyrosine phosphorylated in mature neurons, suggesting that they mediate intracellular signaling (Schalm et al., 2010). Co-immunoprecipitation experiments using a pan-Pcdhg antibody in brain lysate indicated that the A- and B-type Pcdhg isoforms in Pcdhgtcko/tcko mutants still form complexes with Pcdha proteins, they are tyrosine phosphorylated and they interact with Src, suggesting that they are capable of mediating intracellular signaling in the absence of the C-type isoforms (Figures 4D–E). We conclude that Pcdhgtcko/tcko is not a severe hypomorphic or dominant negative mutant, and that expression and function of the remaining A- and B-type Pcdhg isoforms is not appreciably distinct from those of wild type mice.
Figure 4. Expression and function of A- and B-type Pcdhg isoforms are not appreciably affected in the absence of C-type genes.
(A) Schematic representation of Pcdhg genes, transcripts and proteins. Positions of specific primers (red arrows) are indicated. ECD, extracellular domain; ICD, intracellular domain. (B) RT-PCR of individual Pcdhg transcripts in wild type and both lines of mutants. (C) Western blot indicates that the expression level of Pcdhg proteins in the Pcdhgtcko/tcko mutant brain is similar to the wild type level, but higher than that of Pcdhg+/del. Alpha tubulin is used as loading control. (D–E) A-type and B-type Pcdhg isoforms in Pcdhgtcko/tcko mutant brains still form complexes with Pcdha proteins, remain tyrosine phosphorylated, and mediate signaling. Western blots of P0 brain lysate from wild type and both mutants are shown in D, and blots of pan-Pcdhg immunoprecipitates in E. (F) Schematic representation of Pcdh clusters. Three identified cluster-wide enhancers (HS5-1, HS7 and HS16-20) and their respectively regulated regions are indicated. (G) Relative expression levels of individual exons in the Pcdh cluster region in E13.5 wild type and mutant spinal cords based on RNA-Seq analysis. Several genes with extremely low expression levels are excluded but can be found in Table S1. Significance levels are not indicated but can be found in Table S1. The red arrow in F and G indicates the EST gene AK149307.
To determine whether the expression of a common set of genes is altered in the two phenotypically indistinguishable mutants, we carried out deep sequencing (RNA-Seq) studies using embryonic spinal cords at E13.5, a developmental stage when neurogenesis is near completion (Nornes and Carry, 1978), but elevated apoptosis is not yet detected in the mutants (Prasad et al., 2008). Surprisingly, we observed no striking changes in global gene expression in either of the two mutants other than those in the Pcdh gene clusters themselves (Figures S4A-B and Table S1). In the case of Pcdhgdel/del mutants, the majority of Pcdhb genes are significantly upregulated, likely the consequence of the closer proximity of a Pcdhb cluster enhancer (HS16-20) located downstream of the Pcdhg cluster (Yokota et al., 2011), which is now repositioned ~300 kb closer to the Pcdhb cluster (Figures 4F–G). The largest increase in expression is observed with an EST gene (AK149307), which is located immediately upstream of the Pcdhg cluster (Figures 4F–G and Table S1). Sequence analyses revealed that this gene is a relic of the B-type Pcdhg isoforms, and its promoter region also contains a conserved sequence element (CSE) found in most Pcdh genes (Figures S4C–E). The expression levels of most Pcdhg isoforms are not affected by the deletion of C-type genes except for a few neighboring ones which are upregulated, and quantification of constant exon reads indicated that the combinatorial expression levels of the remaining Pcdhg genes in Pcdhgtcko/tcko mice are ~75% of the wild type levels (Figures 4F–G and Table S1). Thus, the loss of function of the C-type isoforms cannot be compensated by other Pcdhg isoforms. Many Pcdhb genes (as well as AK149307) are marginally upregulated in the Pcdhgtcko/tcko mice, likely also due to the action of the Pcdhb cluster enhancer as mentioned above. In addition, no neomorphic Pcdhg variants were detected in Pcdhgtcko/tcko mutants with splice junction analysis of the RNA-Seq data (Table S2). The striking phenotypic similarities in contrast to the vastly distinct Pcdh repertoires in Pcdhgtcko/tcko and Pcdhgdel/del mutants suggest that lack of the C-type Pcdhg isoforms themselves, which is common for both mutants, is the primary cause of the common phenotypes.
