Every chromosome is precious. Each one carries vital genetic information that must not be lost. To ensure chromosome retention and segregation when the cell divides, the spindle apparatus attaches to the chromosomes at the centromere. In higher eukaryotes, the centromere is a big region, often occupying a megabase or more in mammalian chromosomes.
Dedicating large amounts of DNA to centromere function is not a problem for mammalian cells. They carry thousands of megabases of DNA in their genome, the great majority of which does not code for proteins. But what about a virus? Compactness is the viral strategy. If a virus adopts the attractions of latency, remaining in the cell until conditions are right for lytic growth, how can it stably remain in the nucleus? A centromere seems out of the question. A different strategy is outlined in the paper by Lehman and Botchan in this issue of Proceedings (1).
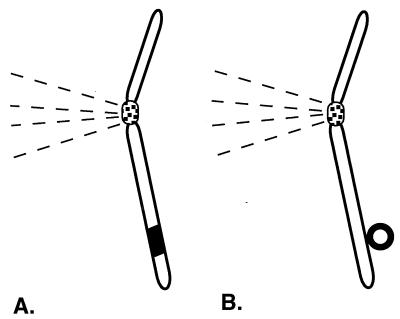
Once in the nucleus, a virus bound for latency must take action to ensure its survival against loss through dissolution of the nuclear membrane upon mitosis or through nuclear pores. An association with the chromosomes would ensure viral retention by taking advantage of chromosomal segregation and stability. Some viruses, like retroviruses and adeno-associated viruses, integrate into the chromosomes, forming covalent bonds that ensure that the viral genome will travel with the chromosomes. A less sure, but adequate, method to colocate with the chromosomes is to rely on a noncovalent association with chromosomes. This is the route of bovine papilloma virus (BPV) (Fig. 1).
Figure 1.
Two ways in which viral genomes can take advantage of the chromosomal centromere to maintain themselves long term in mammalian cells. (A) Integration is a covalent interaction with the chromosomes. It is the maintenance mode of viral genomes such as retroviruses. (B) Genomes such as BPV and EBV maintain a noncovalent close association with chromosomes, mediated by a viral protein, to ensure viral maintenance and segregation. The chromosomal centromere and the spindle attachment are depicted schematically. Viral genomes are shown in bold and, for clarity, are exaggerated greatly in size in relation to the chromosome. (Illustration drawn by Julie Phillips.)
BPV infects dividing basal epithelial cells and remains latent until the infected cells reach the skin surface as terminally differentiated keratinocytes. At that point, lytic replication ensues and viral particles are made (2). During latency, the viral genome exists as a multicopy circular plasmid in the nucleus. It replicates by using the viral origin of replication and the viral E1 and E2 proteins, as well as the host replication apparatus. The E1 and E2 proteins bind to the BPV genome in the origin region. The E1 protein is a helicase required for replication initiation and elongation, and E2 is a transcription factor that also facilitates E1 binding to the origin (3, 4).
Plasmids having the viral origin, E1, and E2 replicate extrachromosomally in mouse tissue culture model systems (2, 5), and it is such a system that Lehman and Botchan used for their studies (1). In mouse cells, BPV plasmids are maintained at a stable copy number for prolonged periods (2). Yet we know that introduced nonviral plasmid DNA is lost rapidly from most mammalian cells, even predominantly nondividing cells (6). How, then, does the extrachromosomal BPV plasmid manage to remain in the cells at a similar copy number for generation after generation?
Lehman and Botchan (1) used a mutant form of E2 to demonstrate that E2 mediates a physical association between the BPV genome and the chromosomes. Fluorescence in situ hybridization studies showed that the E2 A4 mutant produced sectored colonies during growth in mouse cells, such that the BPV genome was entirely lost from parts of the colony. These results suggested that the E2 mutant was losing viral DNA during cell division. A replication defect was not likely because the E2 mutant replicated well in transient assays. The segregation activity of E2 appeared to be regulated by phosphorylation (1, 7).
Striking fluorescence in situ hybridization photographs showed that the E2 mutant did not associate with metaphase chromosomes. Wild-type BPV did associate with chromosomes, with no specific attachment sites apparent. Furthermore, staining with a mAb that recognized wild-type and mutant E2 equally showed that, although wild-type E2 colocalized with chromosomes during mitosis, the mutant E2 did not. These results suggested a direct role for E2 in chromosome attachment. Second site suppressors of the E2 A4 mutant were found in both E2 and E1, suggesting that E1 also was involved in chromosome attachment.
The fluorescence in situ hybridization images of Lehman and Botchan (1) indicate that BPV genomes are distributed among the chromosomes and remain closely associated with them. In this way, when the chromosomes segregate during mitosis, BPV genomes will be distributed to both daughter cells and will not be lost from the nucleus. BPV thus obtains some of the benefits of having a centromere without having to encode this cumbersome structure. By hitchhiking along with chromosomes, BPV ensures its own retention and segregation.
A similar mode of viral maintenance is used by Epstein–Barr virus (EBV), a larger virus that also has a significant latent existence and replicates as an extrachromosomal circle (5). The EBV protein EBNA-1 is found associated with metaphase chromosomes (8), and EBV viral genomes remain tightly associated with chromosomes during flow sorting, although the association is not covalent (9).
The EBV latent origin of replication oriP (10) together with the viral EBNA-1 gene needed to activate the origin define EBV vectors that can replicate stably in human tissue culture cells (11). OriP is bipartite, with a dyad region required for efficient replication and a family of repeats region that also has transcriptional enhancer activity (12, 13). EBNA-1 binds to repeats in each oriP element (14), and both elements are required in cis for efficient replication. Deletion of the dyad part of the origin produces vectors that cannot replicate efficiently but still mediate gene expression for a prolonged time in the cell (12, 13). This effect is due to the ability of EBNA-1 and the family of repeats to cause nuclear retention of linked DNA (15). The retention function mediated by EBNA-1 and the family of repeats is vital to the long term maintenance of EBV plasmids in cells.
