Abstract
Background
Bartonella elizabethae has been reported as a causative agent of human illnesses and strains of this bacterium are commonly isolated from commensal small mammals in Asia.
Methods
Since the zoonotic potential of a pathogen is often related to its host switching ability, we explored the capacity of a B. elizabethae strain to host switch by subcutaneously inoculating groups of Swiss Webster, BALB/c, and C57BL/6 mice with the bacteria at a range of doses.
Results
A low number of mice in each of the three groups showed susceptibility to infection at high doses (105 and 106 bacteria), and developed bacteremias of 6–8 weeks duration.
Conclusion
The capacity of this B. elizabethae strain to switch hosts can have important public health consequences for humans in areas of Asia where many small mammal populations have high bartonellae infection prevalences and live as commensals with humans.
Keywords: Suncus murinus, zoonoses, shrew, rat, Vietnam
Bacteria in the family Bartonellaceae are facultative intracellular microparasites of erythrocytes and endothelial cells. These small, fastidious, pleomorphic bacilli are most closely related to members of the genera Brucella and Agrobacterium. There are two primary human pathogens within the genus, Bartonella quintana, and Bartonella bacilliformis, and 32 other species have been described, 14 of which have been associated with human illnesses (1, 2). With respect to the zoonotic bartonellae, over 22,000 cases of human infection with Bartonella henselae, the ‘cat scratch disease’ agent, are reported annually in the United States alone (3). The burden of human disease from other zoonotically acquired bartonellae infections, both in the United States and on a global scale, has yet to be fully determined.
Over the last several decades detection of bartonellae infections in humans has been associated with a wide spectrum of clinical disease manifestations (3, 4). Symptomatology associated with incidental host infections can include long term headache and myalgia, self-limiting, regional lymphadenopathy, neuroretinitis, and in some cases more serious illnesses such as meningitis and endocarditis (1, 3). While infection of incidental hosts with zoonotic bartonellae can result in disease these bacteria do not normally cause morbidity in their natural hosts (1, 5).
Bartonella infections acquired from peridomestic/commensal small mammals seem to occur at low incidence but this may be due to the inherent difficulties in diagnosing these infections (6, 7). The fastidious requirements of the bacteria in conjunction with low levels of bacteremia in incidentally infected hosts makes detection of this agent complex. Bacterial isolation from incidental host blood is uncommon, and though tissues and blood are often cultured in attempts to obtain an isolate these methods are not often successful (6, 8, 9). Species specific PCR assays and serological tests such as immuno-fluorescent assays, with four-fold changes in titer between acute and convalescent sera, are often relied upon to diagnose infections (9–11).
Whenever possible human cases of zoonotic bartonellae infection have been followed up with epidemiological risk assessments of behavior and environment (9, 12, 13). A consistently identified risk factor seems to be a history of direct and indirect exposure to a broad variety of animals, both wild and domestic, over time, as well as contacts with potential arthropod vectors (6, 14). However, specific exposure events leading to infection are not often identified (7, 9, 13).
In peridomestic and periagricultural environments in many areas of the world human contacts with rodents, lagomorphs, and insectivores can pose heightened risk for acquiring zoonotic bartonellae infections (15–17). In Asia there are several species of rodents and an insectivore species, Suncus murinus, the Asian house shrew, that adapt easily to peridomestic environments and are frequently found infected with diverse bartonellae at high population prevalences (16–18). These commensal small mammals often share a common ectoparasite fauna which may constitute a vector transmission risk to humans for bartonellae infections (19).
Rodents are often the focus of interest as reservoir hosts for bartonellae, but shrews have also been found to harbor both unique (shrew only) and rat-associated bartonellae (17, 20, 21). High nucleotide sequence similarities between shrew isolates from geographically distinct areas in Asia support the assertion that shrews are natural hosts for their own circulating strains of the bacteria, but they are also found infected with B. elizabethae-like strains (17, 20). Infection of shrews with bartonellae closely related to rat strains likely represents spillover events. The proportion of sequence similarity between strains from different hosts in the same geographic location can indicate how recently the bacterial strains may have switched hosts. For example, a bartonella isolate from an Asian house shrew in Vietnam shares 100% sequence identity with B. elizabethae by analysis of a portion of the citrate synthase gene (gltA). B. elizabethae was also isolated from two natural reservoir hosts, Rattus norvegicus and Rattus exulans, in that area as well. This finding points to an ongoing dynamic background of spillovers from rat populations into sympatric shrews. It also demonstrates the adaptability of a B. elizabethae strain in switching hosts, from rats to shrews, and suggests that this bacterium may have an inherent capacity to exploit new hosts.