Genetically blocking apoptosis rescues the neonatal lethality of Pcdhgtcko/tcko mutants but not that of Pcdhgdel/del mutants
Since the primary phenotype observed in both Pcdhgtcko/tcko and Pcdhgdel/del is neuronal cell death, we crossed both mutant lines to Bax knockout mice (Knudson et al., 1995) to compare phenotypes when neuronal apoptosis is genetically blocked. Consistent with previous observations (Weiner et al., 2005), Pcdhgdel/del;Bax−/− pups show improved neurological function as compared with Pcdhgdel/del mutants, yet they still lack voluntary movements, and despite considerable efforts we were unable to recover any Pcdhgdel/del;Bax−/− mutants beyond P0 (Table 1 and Movie S2). Surprisingly, however, while some Pcdhgtcko/tcko;Bax−/− mutants die at P0, many live substantially longer despite being weaker and smaller than wild type and heterozygous pups. By culling littermates we were able to recover a number of Pcdhgtcko/tcko;Bax−/− mutants at weaning age. Some of these animals survived up to 6 months, although their persistent ataxia indicates neurological impairment (Table 1 and Movie S2).
Table 1.
Genetically blocking apoptosis rescues the neonatal lethality of Pcdhgtcko/tcko mutants but not that of Pcdhgdel/del mutants
| Parent genotypes | Age | # Animals Genotyped |
# Single Homozygotes |
# Double Homozygotes |
|---|---|---|---|---|
| Pcdhgtcko/+;Bax+/− | P0 P21 |
127 191 |
29 0 |
16 9 |
| Pcdhgdel/+;Bax+/− | P0 P21 |
89 193 |
11 0 |
9 0 |
Some normal looking littermates (wild type or heterozygous) were not genotyped and therefore correct genotype ratios cannot be calculated based on these numbers. Single homozygotes: Pcdhgtcko/tcko or Pcdhgdel/del mutants that are either Bax+/+ or Bax+/−; double homozygotes: Pcdhgtcko/tcko;Bax−/− or Pcdhgdel/del;Bax−/− mutants.
As described for the Pcdhgdel/del;Bax−/− mutants (Prasad et al., 2008; Weiner et al., 2005), the morphology of spinal cord sections of Pcdhgtcko/tcko;Bax−/− is indistinguishable from that of the Pcdhg+/+;Bax−/− animals, showing no signs of astrogliosis or microglia activation, and the arborization patterns of IaPA terminals appear largely indistinguishable from those of the controls (Figure S5A). Counts of both VGAT+ and VGLUT1+ inputs onto motor neurons were normal in the Bax−/− genetic background, while VGLUT2+ and VAChT+ synapses remain unchanged (Figure S5B). Therefore, the decreased number of VGAT+ contacts, as well as the increased number of VGLUT1+ contacts and IaPA terminals found in both Pcdhgtcko/tcko and Pcdhgdel/del mutants at P0, are both secondary to the cell death of spinal interneurons. These results are reminiscent of findings in the Pcdhg deficient retina in the Bax−/− genetic background, where retinal architecture, cell numbers, and synaptic densities are restored in the absence of neuronal apoptosis (Lefebvre et al., 2008).