Thus, in both BPV and EBV, a viral protein and its binding sites in the origin region of the viral genome seem to mediate a close association with the chromosomes that aids in nuclear retention and segregation of the viral genome. The identity of the chromosomal element to which the viral protein is binding is not known in either case. It could be a chromosomal protein, such as a transcription factor or a scaffold protein, or it may be the chromosomal DNA itself. The EBV nuclear retention function requires domains of EBNA-1 also required for replication and transcription, and it has been suggested that all these functions may be mediated by an overlapping set of cellular proteins (16).
The viral proteins involved in each case, EBNA-1 for EBV and E1 and E2 for BPV, have important roles in viral replication. It is possible that the chromosomal association is involved in enhancing viral replication, as well as providing retention. For both viruses, maintenance of the viral genome in cells can be reinforced by conferring a selective advantage on infected cells through oncogenic transformation. However, replication and retention of the viral genome still are required for its maintenance.
The cleverness of viruses often supplies us with new genetic engineering strategies. To date, many viruses have been used in gene therapy (17), primarily for use of the capsid as a delivery vehicle and, in the case of retroviruses, also for use of the integration mechanism. However, using “whole” viruses in this way also brings many limitations. It is attractive to use only those viral components that are beneficial in the gene therapy scheme. In this regard, the method of nuclear retention used by these extrachromosomal viruses might find use in stabilizing DNA introduced in gene therapy.
A conventional plasmid vector introduced into target cells disappears rapidly from most cells. For example, after lipid-mediated delivery, a peak of plasmid gene expression is seen between 4 and 24 h after transfection, followed by a rapid drop, such that <1% of the DNA is present 4 days after transfection (6). Retention is much better in muscle cells (18), but there are limits on the utility of muscle cells in gene therapy. Gene therapy vectors carrying the EBV family of repeats and EBNA-1 show prolonged retention and prolonged gene expression of linked marker genes in tissue culture cells, suggesting utility in extending the effectiveness of gene therapy (19–21). Papilloma virus vectors carrying the origin, E1, and E2 would be expected to show similar effects. Other DNA viruses that maintain nuclear DNA without integration also might possess special retention sequences and proteins that would be useful to characterize. Whether the viral proteins required by these retention systems will be acceptable in gene therapy and whether these effects can be demonstrated in animals remain to be tested (22).
However, the analogy is good between viruses and gene therapy vectors. Both benefit from compactness, so their options for maintenance are limited similarly. The larger the gene therapy vector, the more difficult the manipulation, manufacture, and delivery, so the challenge is to provide stability with a minimal commitment of DNA. Viruses like BPV and EBV achieve prolonged stability by using a mechanism that encompasses only a few kilobases of DNA.
Acknowledgments
Work in the Calos laboratory is supported by grants from the Cystic Fibrosis Foundation (Z959) and the National Institutes of Health (DK51834).
ABBREVIATIONS
- BPV
bovine papilloma virus
- EBV
Epstein–Barr virus
References
- 1.Lehman C W, Botchan M R. Proc Natl Acad Sci USA. 1998;95:4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howley P M. In: Fundamental Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 947–978. [Google Scholar]
- 3.Seo Y-S, Muller F, Lusky M, Gibbs E, Kim H-Y, Hurwitz J. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M R. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mecsas J, Sugden B. Annu Rev Cell Biol. 1987;3:87–108. doi: 10.1146/annurev.cb.03.110187.000511. [DOI] [PubMed] [Google Scholar]
- 6.Song Y K, Liu F S C, Liu D. Hum Gene Ther. 1997;8:1585–1594. doi: 10.1089/hum.1997.8.13-1585. [DOI] [PubMed] [Google Scholar]
- 7.Lehman C W, King D M, Botchan M R. J Virol. 1997;71:3652–3665. doi: 10.1128/jvi.71.5.3652-3665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grogan E A, Summers, Summers W P, Dowling S, Shedd D, Gradoville L, Miller G. Proc Natl Acad Sci USA. 1983;80:7650–7653. doi: 10.1073/pnas.80.24.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris A, Young B D, Griffin B E. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yates J, Warren N, Reisman D, Sugden B. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yates J L, Warren N, Sugden B. Nature (London) 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 12.Reisman D, Sugden B. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reisman D, Yates J, Sugden B. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawlins D R, Milman G, Hayward D, Hayward G S. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 15.Krysan P J, Haase S B, Calos M P. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middleton T, Sugden B. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verma I M, Somia N. Nature (London) 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 18.Wolff J A, Malone R W, Williams P, Chong W, Acsadi G, Jani A, Felgner P L. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 19.Wohlgemuth J G, Kang S H, Bulboaca G H, Nawotka K A, Calos M P. Gene Ther. 1996;3:503–512. [PubMed] [Google Scholar]
- 20.Lei D C, Kunzelmann K, Koslowsky T, Yezzi M J, Escobar L C, Xu Z, Ellison A R, Rommens J M, Tsui L-C, Tykocinski M, et al. Gene Ther. 1996;3:427–436. [PubMed] [Google Scholar]
- 21.Banerjee S, Livanos E, Vos J-M H. Nat Med. 1995;1:1303–1308. doi: 10.1038/nm1295-1303. [DOI] [PubMed] [Google Scholar]
- 22.Calos M P. Trends Genet. 1996;12:463–466. doi: 10.1016/0168-9525(96)40049-x. [DOI] [PubMed] [Google Scholar]