B. elizabethae and closely related strains have been implicated as agents of human disease (6, 22), and exposure to the bacteria seems high in some human populations as revealed by serological surveys (23–25). B. elizabethae-like strains are most commonly isolated from rats, and rats unquestionably serve as zoonotic sources of human infection for many viral, bacterial, and parasitic diseases (26). The likelihood that incidental hosts can be infected by shrew bartonellae strains is unknown but may be predicted based on the host-specificity of strains obtained from these animals (27). Some bartonellae have been shown to infect a variety of hosts (28), while others display a high level of host specificity (29).
Shrews are mammals in the Order Soricomorpha and infection of laboratory mice (Order Rodentia) with a shrew bartonella strain could demonstrate the host switching capacity of this bacteria. B. elizabethae is an emerging bacterial agent in Asia, and information about the bacterium's natural history and zoonotic potential is lacking. Therefore we designed a study to evaluate the ability of our Asian house shrew B. elizabethae strain to switch from its host of origin to the laboratory mouse, Mus musculus.
Three laboratory mouse stocks were inoculated with a range of bacterial doses, and susceptibility to infection and bacteremia kinetics of infected mice were observed. In this study we used three mouse stocks representing different genetic backgrounds, thus allowing for a broad range of observations and interpretation of response to infection (30). Recognizing the importance of individual host immune status in susceptibility to infection we chose inbred BALB/c mice, which tend toward a TH2 type immune response, and inbred C57BL/6 mice which display a relatively robust TH1 response for the study (30). We also included genetically heterogeneous, outbred Swiss Webster mice in our design to assess possible variability in individual host susceptibility with respect to the other two mouse stocks.
Materials and methods
Mice
Four-to-six-week old Swiss Webster (SW) female mice were obtained from an outbred closed colony at the Division of Vector-Borne Diseases (DVBD), Fort Collins, Colorado, USA. Age matched BALB/c and C57BL/6 female mice were ordered from Jackson Laboratories (Sacramento, California, USA). Mice were group caged according to stock identity and bacterial dose for the study duration. All work on the mice was approved by and conducted under the supervision of DVBD's Institutional Animal Care and Use Committee (protocol #07-004), in accordance with the United States Public Health Service standards for the humane care of laboratory animals.
Bacteria
The B. elizabethae strain, Sm6145vi, used in this study was originally isolated from an Asian house shrew captured in Vietnam. Passage history and other information about Sm6145vi are detailed in Table 1, and does not include growth of bacteria for this study. We used a low passage bacterial strain to preserve fidelity to the original isolate to the greatest extent possible. To grow stock to infect mice, bacteria were inoculated onto heart infusion agar (HIA) plates supplemented with 5% rabbit blood. Following 5 days growth at 35 °C in a 5% CO2 incubator bacteria were harvested in physiological saline and frozen at −80 °C until used for mouse inoculations. To calculate bacterial stock concentration an aliquot of frozen stock was thawed, diluted 10-fold, and plated as above to permit enumeration of colony forming units (cfu).
Table 1.
The low passage Bartonella elizabethae strain used in this study was originally isolated from the Asian house shrew, Suncus murinus
| Asian house shrew Bartonella elizabethae strain | Sm6145vi |
| Place of origin | Dong Nai Province, Vietnam (2003) |
| Primary isolation | Whole blood inoculated onto BHIa supplemented with 5% rabbit blood |
| Subsequent passage history (n=number of passages) | BHI supplemented with 5% rabbit blood, n=3; BALB/c in vivob, n=1; BHI supplemented with 5% rabbit blood, n=2 |
| Nucleotide sequence [GenBank accession number] | gltA [JF523414 ] |
BHI = Brain heart infusion agar plates.
BALB/c mouse inoculated intraperitoneally and subcutaneously with a divided dose of 107 cfu bacteria.
Experimental design
Groups of 36 mice of each mouse stock were used in the study for a total of 118 experimental mice. Six mice were inoculated at each dose. Mice were inoculated only once, subcutaneously in the scruff, with bacterial doses ranging from 101 to 106 cfu diluted in saline to 350 µl. Mice were weighed weekly and assessed during handling to evaluate whether weight loss, decreased grooming, or altered behavior might occur following infection.