DISCUSSION
The role of the Pcdhg gene cluster in neuronal survival is mediated by the C-type isoforms
Previous genetic studies using full cluster deletion mutants revealed that Pcdhgs are required for neuronal survival, but the underlying mechanism remains elusive. In this study, we generated mice lacking subsets of Pcdhg genes and performed quantitative analyses on specific types of neurons and synapses. Mice lacking C-type Pcdhg isoforms are phenotypically indistinguishable from Pcdhg null mutants, and the cellular and synaptic changes examined in both the spinal cord and retina are essentially identical. By contrast, mice lacking a subset of A-type isoforms are viable and fertile, revealing at least some level of functional redundancy among the alternative Pcdhg isoforms. Molecular and biochemical analyses demonstrate that deletion of C-type isoforms does not appreciably alter the expression or function of the A-type and B-type isoforms, indicating that the C-type isoform knockouts are not simply hypomorphic or dominant negative for Pcdhg function. Furthermore, transcriptome profiling shows that the Pcdh repertoires of the two mutants differ significantly, but no neomorphic Pcdhg variants are generated. Therefore, the loss of function of C-type isoforms themselves is most likely responsible for the identical phenotypes observed in both the C-type isoform knockouts and the Pcdhg null mutants.
The most remarkable difference between the two types of mutants is that, the neonatal lethality of C-type isoform knockouts can be rescued by genetically blocking apoptosis, while that of the full cluster Pcdhg deletion mutants cannot be rescued. The persistence of neonatal lethality in Pcdhgdel/del;Bax−/− mutants reveals an additional, independent role of Pcdhg isoforms that is required for postnatal development. Therefore, the role of Pcdhg cluster in neuronal survival is primarily, if not specifically mediated by the C-type isoforms, whereas the requirement of Pcdhg cluster for postnatal development appears to be the collective function of all 22 isoforms in neuronal wiring. Indeed, in a parallel study, we have found dendritic arborization defects in Pcdhg null mutants that are not observed in the C-type isoform knockouts (Lefebvre et al., 2012). Hence similar phenotypes are observed in the Pcdhgtcko/tcko and Pcdhgdel/del mutants because the C-type genes are deleted in both lines, and the resulting neuronal cell loss dominates the phenotypes. In the absence of apoptosis, however, the neonatal lethality in C-type isoform knockouts is rescued since neural circuitries essential for postnatal survival are largely preserved by the remaining 19 A-type and B-type Pcdhgs. By contrast, Pcdhgdel/del;Bax−/− compound mutants still die at P0 since the entire Pcdhg cluster is deleted, which severely compromises critical wiring and synaptic function carried out by all isoforms synergistically. The dual role of Pcdhg cluster in neuronal survival and neuronal wiring is thus elegantly accomplished by functional and regulatory diversification of its isoforms.
Neuronal apoptosis occurs independently of synaptic defects in Pcdhg deficient mice
Because of their differential expression, homophilic affinity and synaptic localization, the clustered Pcdhs have been proposed to be the “synaptic adhesive code” that specifies neuronal connectivity (Junghans et al., 2005; Serafini, 1999; Shapiro and Colman, 1999). Therefore, an intuitively attractive hypothesis for the concurrent neuronal apoptosis and synaptic defects in Pcdhg deficient mice is that the loss of function of Pcdhgs leads to synaptic loss, which in turn compromises neuronal survival (Junghans et al., 2005; Prasad et al., 2008). However, several observations reported here strongly argue against this possibility. 1) Deletion of Pcdhg cluster does not lead to a general loss of synapses. Instead, we found that Pcdhg deficiency differentially influences cholinergic, glutamatergic, GABAergic and glycinergic synapses on motor neurons. Therefore, the synapse loss observed using generic synaptic markers only reflects the additive effects of alterations in multiple types of synapses, and loss of synaptic contacts, at the very least, cannot explain the loss of neurons in all cases; 2) Consistent with the observations in the retina, the Pcdhgdel/del;Bax−/− and Pcdhgtcko/tcko;Bax−/− compound mutants showed preservation of stretch reflex circuits and major synaptic inputs onto motor neurons, indicating that these severe synaptic defects observed are secondary to interneuron loss; 3) Neural circuitries and synaptic functions are restored to a substantial extent in Pcdhgtcko/tcko;Bax−/− mutants, as shown by the rescue of neonatal lethality. If the synaptic defects are primary, they should remain when neuronal apoptosis is blocked by Bax deficiency (Buss et al., 2006). This is most likely the case in Pcdhgdel/del;Bax−/− mutants, which could not be rescued.
Taken together, these observations strongly suggest that neuronal cell death in Pcdhgtcko/tcko and Pcdhgdel/del mutants does not result from synaptic defects, but occurs independently due to the lack of C-type genes. Although we cannot rule out the possibility that synaptic or wiring defects may have contributed to the apoptosis of certain types of neurons, it cannot account for the massive cell death observed. In fact, loss of synaptic partners does not usually lead to apoptosis, and even grossly aberrant synaptic connections created experimentally are sometimes maintained without affecting the survival of source neurons (Buss et al., 2006; Oppenheim, 1991).
Functional similarities and differences between vertebrate Pcdhs and invertebrate Dscams
The vertebrate-specific Pcdh gene cluster shares remarkable resemblance with Drosophila Dscam1 gene in that they both have a complex genomic structure, which encodes a large number of distinct isoforms of cell adhesion molecules with homophilic binding affinity. These parallels have led to the hypothesis that Pcdhs, like Dscams, may provide a source of cell surface diversity for neurite self-recognition and self-avoidance (Zipursky and Sanes, 2010). This possibility is supported by our recent finding of dendritic self-avoidance defects in Pcdhg deficient mice (Lefebvre et al., 2012). A fundamental difference, however, resides in the fact that each Dscam1 isoform appears to be functionally equivalent, whereas certain Pcdhs, such as the C-type isoforms, have unique roles as we show here. Unlike Dscam1 where isoform diversity is generated by alternative splicing, differential expression of Pcdh isoforms is regulated by alternative promoter choice (Tasic et al., 2002; Wang et al., 2002a), which provides precise spatial and temporal controls over gene expression. As noted earlier, the C-type isoforms are phylogenetically unique among Pcdhs and exhibit distinct expression patterns. It remains to be seen whether just one of the three C-type genes is solely responsible for this function, or whether they work synergistically. We note that the two other C-type isoforms in the Pcdha cluster (Pcdhac1 and Pcdhac2) are dispensable for neuronal survival (Hasegawa et al., 2008; Katori et al., 2009), but they may play other specific roles yet to be identified. Further functional studies specifically targeting each of these C-type isoforms would be required to address these possibilities.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rick Myers, Rolf Kemler, Greg Philips, Andreas Kolb and Philippe Soriano for providing essential reagents, Monica Carrasco, Flo Pauli, Jiangwen Zhang, Hilary Bowden, and Amy Kirner for technical assistance, and Tom Jessell, George Mentis, Angel de Blas, Larry Shapiro, and members of the Maniatis laboratory for advice and discussion. This work is funded by NIH grants NS047357 to F. J. A., R01NS029169 to J. R. S., and R01NS043915 to T. M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
The Supplemental Data include the Supplemental Experimental Procedures, 5 Figures, 2 Tables, and 2 Movies and can be found with this article online.