Beginning 1 week post-inoculation through week 13 blood was collected from mice by submandibular vessel plexus puncture. Mice were first intraperitoneally injected with a ketamine–xylazine solution (100 mg/kg ketamine + 50 mg/kg xylazine), then the cheek was shaved and the skin cleansed to reduce contamination of the blood during collection. Volumes of 75–125 µl of blood were collected from each mouse weekly. Samples were stored at −80 °C until tested for bacteremia.
Testing of blood for bacteremia
Frozen whole blood samples were thawed and diluted (50 µl blood:150 µl brain heart infusion diluent with 8% amphotericin B added). Heart infusion agar plates enriched with 5% rabbit blood were inoculated with 100 µl of the diluted samples and placed in a CO2 incubator at 35 °C for 10–21 days to permit bacterial growth. Bacteremic blood samples were subsequently diluted 1:100 and 1:1000 to allow for cfu enumeration. Positive samples were confirmed as bartonella by colony morphology and microscopic appearance as small, Gram negative bacilli following Gram staining.
Results
Infection of Swiss Webster mice
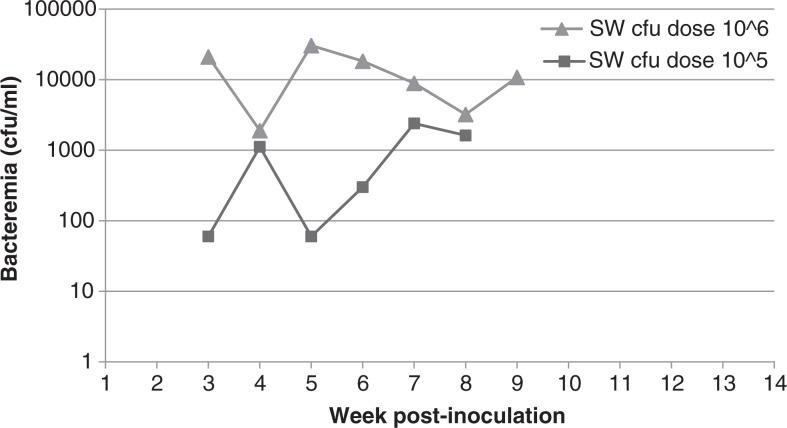
Following inoculation with B. elizabethae Sm6145vi, 2 of 36 SW mice became infected and developed bacteremia: 1 of 6 at the 106 cfu dose and 1 of 6 at the 105 cfu dose (Table 2, Fig. 1). Bacteremia lasted 7 weeks in the 106 cfu dose SW mouse and 6 weeks in the 105 cfu dose mouse. Onset of bacteremia occurred 3 weeks post-inoculation for both mice, but level at onset for the 106 cfu dose mouse was 2.1×104 cfu/ml blood whereas level at onset for the 105 cfu dose mouse was 60 cfu/ml blood. The 106 cfu dose SW mouse attained the highest level bacteremia of any mouse in the study (3.1×104 cfu/ml blood). Swiss Webster mice inoculated with 104 cfu or less of Sm6145vi failed to develop bacteremias.
Table 2.
Three mouse stocks were inoculated with Bartonella elizabethae strain Sm6145vi. Only mice inoculated at the highest two doses, 105 and 106 bacteria/mouse, became infected and manifested bacteremias
| Mouse stock inoculated (infective dose in cfua) | Number bacteremic at each dose/number inoculated at each dose |
|---|---|
| Swiss Webster (106) | 1/6 (16.7%) |
| Swiss Webster (105) | 1/6 (16.7%) |
| Swiss Webster (104) | 0/6 |
| Swiss Webster (103) | 0/6 |
| Swiss Webster (102) | 0/6 |
| Swiss Webster (101) | 0/6 |
| BALB/c (106) | 1/6 (16.7%) |
| BALB/c (105) | 0/6 |
| BALB/c (104) | 0/6 |
| BALB/c (103) | 0/6 |
| BALB/c (102) | 0/6 |
| BALB/c (101) | 0/6 |
| C57BL/6 (106) | 2/6 (33%) |
| C57BL/6 (105) | 0/6 |
| C57BL/6 (104) | 0/6 |
| C57BL/6 (103) | 0/6 |
| C57BL/6 (102) | 0/6 |
| C57BL/6 (101) | 0/6 |
Colony forming units of bacteria.
Fig. 1.
Bacteremia kinetics of Swiss Webster mice infected with B. elizabethae Sm6145vi. Only 2 of 36 total Swiss Webster mice became bacteremic following inoculation. Those were mice inoculated at the two highest doses.
Infection of BALB/c mice
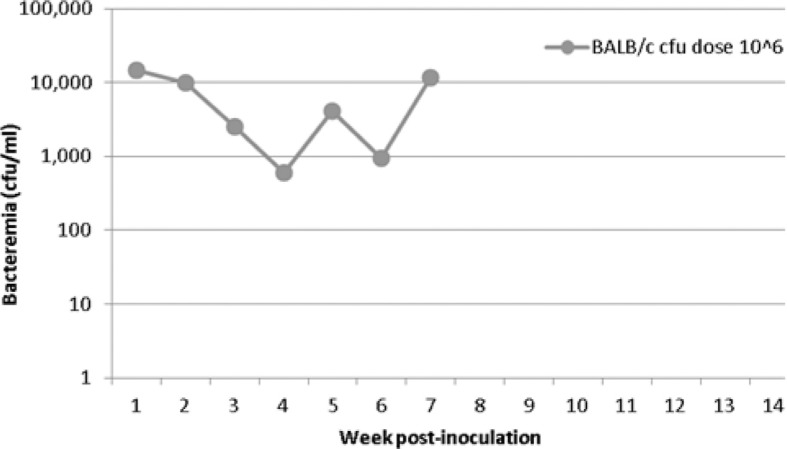
One of six BALB/c mice became infected following inoculation with a 106 cfu dose of the bacteria. The duration of bacteremia for this mouse was 7 weeks, with onset at week 1 post-inoculation (Table 2, Fig. 2). The level of bacteremia at onset was 1.5×104 cfu/ml blood, which was also the highest level of bacteremia observed in this mouse over the course of infection. This mouse had the earliest onset of bacteremia (week 1) of any bacteremic mouse in the study. BALB/c mice inoculated with 105 cfu or less of B. elizabethae Sm6145vi failed to develop bacteremia.
Fig. 2.
Bacteremia kinetics of a BALB/c mouse infected with B. elizabethae Sm6145vi. Only 1 of 36 BALB/c mice became bacteremic following inoculation of the bacteria. That one mouse was inoculated at the highest dose.
Infection of C57BL/6 mice
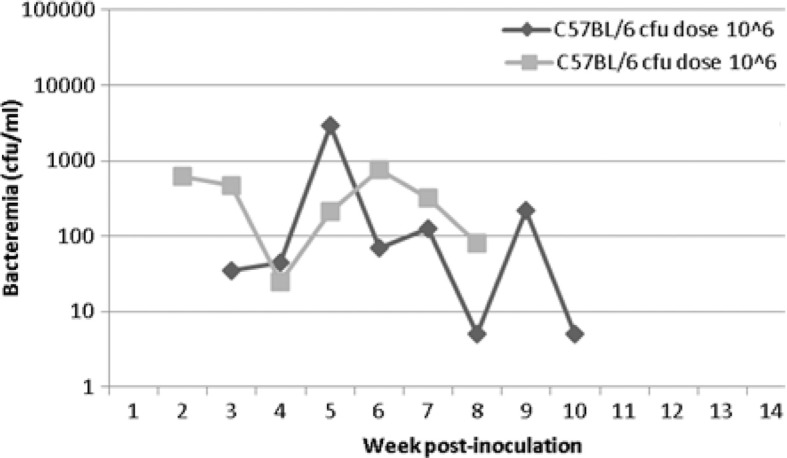
Two of six C57BL/6 mice became infected at the 106 cfu dose and developed bacteremias lasting 7 and 8 weeks in length (Table 2, Fig. 3). These two mice had overall lower levels of bacteremia than the other mice in the study. Onset of bacteremia was observed at weeks 2 and 3 for the mice, with levels at onset of 615 and 35 cfu/ml blood, respectively. The highest levels of bacteremia observed for the mice were 2.9×103 and 755 cfu/ml blood, again respectively. C57BL/6 mice inoculated with 104 cfu or less of B. elizabethae Sm6145vi failed to develop bacteremias.
Fig. 3.