REFERENCES
- Asada H, Kawamura Y, Maruyama K, Kume H, Ding RG, Kanbara N, Kuzume H, Sanbo M, Yagi T, Obata K. Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci U S A. 1997;94:6496–6499. doi: 10.1073/pnas.94.12.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss RR, Sun W, Oppenheim RW. Adaptive roles of programmed cell death during nervous system development. Annu Rev Neurosci. 2006;29:1–35. doi: 10.1146/annurev.neuro.29.051605.112800. [DOI] [PubMed] [Google Scholar]
- Chen HH, Hippenmeyer S, Arber S, Frank E. Development of the monosynaptic stretch reflex circuit. Curr Opin Neurobiol. 2003;13:96–102. doi: 10.1016/s0959-4388(03)00006-0. [DOI] [PubMed] [Google Scholar]
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet. 2005;37:171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- Feng G, Tintrup H, Kirsch J, Nichol MC, Kuhse J, Betz H, Sanes JR. Dual requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science. 1998;282:1321–1324. doi: 10.1126/science.282.5392.1321. [DOI] [PubMed] [Google Scholar]
- Fukuda E, Hamada S, Hasegawa S, Katori S, Sanbo M, Miyakawa T, Yamamoto T, Yamamoto H, Hirabayashi T, Yagi T. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur J Neurosci. 2008;28:1362–1376. doi: 10.1111/j.1460-9568.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- Han MH, Lin C, Meng S, Wang X. Proteomics analysis reveals overlapping functions of clustered protocadherins. Mol Cell Proteomics. 2010;9:71–83. doi: 10.1074/mcp.M900343-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa S, Hamada S, Kumode Y, Esumi S, Katori S, Fukuda E, Uchiyama Y, Hirabayashi T, Mombaerts P, Yagi T. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol Cell Neurosci. 2008;38:66–79. doi: 10.1016/j.mcn.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Jankowska E. Interneuronal relay in spinal pathways from proprioceptors. Prog Neurobiol. 1992;38:335–378. doi: 10.1016/0301-0082(92)90024-9. [DOI] [PubMed] [Google Scholar]
- Junghans D, Haas IG, Kemler R. Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr Opin Cell Biol. 2005;17:446–452. doi: 10.1016/j.ceb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, Yagi T. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J Biol Chem. 2006;281:30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, Hasegawa S, Yagi T. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci. 2009;29:9137–9147. doi: 10.1523/JNEUROSCI.5478-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson CM, Tung KS, Tourtellotte WG, Brown GA, Korsmeyer SJ. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012 doi: 10.1038/nature11305. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Zhang Y, Meister M, Wang X, Sanes JR. gamma-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development. 2008;135:4141–4151. doi: 10.1242/dev.027912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Hamada S, Morishita H, Mutoh T, Yagi T. Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J Biol Chem. 2004;279:49508–49516. doi: 10.1074/jbc.M408771200. [DOI] [PubMed] [Google Scholar]
- Nornes HO, Carry M. Neurogenesis in spinal cord of mouse: an autoradiographic analysis. Brain Res. 1978;159:1–6. doi: 10.1016/0006-8993(78)90105-1. [DOI] [PubMed] [Google Scholar]
- Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- Prasad T, Wang X, Gray PA, Weiner JA. A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: insights from genetic analyses of the protocadherin-gamma gene cluster. Development. 2008;135:4153–4164. doi: 10.1242/dev.026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalm SS, Ballif BA, Buchanan SM, Phillips GR, Maniatis T. Phosphorylation of protocadherin proteins by the receptor tyrosine kinase Ret. Proc Natl Acad Sci U S A. 2010;107:13894–13899. doi: 10.1073/pnas.1007182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci U S A. 2010;107:14893–14898. doi: 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T. Finding a partner in a crowd: neuronal diversity and synaptogenesis. Cell. 1999;98:133–136. doi: 10.1016/s0092-8674(00)81008-9. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 2002a;16:1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR. Gamma protocadherins are required for survival of spinal interneurons. Neuron. 2002b;36:843–854. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- Weiner JA, Wang X, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proc Natl Acad Sci U S A. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik SM, Katsurabayashi S, Guillemin I, Friauf E, Rosenmund C, Brose N, Rhee JS. A shared vesicular carrier allows synaptic corelease of GABA and glycine. Neuron. 2006;50:575–587. doi: 10.1016/j.neuron.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, Hirabayashi M, Hirabayashi T, Yagi T. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem. 2011;286:31885–31895. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky SL, Sanes JR. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 2010;143:343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.