Bacteremia kinetics of C57BL/6 mice infected with B. elizabethae Sm6145vi. Only 2 of 36 C57BL/6 mice inoculated became bacteremic. Those two mice were each inoculated with 106 cfu/mouse.
Bacteremic mice of all three stocks in the study showed fluctuations in their levels of bacteremia which could exceed an order of magnitude from 1 week to the next. No mice showed weight loss during this study, or other adverse health effects as assessed during handling. Indeed, all mice gained weight as expected for their age.
Discussion
In this study an Asian house shrew B. elizabethae strain (Sm6145vi) infected laboratory mice, a demonstration of cross-order host switching by the bacteria (from Order Soricomorpha to Order Rodentia). This is to some extent an unusual finding as bartonella bacteria generally only infect and produce bacteremias in hosts taxonomically close to their natural reservoir hosts (28, 29, 31, 32). The ability of this strain to host switch across mammalian orders may make it more likely to undergo zoonotic transmission to humans (27). Though this B. elizabethae strain was isolated from a shrew, its close phylogenetic relatedness to rat isolates of B. elizabethae in the same location means it is likely a spillover from rats to sympatric shrews in Vietnam. Therefore, unlike some other bartonellae this strain seems to have an inherent capacity to infect diverse hosts (29). However, bacteremia occurred only in mice inoculated with high doses of B. elizabethae Sm6145vi (105 and 106 cfu), and it remains uncertain how many bartonella bacteria are required to establish infection under natural conditions.
B. elizabethae has been reported as the causative agent of several cases of human illness (6, 22), and serologic evidence of human infection with the bacterium has been reported from Thailand (10, 33). Still, the zoonotic potential of this bacterium is not well understood. Since B. elizabethae, and strains phylogenetically close to B. elizabethae have been found in numerous commensal small mammal populations in Asia (15, 17), an understanding of the risk to humans for acquiring infections from these hosts is desirable. Given that the level of host specificity of an infectious agent is generally considered a predictor for the likelihood that the agent can switch hosts, and potentially cause illness in those hosts, our findings help define the zoonotic potential of this bacteria (27).
Previously, B. elizabethae isolated from a human endocarditis patient (the type strain) was evaluated for its ability to infect several different rodent species, among them R. norvegicus, a natural reservoir host for the bacteria (22, 29). In that study the bacteria failed to infect Wistar rats (R. norvegicus), cotton rats (Sigmodon hispidus), BALB/c (M. musculus), and white-footed deer mice (Peromyscus leucopus), though inoculated doses were as high as 107 bacteria (29). It remains unknown whether the bacteria had undergone adaptation to the human host resulting in a high level of host specificity, or whether the isolate's passage history might have influenced the outcome (34). An additional unanswered question was whether related isolates from natural reservoirs would also display a narrow host range, or a host specific phenotype.
In contrast to those findings, we observed susceptibility of three stocks of M. musculus (SW, BALB/c, and C57BL/6) to infection with B. elizabethae Sm6145vi at doses of 105–106 bacteria. Mice developed bacteremia levels potentially high enough to infect ectoparasites such as fleas feeding on a host during the course of bacteremia (35). The bacteremia levels observed in our incidental host mouse model may be sufficient to promote some secondary infections of susceptible hosts, especially if high enough levels of bacterial exposure exist in terms of transmissible contacts between animals.
Subjective differences in bacteremia levels between mice inoculated at the same dose may be due to individual host response, with respect to the particular mouse stock (Fig. 1). Likewise, differences between bacteremia levels of infected mice of the three stocks could be explained by individual mouse responses, stock differences, or dose differences (Fig. 1, SW mice infected at two different doses). With such small numbers of bacteremic mice it is difficult to speculate or draw conclusions about the bacteremia levels.
Bacteremias of several months duration or more are commonly observed in natural reservoir hosts infected with their co-adapted bartonellae (5, 31, 36). The bacteremia duration of mice in our study is shorter than that observed during such natural host infections, but is consistent with bacteremia kinetics of laboratory mice experimentally infected with non-homologous host source bartonellae (37, 38). Truncations in bacteremia duration are likely due to the bacteria not being optimally adapted to laboratory mice, and are probably characteristic of non-natural host infections (32, 37, 38).
Similar, small proportions of mice of each stock used in this study were susceptible to infection and developed bacteremias following inoculation of B. elizabethae strain Sm6145vi. The differing genetic backgrounds among the three stocks did not appear to affect susceptibility of mice to infection, at least not with the dose range assayed. It is possible that inoculation of larger group sizes would reveal more apparent differences between stocks. We did not attempt to assess for differential response to infection for the three mouse stocks at different exposure doses, as the number of bacteremic mice in each dose group was low. It would be difficult to assign a biologically significant interpretation to such slight differences in infection rates and level and duration of bacteremia without knowing that the observed differences are relevant to the transmission dynamics of the bacteria. Bacteremia duration in infected hosts almost undoubtedly affects the transmission likelihood of the bacteria. Simply put, long bacteremias increase the size of the temporal window for potentially transmissible contacts to occur between infected hosts and susceptibles, or for arthropod vectors to acquire the agent (27). Likewise high bacteremia levels can influence the probability that contacts with infected hosts result in bacterial transmission or arthropod vectors become infected following ingestion of infectious blood (27). However, to reasonably extrapolate our laboratory based findings to the natural transmission cycle of B. elizabethae, additional studies need to be done to define the transmission dynamics of the bacteria in its normal hosts.
Further evaluations of B. elizabethae Sm6145vi could yield more information about this bacterium's host switching ability, adaptive capacity, and zoonotic potential. Additional in vivo passage(s) of the bacteria in laboratory mice followed by another experimental infection study might reveal differences in the ability of a mouse adapted clone to infect different mouse stocks. Such an experiment could also provide insight into B. elizabethae's rate of adaptation to a new host. Alternatively, experimental infection studies could be done to evaluate the ability of this bacterium to infect R. norvegicus and R. rattus, natural reservoir hosts of B. elizabethae (26, 39), to determine if adaptation to other hosts has altered its capacity to infect its natural hosts (29). Knowledge gained about the zoonotic potential of B. elizabethae strains can aid us in implementing measures to reduce human infection risk in areas of the world where these strains circulate in small mammal populations.
Conflict of interest and funding
The authors declare they have no conflicts of interest.
References
- 1.Vayssier-Taussat M, Le Rhun D, Bonnet S, Cotte V. Insights in Bartonella host specificity. Ann N Y Acad Sci. 2009;1166:127–32. doi: 10.1111/j.1749-6632.2009.04531.x. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser P, Riess T, O'Rourke F, Linke D, Kempf VAJ. Bartonella spp.: throwing light on uncommon human infections. Int J Med Microbiol. 2011;301:7–15. doi: 10.1016/j.ijmm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Chomel BB, Kasten RW. Bartonellosis, an increasingly recognized zoonosis. J Appl Microbiol. 2010;109:743–50. doi: 10.1111/j.1365-2672.2010.04679.x. [DOI] [PubMed] [Google Scholar]
- 4.Breitschwerdt E, Maggi R, Chomel B, Lappin M. Bartonellosis: an emerging infectious disease of zoonotic importance to animals and human beings. J Vet Emerg Crit Care. 2010;20:8–30. doi: 10.1111/j.1476-4431.2009.00496.x. [DOI] [PubMed] [Google Scholar]
- 5.Birtles RJ. Bartonellae as elegant hemotropic parasites. Ann N Y Acad Sci. 2005;1063:270–9. doi: 10.1196/annals.1355.044. [DOI] [PubMed] [Google Scholar]
- 6.Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney S, et al. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am J Trop Med Hyg. 2010;82:1140–5. doi: 10.4269/ajtmh.2010.09-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeanclaude D, Godmer P, Leveiller D, Pouedras P, Fournier PE, Raoult D, et al. Bartonella alsatica endocarditis in a French patient in close contact with rabbits. Clin Microbiol Infect. 2009;15(Suppl 2):110–1. doi: 10.1111/j.1469-0691.2008.02187.x. [DOI] [PubMed] [Google Scholar]
- 8.Zangwill KM, Hamilton DH, Perkins BA, Regnery RL, Plikaytis BD, Hadler JL, et al. Cat scratch disease in Connecticut. Epidemiology, risk factors, and evaluation of a new diagnostic test. N Engl J Med. 1993;329:8–13. doi: 10.1056/NEJM199307013290102. [DOI] [PubMed] [Google Scholar]
- 9.Breitschwerdt EB, Maggi RG, Duncan AW, Nicholson WL, Hegarty BC, Woods CW. Bartonella species in blood of immunocompetent persons with animal and arthropod contact. Emerg Infect Dis. 2007;13:938–41. doi: 10.3201/eid1306.061337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhengsri S, Baggett H, Peruski L, Morway C, Bai Y, Fisk T, et al. Bartonella spp. infections, Thailand. Emerg Infect Dis. 2010;16:743–5. doi: 10.3201/eid1604.090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitschwerdt E, Maggi R, Mozayeni BR, Hegarty B, Bradley J, Mascarelli P. PCR amplification of Bartonella koehlerae from human blood and enrichment blood cultures. Parasit Vectors. 2010;3:76. doi: 10.1186/1756-3305-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosoy M, Murray M, Gilmore RD, Jr, Bai Y, Gage KL. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol. 2003;41:645–50. doi: 10.1128/JCM.41.2.645-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maggi R, Kosoy M, Mintzer M, Breitschwerdt E. Isolation of Candidatus Bartonella melophagi from human blood. Emerg Infect Dis. 2009;15:66–8. doi: 10.3201/eid1501.081080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGill S, Wesslen L, Hjelm E, Holmberg M, Rolf C, Friman G. Serological and epidemiological analysis of the prevalence of Bartonella spp. antibodies in Swedish elite orienteers 1992–93. Scand J Infect Dis. 2001;33:423–8. doi: 10.1080/00365540152029882. [DOI] [PubMed] [Google Scholar]
- 15.Harrus S, Bar-Gal G, Golan A, Elazari-Volcani R, Kosoy M, Morick D, et al. Isolation and genetic characterization of a Bartonella strain closely related to Bartonella tribocorum and Bartonella elizabethae in Israeli commensal rats. Am J Trop Med Hyg. 2009;81:55–8. [PubMed] [Google Scholar]
- 16.Liu Q, Sun J, Lu L, Fu G, Ding G, Song X, et al. Detection of Bartonella species in small mammals from Zheijiang province. China J Wildl Dis. 2010;46:179–85. doi: 10.7589/0090-3558-46.1.179. [DOI] [PubMed] [Google Scholar]
- 17.Gundi VAKB, Kosoy M, Myint KSA, Shrestha S, Shrestha M, Pavlin J, et al. Prevalence and genetic diversity of Bartonella species detected in different tissues of small mammals in Nepal. Appl Environ Microbiol. 2010;76:8247–54. doi: 10.1128/AEM.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai Y-L, Chuang S-T, Chang C-C, Kass PH, Chomel BB. Bartonella species in small mammals and their ectoparasites in Taiwan. Am J Trop Med Hyg. 2010;83:917–23. doi: 10.4269/ajtmh.2010.10-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chenchittikul M, Daengpium S, Hasegawa M, Itoh T, Phanthumachinda B. A study of commensal rodents and shrews with reference to the parasites of medical importance in Chanthaburi Province, Thailand. Southeast Asian J Trop Med Public Health. 1983;14:255–9. [PubMed] [Google Scholar]
- 20.Bai Y, Montgomery SP, Sheff KW, Chowdhury MA, Breiman RF, Kabeya H, et al. Bartonella strains in small mammals from Dhaka, Bangladesh, related to bartonella in America and Europe. Am J Trop Med Hyg. 2007;77:567–70. [PubMed] [Google Scholar]
- 21.Hsieh JW, Tung KC, Chen WC, Lin JW, Chien LJ, Hsu YM, et al. Epidemiology of Bartonella infection in rodents and shrews in Taiwan. Zoonoses Public Health. 2010;57:439–46. doi: 10.1111/j.1863-2378.2009.01234.x. [DOI] [PubMed] [Google Scholar]
- 22.Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, et al. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–81. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comer J, Diaz T, Vlahov D, Monterroso E, Childs J. Evidence of rodent-associated Bartonella and Rickettsia infections among intravenous drug users from Central and East Harlem, New York City. Am J Trop Med Hyg. 2001;65:855–60. doi: 10.4269/ajtmh.2001.65.855. [DOI] [PubMed] [Google Scholar]
- 24.Smith H, Reporter R, Rood M, Linscott A, Mascola L, Hogrefe W, et al. Prevalence study of antibody to ratborne pathogens and other agents among patients using a free clinic in downtown Los Angeles. J Infect Dis. 2002;186:1673–6. doi: 10.1086/345377. [DOI] [PubMed] [Google Scholar]
- 25.Ehrenborg C, Bystrm R, Hjelm E, Friman G, Holmberg M. High Bartonella spp. seroprevalence in a Swedish homeless population but no evidence of trench fever. Scand J Infect Dis. 2008;40:208–15. doi: 10.1080/00365540701632972. [DOI] [PubMed] [Google Scholar]
- 26.Childs JE, Ellis BA, Nicholson WL, Kosoy M, Sumner JW. Shared vector-borne zoonoses of the Old World and New World: home grown or translocated? Schweizerische medizinische Wochenschrift. 1999;129:1099–105. [PubMed] [Google Scholar]
- 27.Cleaveland S, Haydon DT, Taylor L. Overviews of pathogen emergence: which pathogens emerge, when and why? Curr Top Microbiol Immunol. 2007;315:85–111. doi: 10.1007/978-3-540-70962-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Telfer S, Clough HE, Birtles LR, Bennett M, Carslake D, Helyar S, et al. Ecological differences and coexistence in a guild of microparasites: Bartonella in wild rodents. Ecology. 2007;88:1841–9. doi: 10.1890/06-1004.1. [DOI] [PubMed] [Google Scholar]
- 29.Kosoy MY, Saito EK, Green D, Marston EL, Jones DC, Childs JE. Experimental evidence of host specificity of Bartonella infection in rodents. Comp Immunol Microbiol Infect Dis. 2000;23:221–38. doi: 10.1016/s0147-9571(99)00075-2. [DOI] [PubMed] [Google Scholar]
- 30.Scott P, Scharton T. Interaction between the innate and the acquired immune system following infection of different mouse strains with Leishmania major. Ann N Y Acad Sci. 1994;730:84–92. doi: 10.1111/j.1749-6632.1994.tb44241.x. [DOI] [PubMed] [Google Scholar]
- 31.Kosoy M, Mandel E, Green D, Marston E, Childs J. Prospective studies of Bartonella of rodents. Part I. Demographic and temporal patterns in population dynamics. Vector Borne Zoonotic Dis. 2004;4:285–95. doi: 10.1089/vbz.2004.4.285. [DOI] [PubMed] [Google Scholar]
- 32.Colton L, Zeidner N, Kosoy MY. Experimental infection of Swiss webster mice with four rat bartonella strains: host specificity, bacteremia kinetics, dose dependent response, and histopathology. Comp Immunol Microbiol Infect Dis. 2011;34:465–73. doi: 10.1016/j.cimid.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Bhengsri S, Baggete H, Peruski L, Morway C, Bai Y. Bartonella seroprevalence in rural Thailand. Southeast Asian J Trop Med Public Health. 2011;42:687–92. [PubMed] [Google Scholar]
- 34.Kyme P, Dillon B, Iredell J. Phase variation in Bartonella henselae. Microbiology. 2003;149:621–9. doi: 10.1099/mic.0.26014-0. [DOI] [PubMed] [Google Scholar]
- 35.Chomel BB, Kasten RW, Floyd-Hawkins K, Chi B, Yamamoto K, Roberts-Wilson J, et al. Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol. 1996;34:1952–6. doi: 10.1128/jcm.34.8.1952-1956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai Y, Calisher CH, Kosoy MY, Root JJ, Doty JB. Persistent infection or successive re-infection of deer mice with Bartonella vinsonii subsp. arupensis. Appl Environ Microbiol. 2011;77:1728–31. doi: 10.1128/AEM.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marignac G, Barrat F, Chomel B, Vayssier-Taussat M, Gandoin C, Bouillin C, et al. Murine model for Bartonella birtlesii infection: new aspects. Comp Immunol Microbiol Infect Dis. 2010;33:95–107. doi: 10.1016/j.cimid.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Koesling J, Aebischer T, Falch C, Schulein R, Dehio C. Cutting edge: antibody-mediated cessation of hemotropic infection by the intraerythrocytic mouse pathogen Bartonella grahamii. J Immunol. 2001;167:11–4. doi: 10.4049/jimmunol.167.1.11. [DOI] [PubMed] [Google Scholar]
- 39.Ellis BA, Regnery RL, Beati L, Bacellar F, Rood M, Glass GG, et al. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–4. doi: 10.1086/314824. [DOI] [PubMed] [Google Scholar]